Q Fever in humans, domestic animals and wildlife
Anita Tolpinrud A , Anne-Lise Chaber B , Anke K. Wiethoelter A , Joanne M. Devlin A , John Stenos C , Simon M. Firestone A and Mark A. Stevenson A *A
B
C

Anita Tolpinrud is a wildlife veterinarian and researcher focusing on wildlife epidemiology, zoonotic disease reservoirs, One Health and conservation medicine. Her current research and recently completed PhD thesis explore the wildlife reservoirs and epidemiology of Q fever in Australia. |

Dr Anne-Lise Chaber is a One Health practitioner and academic. Her research focuses on the anthropogenic drivers of (re)emerging infectious zoonotic diseases, examining the impact of human activities on disease transmission at the wildlife–livestock–human interface. Her work integrates human, animal and environmental health to enhance global disease prevention, surveillance, management and response strategies. She holds a joint appointment at the School of Public Health and the School of Animal and Veterinary Sciences at the University of Adelaide, Australia. |

Anke Wiethoelter is an associate professor in veterinary epidemiology and One Health at the Melbourne Veterinary School with a research focus on infectious diseases at the wildlife–livestock–human interface and determinants of health behaviour. She teaches epidemiology, evidence-based practice and One Health to both undergraduate and graduate students. |

Prof. Joanne Devlin researches the pathogenesis of a range of veterinary infectious diseases, and she has a particular interest in disease control. Her work includes diseases of domestic animals and wildlife, including birds, horses and marsupials. Her research includes fundamental research as well as more applied research, including vaccine development and testing. She was awarded an Australian Research Council (ARC) Postdoctoral Fellowship in 2008 and an ARC Future Fellowship in 2014. She is a current member of the ARC College of Experts and was appointed as the head of school for Melbourne Veterinary School in 2023. |

John Stenos is the senior scientist of the Australian Rickettsial Reference Laboratory (ARRL). John completed a postdoctoral fellowship in the world leading laboratory for rickettsial diseases (Center for Biodefense and Emerging Infectious Diseases, University of Texas Medical Branch, USA) in 1998. He has 29 years’ experience in the microbial culture of bacterial pathogens, especially rickettsia, and the development of new serological tests, particularly to detect vector-borne diseases. |

Simon M. Firestone is an associate professor in veterinary epidemiology and public health in the Melbourne Veterinary School. His research focuses on modelling infectious disease outbreaks, Bayesian diagnostic test validation, zoonoses surveillance, outbreak investigation and control, with projects on COVID-19, Q fever, foot-and-mouth disease, African swine fever, Mycoplasma bovis, foodborne disease, influenzas and arboviruses. |

Mark Stevenson is professor of veterinary epidemiology at The University of Melbourne where he leads a group working on applied epidemiological research with an emphasis on transboundary and endemic animal infectious diseases such as bovine spongiform encephalopathy, bovine tuberculosis, foot-and-mouth disease, Q fever and African swine fever. |
Abstract
Q Fever is a zoonotic disease caused by Coxiella burnetii, which can infect a wide range of host species, including humans, domestic animals and wild animals. Domestic livestock are the primary reservoir for human infections and humans are usually considered incidental hosts, with human-to-human infection being exceedingly rare. Because livestock are reservoirs, at-risk groups for Q fever have been considered to be abattoir workers, veterinary personnel, farm workers, livestock handlers and wool shearers. However, there has been an increasing prevalence of human cases that have a history of direct or indirect exposure to wildlife, pointing towards likely wildlife reservoirs. Coxiellosis can be diagnosed using indirect and direct methods. Specific antibodies against C. burnetii are usually detectable within 1–3 weeks of infection in humans and experimental animal models. Anti-phase 2 immunoglobin M (IgM) and immunoglobin G (IgG) antibodies appear first in the early stages of acute infections, followed by a delayed and less pronounced phase 1 IgM and IgG antibody response. Interpretation of polymerase chain reaction (PCR) and serology test results are useful for estimating the date of onset of symptoms or clinical signs and the date of exposure. This allows a time window of exposure to be determined and may assist with identification of likely sources of infection.
Keywords: Coxiella burnetii, diagnosis, outbreak, PCR, Q fever, rickettsia, serology, zoonosis.
Introduction
Q Fever is a zoonotic bacterial disease caused by Coxiella burnetii. In humans, Q fever can manifest as an acute debilitating illness (typically lasting 2–4 weeks), with a proportion of affected individuals going on to develop chronic syndromes. Inhalation of dust particles contaminated with C. burnetii is the main route of infection in humans1 and the infectivity of C. burnetii has been shown to be high enough to pose a serious risk to humans even when it is present in very small numbers in the environment.2 In 2007–10, there was a large outbreak of Q fever (4000 confirmed human cases) in the Netherlands, arising from the spread of C. burnetii from endemically infected dairy goat herds.3
Coxiella burnetii can infect a wide range of host species, including humans, domestic animals and wild animals. As increasing urbanisation brings human populations and farmed livestock and wildlife in closer proximity, there is a risk that Q fever outbreaks, similar to that which occurred in the Netherlands in 2007–10, could occur elsewhere. In addition, the effect of climate change (which leads to hot, dry and windy conditions in some areas) provides ideal conditions for airborne spread of Q fever.
This paper provides a review of the key features of coxiellosis in humans, domestic animals and wildlife, with a focus on available diagnostic procedures and their interpretation.
Aetiology
Coxiella burnetii are obligate intracellular coccobacilli that morphologically appear as small (0.2–0.4 µm by 0.4–1 µm) pleomorphic rods.4,5 The genus Coxiella is currently classified as a member of the family Coxiellaceae in the order Legionellales (class Gammaproteobacteria), which contains Legionella and Francisella species.5 Coxiella burnetii have a biphasic development cycle, consisting of a large cell variant (LCV) and small cell variant (SCV). Although the LCV is a vegetative and exponentially replicating form, the SCV is spore-like and extremely environmentally stable, enabling persistence outside a host for months or even years.6 Coxiella burnetii also display two antigenic phases (phase 1 and phase 2), generated by mutational variation in its surface lipopolysaccharide.1 They are among the most infective bacteria known, which, together with their potential for environmental persistence, have resulted in recognition of them as potential biological warfare agents and category B biological terrorism agents.2,6
Epidemiology
Although C. burnetii have a near global distribution, the epidemiology of Q fever is highly variable geographically, which may reflect differences between strains, host biology and ecology, and environmental conditions.4 Coxiella burnetii have an unusually wide range of potential hosts and animal reservoirs, involving several taxa of vertebrate and invertebrate species, as well as amoebae.1,4,5 Their ability to adapt to and persist in so many host types and under varied environmental conditions has probably contributed to their widespread geographical distribution. Domestic livestock are the primary reservoir for human infections and humans are usually considered incidental hosts, with human-to-human infection exceedingly rare.1,4,5,7 Although most human cases are sporadic, with outbreaks typically small and limited, with a common point source, large-scale outbreaks can occur.7,8
Shedding of C. burnetii has been demonstrated in the birth products, milk, urine, faeces, semen and vaginal secretions of infected animals.1,4,5,9,10 Inhalation of contaminated aerosols is the primary mode of transmission to humans, most commonly after parturition or abortion in infected domestic animals.1,8 Other sources include the slaughter and dressing of infected carcasses, wool shearing or aerosolisation of contaminated faecal material.11,12 Air samples have been found to contain C. burnetii for several weeks or months after parturition in infected herds10,13 and aerosols can be carried by wind over several kilometres.14,15 Other possible routes of transmission that have been explored experimentally include arthropod vectors, ingestion, sexual contact and vertical transmission to offspring, but the epidemiological significance of these routes is likely to be low or negligible.16,17 Because livestock are a common reservoir, the groups at risk of acquiring Q fever include abattoir workers, veterinary personnel, farm workers, livestock handlers and wool shearing staff,1,18 whereas large outbreaks have been reported in communities living in relative proximity to high-density livestock operations.7,8,15,19 Cases of Q fever have also been reported in those assisting cats during parturition20 and in Australia, there have been increasing numbers of human cases associated with direct or indirect exposure to wildlife, rather than the traditional risk factors, pointing towards the likelihood of wildlife reservoirs of infection.21
Clinical aspects
Most infections with C. burnetii in humans and animals are clinically inapparent. Approximately 60% of primary human infections are asymptomatic, and the majority of symptomatic infections result in a mild and self-limiting febrile or flu-like illness. Only 2–5% of acute infections are serious enough to result in hospitalisation, usually due to pneumonia, hepatitis or prolonged fever. Rarely, other localised inflammatory processes have been reported, including pericarditis, myocarditis, meningitis, meningoencephalitis, bone marrow necrosis, pancreatitis or orchitis.1,4 In some individuals (less than 5% of human infections), the immune response is insufficient and fails to eliminate the organism, leading to persistent or chronic Q fever, despite high concentrations of serum antibodies against C. burnetii. Chronic infections most commonly result in infective endocarditis or vascular lesions.1,4 Infection can also result in a presentation known as Q fever fatigue syndrome (QFS) in 20–30% of patients, which may last up to 10 years.22
Clinically relevant signs in naturally infected animals are rare, with the notable exception being reproductive failure and late-stage abortions (due to placentitis and metritis) in some placental mammals, particularly small domestic ruminants.1 Following infection, these clinical signs usually persist in subsequent reproductive cycles and can cause ongoing economic losses.9
The host defence against C. burnetii is primarily dependent on systemic cell-mediated immunity, with the humoral response probably only playing an adjunctive role.4,23 Specific antibodies probably facilitate a rapid cellular response through accelerated macrophage activation.23 However, high antibody titres are not sufficient to achieve clearance of C. burnetii, as is evident in patients with chronic Q fever.4,23
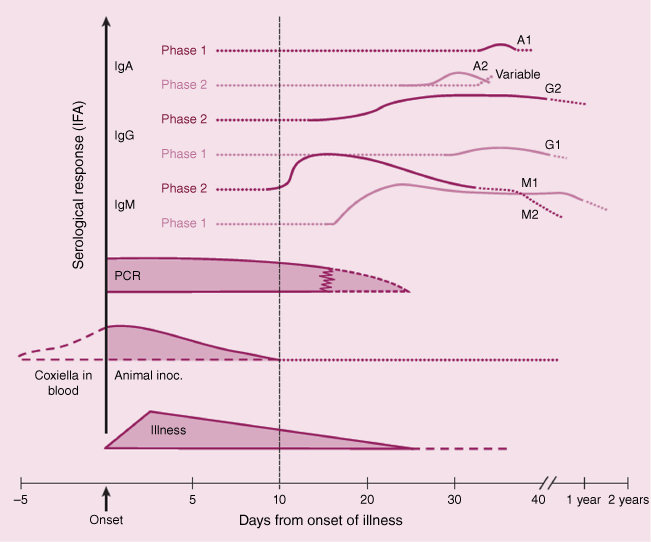
The production of specific antibodies against C. burnetii is usually evident within 1–3 weeks after infection in humans and experimental animal models, although the antibody response can vary widely between individual hosts and strains of C. burnetii.1,4,5,24 Most commonly, immunoglobin M (IgM) and immunoglobin G (IgG) antibodies against phase 2 antigens appear first in the early stages of acute infections. These antibodies rise sharply in the first weeks after exposure, followed by a delayed and less pronounced response to phase 1 antigens (Fig. 1).24 After a primary acute infection, specific IgG antibodies may persist for years, whereas the IgM response generally declines rapidly.24 Persistent infections are characterised by very high titres of IgG and immunoglobin A (IgA) antibodies against both phase 1 and 2 antigens, with the IgG response against phase 1 antigens predominating.1
Relative change in quantitative PCR results and serological (IgA, IgG and IgM) responses to Coxiella burnetii, as a function of the number of days after the onset of symptoms or clinical signs (reproduced with permission from Marmion46). The values shown on the vertical axis are qualitative, relative estimates. A patient presenting for diagnosis at 5 days after the onset of symptoms or clinical signs would be likely to be positive by PCR and negative by serology. At day 15, the same patient would be likely to be positive by PCR and positive for IgM against phase 2 antigens, and at day 25, PCR negative and positive for IgM against phase 1 and IgA phase 2 antigens. Patients presenting for diagnosis at 40+ days (i.e. 6 weeks or longer) are likely to be PCR negative, and positive for IgM against phase 1 antibodies and IgG against phase 2 antigens.
Diagnosis
Indirect methods
Serology is the easiest way to detect exposure to C. burnetii in unvaccinated individuals or animal populations, as antibodies tend to persist for extended periods after infection. Serological assays also have the benefit of being cheap and reasonably easy to perform. The main disadvantage of serology is that a positive result merely indicates historical exposure to the pathogen, which may or may not reflect the current disease status. Serial testing to demonstrate a rising antibody titre is necessary to differentiate a recent infection from a historical one, which can be logistically challenging in animals, particularly in wildlife.
The earliest techniques used to identify antibodies against C. burnetii were complement fixation tests (CFTs) and agglutination (AG) tests. These were commonly used for screening animal sera, regardless of species, but are insensitive, particularly in birds.6,25,26 As a result, they have largely been superseded by more sensitive tests, such as enzyme-linked immunosorbent assays (ELISAs) and indirect immunofluorescence assays (IFAs).6 IFAs are the preferred (reference) test for detecting antibodies against C. burnetii in humans. Although they are more laborious to perform than ELISAs and require a skilled technician for accurate interpretation, IFAs have frequently been shown to have higher diagnostic accuracy in human studies.27,28 Conversely, the World Organisation for Animal Health recommends the ELISA for veterinary use,6 although findings and recommendations about their diagnostic accuracy vary considerably between kits and host species in different studies.29–31 Although there are several commercially available ELISA kits available for use in domestic ruminants, few have been validated for use in other species.
Direct methods
Tests to detect the pathogen directly are the most reliable method to demonstrate an active (current) infection. However, the sensitivity of these tests may be limited if the pathogen is localised in specific tissues or if the duration of bacteraemia or infection is short, necessitating careful selection and timing of sample collection.
Although bacterial culture and isolation could be considered the most definitive test for diagnosis of infection, culture of C. burnetii is difficult and laborious. Its high infectivity restricts culture and isolation to facilities with appropriate levels of biological containment. Culture has historically relied on inoculation of cell cultures, embryonated chicken eggs or laboratory animals, but more recent advances have facilitated in vitro growth in axenic medium.32
Visualisation of bacteria in situ may be possible in smears from clinical samples or in histopathological sections. Although C. burnetii are Gram-negative organisms, they stain poorly with Gram stain.33 However, they can be well visualised in clinical samples and cultures using Gimenez, Macchiavello or Giemsa stains, providing the bacterial load is sufficient (>105 bacteria mL–1).6,33 Immunohistochemistry (IHC) or, less commonly, fluorescence in situ hybridisation (FISH) techniques are more specific methods for confirming the presence of C. burnetii in clinical samples, although the stains and controls required may be less readily available.34,35
Polymerase chain reaction (PCR) assays are the direct method most commonly used to demonstrate active infection with C. burnetii, because of their greater sensitivity.6 In addition, PCR assays can have a high throughput, a quick turnaround time and have a relatively greater ease of use than other methods. A wide range of protocols have been described, targeting a variety of plasmid and chromosomal genes, with varying levels of sensitivity and specificity. Common targets include the transposase-like insertion sequence (IS1111), the isocitrate dehydrogenase gene (icd), the superoxide dismutase gene (sodB), the outer membrane protein 1 gene (com1), the outer membrane protein A gene (OmpA), heat shock operon genes (htpA and htpB) and the 16S ribosomal RNA gene (16S rRNA).4,36,37 In addition, use of real-time PCR assays enables quantification of the bacterial load in a sample, although the high genetic diversity between isolates may complicate quantification for some targets. For instance, the copy numbers of IS1111, which is frequently the target used in screening tests because it yields assays with a higher sensitivity, has been shown to vary from zero to over 100 between different isolates, making quantification based on this target unreliable.37,38 Single copy targets (such as com1) are therefore more useful for quantification, although the sensitivity of these tests may be significantly reduced. However, care is needed because of the potential for cross-reactivity with Coxiella-like tick endosymbionts, many of which have similar genes to C. burnetii and are still poorly characterised.16
Table 1 provides details of the sensitivity and specificity of each of the diagnostic tests described above. Fig. 1 shows the change in IgA, IgG and IgM responses and quantitation of C. burnetii by real time PCR, as a function of the number of days since onset of symptoms or clinical signs in in infected humans and animals.
| Species | Tissue | Test | Diagnostic sensitivity (%) | Diagnostic specificity (%) | |
|---|---|---|---|---|---|
| Human | Serum | IFA (IgG phase 1) 39 | 87.2 | 90.0 | |
| IFA (IgG phase 2) 39 | 97.7 | 100.0 | |||
| IFA (IgG phase 2) 40 | 100.0 | 95.3 | |||
| IFA (IgM phase 1) 39 | 60.0 | 64.7 | |||
| IFA (IgM phase 2) 39 | 66.7 | 75.9 | |||
| ELISA (IgM phase 2) 40 | 85.7 | 97.6 | |||
| CFT 41 | 72.9 | 89.9 | |||
| Cattle | Serum | IFA 31 | 73.6 | 98.2 | |
| cELISA 1 42 | 87.0 | 99.1 | |||
| cELISA 2 42 | 98.6 | 97.1 | |||
| cELISA 3 42 | 55.7 | 99.3 | |||
| CFT 43 | 26.6 | 99.7 | |||
| CFT 44 | 68.0 | 100.0 | |||
| CFT 42 | 36.2 | 98.3 | |||
| Sheep and goats | Serum | IFA (IgG) 29 | 94.8 | 92.5 | |
| IFA (IgM) 29 | 88.8 | 92.4 | |||
| ELISA 29 | 70.0 | 96.0 | |||
| CFT 43 | 10.0 | 99.9 | |||
| CFT 42 | 36.2 | 98.3 | |||
| Milk | com1 PCR 29 | 56.0 | 100.0 A | ||
| ELISA 45 | 89.0 | 100.0 | |||
| Macropods | Serum | IFA 30 | 97.6 | 98.5 | |
| ELISA 30 | 42.1 | 99.2 |
Interpretation of PCR and serology test results using the guidelines listed above may assist in providing an estimate of the date of onset of clinical signs or exposure, where this is unknown by the human patients or animal owners. This may allow a time window of infection to be determined and assist with the identification of a likely source. In turn, this can assist in implementation of control measures to manage or eliminate spread infection.
Data availability
This paper does not use any original data. All information and analyses presented are based on existing literature and publicly available sources.
References
1 Maurin M, Raoult D (1999) Q Fever. Clin Microbiol Rev 12, 518-553.
| Crossref | Google Scholar | PubMed |
2 Jones RM et al. (2006) The infectious dose of Coxiella burnetii (Q fever). Appl Biosaf 11, 32-41.
| Crossref | Google Scholar |
3 Schneeberger P et al. (2014) Q Fever in the Netherlands 2007–2010: what we learned from the largest outbreak ever. Med Mal Infect 44, 339-353.
| Crossref | Google Scholar | PubMed |
4 Eldin C et al. (2017) From Q fever to Coxiella burnetii infection: a paradigm change. Clin Microbiol Rev 30, 115-190.
| Crossref | Google Scholar | PubMed |
5 Roest HIJ et al. (2013) Clinical microbiology of Coxiella burnetii and relevant aspects for the diagnosis and control of the zoonotic disease Q fever. Vet Q 33, 148-160.
| Crossref | Google Scholar | PubMed |
6 Rousset E, et al. (2018) Chapter 3.1.18. Q Fever. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 8th edn. World Organisation for Animal Health. https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.01.18_Q_FEVER.pdf
7 Tan T et al. (2024) A systematic review of global Q fever outbreaks. One Health 18, 100667.
| Crossref | Google Scholar | PubMed |
8 Vellema P, van den Brom R (2014) The rise and control of the 2007–2012 human Q fever outbreaks in the Netherlands. Small Rumin Res 118, 69-78.
| Crossref | Google Scholar |
9 Canevari JT et al. (2018) The prevalence of Coxiella burnetii shedding in dairy goats at the time of parturition in an endemically infected enterprise and associated milk yield losses. BMC Vet Res 14, 353.
| Crossref | Google Scholar | PubMed |
10 Joulié A et al. (2015) Circulation of Coxiella burnetii in a naturally infected flock of dairy sheep: shedding dynamics, environmental contamination, and genotype diversity. Appl Environ Microbiol 81, 7253-7260.
| Crossref | Google Scholar | PubMed |
11 Christen J-R et al. (2020) Capybara and brush cutter involvement in Q fever outbreak in remote area of Amazon rain forest, French Guiana, 2014. Emerg Infect Dis 26, 993-997.
| Crossref | Google Scholar | PubMed |
12 Hellenbrand W et al. (2001) Changing epidemiology of Q fever in Germany, 1947–1999. Emerg Infect Dis 7, 789-796.
| Crossref | Google Scholar | PubMed |
13 Kersh GJ et al. (2013) Presence and persistence of Coxiella burnetii in the environments of goat farms associated with a Q fever outbreak. Appl Environ Microbiol 79, 1697-1703.
| Crossref | Google Scholar | PubMed |
14 Hawker JI et al. (1998) A large outbreak of Q fever in the West Midlands: windborne spread into a metropolitan area? Commun Dis Public Health 1, 180-187.
| Google Scholar |
15 Tissot-Dupont H et al. (2004) Wind in November, Q fever in December. Emerg Infect Dis 10, 1264-1269.
| Crossref | Google Scholar | PubMed |
16 Duron O et al. (2015) The importance of ticks in Q fever transmission: what has (and has not) been demonstrated? Trends Parasitol 31, 536-552.
| Crossref | Google Scholar | PubMed |
17 Gale P et al. (2015) Q Fever through consumption of unpasteurised milk and milk products – a risk profile and exposure assessment. J Appl Microbiol 118, 1083-1095.
| Crossref | Google Scholar | PubMed |
18 Sloan-Gardner TS et al. (2017) Trends and risk factors for human Q fever in Australia, 1991–2014. Epidemiol Infect 145, 787-795.
| Crossref | Google Scholar | PubMed |
19 Smit LA et al. (2012) Q Fever and pneumonia in an area with a high livestock density: a large population-based study. PLoS ONE 7, e38843.
| Crossref | Google Scholar | PubMed |
20 Shapiro A et al. (2017) Q Fever (Coxiella burnetii) knowledge and attitudes of Australian cat breeders and their husbandry practices. Zoonoses Public Health 64, 252-261.
| Crossref | Google Scholar | PubMed |
21 Massey PD et al. (2009) Enhanced Q fever risk exposure surveillance may permit better informed vaccination policy. Commun Dis Intell Q Rep 33, 41-45.
| Google Scholar | PubMed |
22 Morroy G et al. (2016) Fatigue following acute Q-fever: a systematic literature review. PLoS ONE 11, e0155884.
| Crossref | Google Scholar | PubMed |
23 Humphres RC, Hinrichs DJ (1981) Role of antibody in Coxiella burnetii infection. Infect Immun 31, 641-645.
| Crossref | Google Scholar | PubMed |
24 Wielders CC et al. (2015) Kinetics of antibody response to Coxiella burnetii infection (Q fever): estimation of the seroresponse onset from antibody levels. Epidemics 13, 37-43.
| Crossref | Google Scholar | PubMed |
25 Lucchese L et al. (2016) Evaluation of serological tests for Q fever in ruminants using the latent class analysis. Clin Res Infect Dis 3, 1030.
| Crossref | Google Scholar |
26 Sidwell RW et al. (1964) The occurrence of a possible epizootic of Q fever in fauna of the Great Salt Lake Desert of Utah. Am J Trop Med Hyg 13, 754-762.
| Crossref | Google Scholar | PubMed |
27 França DA et al. (2023) Comparison of three serologic tests for the detection of anti-Coxiella burnetii antibodies in patients with Q fever. Pathogens 12, 873.
| Crossref | Google Scholar | PubMed |
28 Stephen S et al. (2017) Unreliability of three commercial Coxiella burnetii phase II IgM ELISA kits for the seroscreening of acute Q fever in human cases. Indian J Med Res 146, 386-391.
| Crossref | Google Scholar | PubMed |
29 Muleme M et al. (2016) Bayesian validation of the indirect immunofluorescence assay and its superiority to the enzyme-linked immunosorbent assay and the complement fixation test for detecting antibodies against Coxiella burnetii in goat serum. Clin Vaccine Immunol 23, 507-514.
| Crossref | Google Scholar | PubMed |
30 Tolpinrud A et al. (2022) Validation of an indirect immunofluorescence assay and commercial Q fever enzyme-linked immunosorbent assay for use in macropods. J Clin Microbiol 60, e0023622.
| Crossref | Google Scholar | PubMed |
31 Wood C et al. (2019) Validation of an indirect immunofluorescence assay (IFA) for the detection of IgG antibodies against Coxiella burnetii in bovine serum. Prev Vet Med 169, 104698.
| Crossref | Google Scholar | PubMed |
32 Omsland A et al. (2008) Sustained axenic metabolic activity by the obligate intracellular bacterium Coxiella burnetii. J Bacteriol 190, 3203-3212.
| Crossref | Google Scholar | PubMed |
33 Giménez DF (1965) Gram staining of Coxiella burnetii. J Bacteriol 90, 834-835.
| Crossref | Google Scholar | PubMed |
34 Jensen TK et al. (2007) Application of fluorescent in situ hybridisation for demonstration of Coxiella burnetii in placentas from ruminant abortions. APMIS 115, 347-353.
| Crossref | Google Scholar | PubMed |
35 Lepidi H et al. (2009) Immunohistochemical detection of Coxiella burnetii in chronic Q fever hepatitis. Clin Microbiol Infect 15, 169-170.
| Crossref | Google Scholar | PubMed |
36 Khademi P et al. (2024) Molecular and genotyping techniques in diagnosis of Coxiella burnetii: an overview. Infect Genet Evol 123, 105655.
| Crossref | Google Scholar | PubMed |
37 Klee SR et al. (2006) Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol 6, 2.
| Crossref | Google Scholar | PubMed |
38 Gardner BR et al. (2023) A novel marine mammal Coxiella burnetii – genome sequencing identifies a new genotype with potential virulence. Pathogens 12, 893.
| Crossref | Google Scholar | PubMed |
39 Slabá K et al. (2005) Efficiency of various serological techniques for diagnosing Coxiella burnetii infection. Acta Virol 49, 123-127.
| Google Scholar | PubMed |
40 Meekelenkamp J et al. (2012) Comparison of ELISA and indirect immunofluorescent antibody assay detecting Coxiella burnetii IgM phase II for the diagnosis of acute Q fever. Eur J Clin Microbiol Infect Dis 31, 1267-1270.
| Crossref | Google Scholar | PubMed |
41 Field P et al. (2000) Comparison of a commercial enzyme-linked immunosorbent assay with immunofluorescence and complement fixation tests for detection of Coxiella burnetii (Q fever) immunoglobulin M. J Clin Microbiol 38, 1645-1647.
| Crossref | Google Scholar | PubMed |
42 Horigan M et al. (2011) Q Fever diagnosis in domestic ruminants comparison between complement fixation and commercial enzyme-linked immunosorbent assays. J Vet Diagn Invest 23, 924-931.
| Crossref | Google Scholar | PubMed |
43 Natale A et al. (2012) Old and new diagnostic approaches for Q fever diagnosis: correlation among serological (CFT, ELISA) and molecular analyses. Comp Immunol Microbiol Infect Dis 35, 375-379.
| Crossref | Google Scholar | PubMed |
44 Kittelberger R et al. (2009) Comparison of the Q-fever complement fixation test and two commercial enzyme-linked immunosorbent assays for the detection of serum antibodies against Coxiella burnetti (Q-fever) in ruminants: recommendations for use of serological tests on imported animals in New Zealand. NZ Vet J 57, 262-268.
| Crossref | Google Scholar | PubMed |
45 Muleme M et al. (2017) Peripartum dynamics of Coxiella burnetii infections in intensively managed dairy goats associated with a Q fever outbreak in Australia. Prev Vet Med 139, 58-66.
| Crossref | Google Scholar | PubMed |
 Anita Tolpinrud is a wildlife veterinarian and researcher focusing on wildlife epidemiology, zoonotic disease reservoirs, One Health and conservation medicine. Her current research and recently completed PhD thesis explore the wildlife reservoirs and epidemiology of Q fever in Australia. |
 Dr Anne-Lise Chaber is a One Health practitioner and academic. Her research focuses on the anthropogenic drivers of (re)emerging infectious zoonotic diseases, examining the impact of human activities on disease transmission at the wildlife–livestock–human interface. Her work integrates human, animal and environmental health to enhance global disease prevention, surveillance, management and response strategies. She holds a joint appointment at the School of Public Health and the School of Animal and Veterinary Sciences at the University of Adelaide, Australia. |
 Anke Wiethoelter is an associate professor in veterinary epidemiology and One Health at the Melbourne Veterinary School with a research focus on infectious diseases at the wildlife–livestock–human interface and determinants of health behaviour. She teaches epidemiology, evidence-based practice and One Health to both undergraduate and graduate students. |
 Prof. Joanne Devlin researches the pathogenesis of a range of veterinary infectious diseases, and she has a particular interest in disease control. Her work includes diseases of domestic animals and wildlife, including birds, horses and marsupials. Her research includes fundamental research as well as more applied research, including vaccine development and testing. She was awarded an Australian Research Council (ARC) Postdoctoral Fellowship in 2008 and an ARC Future Fellowship in 2014. She is a current member of the ARC College of Experts and was appointed as the head of school for Melbourne Veterinary School in 2023. |
 John Stenos is the senior scientist of the Australian Rickettsial Reference Laboratory (ARRL). John completed a postdoctoral fellowship in the world leading laboratory for rickettsial diseases (Center for Biodefense and Emerging Infectious Diseases, University of Texas Medical Branch, USA) in 1998. He has 29 years’ experience in the microbial culture of bacterial pathogens, especially rickettsia, and the development of new serological tests, particularly to detect vector-borne diseases. |
 Simon M. Firestone is an associate professor in veterinary epidemiology and public health in the Melbourne Veterinary School. His research focuses on modelling infectious disease outbreaks, Bayesian diagnostic test validation, zoonoses surveillance, outbreak investigation and control, with projects on COVID-19, Q fever, foot-and-mouth disease, African swine fever, Mycoplasma bovis, foodborne disease, influenzas and arboviruses. |
 Mark Stevenson is professor of veterinary epidemiology at The University of Melbourne where he leads a group working on applied epidemiological research with an emphasis on transboundary and endemic animal infectious diseases such as bovine spongiform encephalopathy, bovine tuberculosis, foot-and-mouth disease, Q fever and African swine fever. |


