A One Health approach for the genomic surveillance of AMR
Veronica M. Jarocki A B # , Max L. Cummins
A B # , Max L. Cummins  A C # , Celeste M. Donato
A C # , Celeste M. Donato  D E # , Benjamin P. Howden
D E # , Benjamin P. Howden  D F * and Steven P. Djordjevic
D F * and Steven P. Djordjevic  A C *
A C *
A
B
C
D
E
F

Dr Veronica Jarocki is a CRC SAAFE Foundational Postdoctoral Fellow based at the Australian Institute of Microbiology and Infection (AIMI) at the University of Technology—Sydney. Her research interests include genomic AMR surveillance in wastewater, humans and animals and characterising the mechanisms behind AMR dissemination between sources. She has a background in subunit vaccine design and development against animal pathogens. |

Dr Max Cummins is a researcher at AIMI at the University of Technology—Sydney, whose research interests include microbial genomics, antimicrobial resistance, bioinformatics, big data, AI and data visualisation. Max co-leads the Australian Pathogen Genomics Program’s One Health theme, utilising bioinformatic tools to generate meaningful and informative insights into bacterial populations found across humans, animals, food and the environment. |

Dr Celeste Donato is a postdoctoral researcher at the Doherty Institute and her research interests include pathogen genomics, molecular and genomic epidemiology with expertise in viral enteric pathogens. Celeste co-leads the One Health and Candida auris projects within the Australian Pathogen Genomics program. |

Benjamin Howden is Infectious Diseases Physician and Public Health Microbiologist. He is Professor and Director of the Microbiological Diagnostic Unit Public Health Laboratory at The University of Melbourne (Doherty Institute). His research interests include genomics for understanding and responding bacterial pathogens of public health significance and antimicrobial resistance. He is lead of the Australian Pathogen Genomics Program (AusPathoGen). |

Steven Djordjevic is a Distinguished Professor of Infectious Disease in the Australian Institute for Microbiology & Infection at the University of Technology—Sydney. His research interests are in antimicrobial resistance and pathogen evolution through a One Health lens. His laboratory conducts genomic epidemiological analyses of a broad range of Gram-negative bacterial pathogens, and the mobile elements they carry, from hospital and veterinary clinical environments, food production animals, urban and native wildlife, wastewater and natural water systems. |
Abstract
In the face of an escalating antimicrobial resistance (AMR) crisis, genomic technologies have emerged as indispensable allies, providing innovative tools for a nuanced understanding of the abundance, persistence and mobilisation of antimicrobial resistance genes within microbial populations. This article explores advancements in genomic surveillance, including the integration with advanced computational tools to enhance our ability to predict AMR trends, detect outbreaks, and inform mitigation strategies. It highlights the critical role of a One Health approach, emphasising the importance of cross-sectoral collaboration among scientists, health care professionals, industry and policymakers to leverage genomic data for AMR management. The article showcases pioneering initiatives in Australia, such as the Melbourne Genomics Alliance’s Controlling Superbugs Clinical Flagship, the Australian Centre for Genomic Epidemiological Microbiology and AusTrakka, and discusses the need to both build global genomic databases that promote equitable analytics, and secure data-sharing platforms that support comprehensive surveillance networks. Through national and international collaborative efforts, One Health genomic surveillance represents a key strategy in enhancing our understanding and control of AMR and should be integrated into public health frameworks to safeguard against ever emerging AMR threats.
Keywords: antimicrobial resistance, genomics, One Health, WGS, whole-genome sequencing.
Introduction
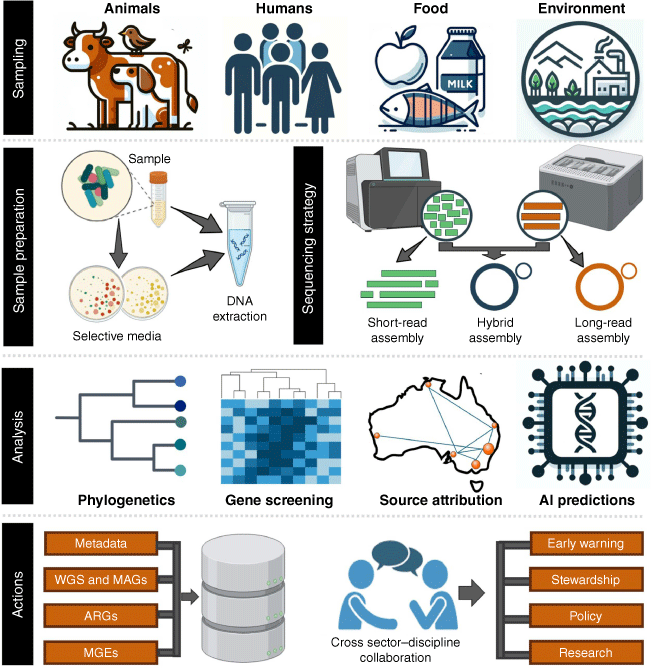
Through whole-genome sequencing (WGS) and metagenomics, researchers can dissect the complex dynamics of antimicrobial resistance (AMR) with unprecedented precision.1 A key offering of genomic surveillance is the ability to reveal the intricate genetic underpinnings of AMR, such as the variety of mutations, plasmids, integrons, insertion sequences, and transposons that contribute to the propagation and evolution of resistance. By enabling a more detailed characterisation of AMR alongside an in-depth examination of the mechanisms driving resistance, genomic surveillance marks a significant advancement in our ability to monitor AMR trends and predict emerging pathogenic and AMR lineages.
The digital nature of genomic data facilitates integration into machine learning and geospatial information systems, enabling sophisticated analyses that can predict resistance phenotypes, identify AMR outbreaks and attribute antimicrobial resistance genes (ARGs) to source. Artificial intelligence models have shown promise in deciphering complex bacterial transmission networks and this synergy between genomics and advanced computational tools is revolutionising our approach to tackling AMR, making it possible to trace the origins of resistance, predict its spread, and formulate targeted response strategies.2,3
As genomic technologies continue to evolve, they must remain at the forefront of our surveillance efforts. The detailed insights provided by WGS and metagenomics into the mechanisms of AMR, coupled with the analytical power of computational models, are essential for informing stewardship practices, defining baseline AMR data, and guiding mitigation strategies. The integration of a One Health approach is paramount for a comprehensive genomic surveillance and AMR management strategy (Fig. 1). Collaboration and data sharing across disciplines and sectors are critical to harnessing the full potential of genomic surveillance. Partnerships between microbiologists, epidemiologists, ecologists, healthcare professionals, industry, and policymakers facilitates the flow of information and resources necessary for timely and effective actions. The commitment to sharing data and insights through national and international networks enhances local and global efforts to monitor and respond to AMR threats, reflecting a collective endeavour to safeguard planetary health.
Integrating genomics in One Health: from national surveillance to global pandemic preparedness
Historically, the application of genomic epidemiology to AMR has largely focused on health-care and public health settings such as gaining enhanced resolution of nosocomial-acquired infection and outbreak epidemiology.4 In Australia, the Melbourne Genomics Health Alliance Controlling Superbugs Clinical Flagship (2017–2019) was the first project worldwide to incorporate multiple hospitals and target multiple organisms to investigate the prospective role of genomics in identifying and preventing superbug transmission. This project established the feasibility and impact of using genomic sequencing to detect patient-to-patient superbug transmissions and outbreaks occurring in, and between, hospitals in real time.5 Such initiatives have highlighted the significant opportunities afforded by pathogen genomics. The Australian Centre for Genomic Epidemiological Microbiology (AusGEM), a collaborative partnership between NSW Department of Primary Industries and University of Technology—Sydney, came into existence in 2014. The program supported WGS of antibiotic resistant Escherichia coli from diverse hosts and environments, generating evidence of numerous multi-host transmission events and establishing collaborations across the One Health space within Australia and internationally.
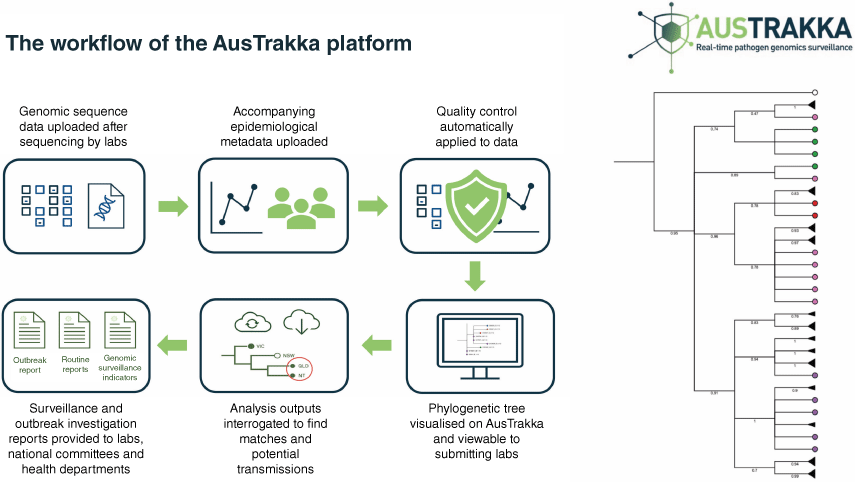
Primarily in high-income settings, there has been substantial growth in the development and implementation of pathogen genomics-enabled surveillance and genomic epidemiology. Such examples include the formation of the GenomeTrakr network in 2010, which was established by the US Food and Drug Administration. The GenomeTrakr network was the first attempt at large-scale, real-time genomic data sharing on foodborne pathogens on a national level.6 Similarly, Public Health England (PHE) launched the PHE Pathogen Genomics Service in 2014, facilitating national integration of genomic and epidemiological data, and the UK Health Security Agency (UKHSA) has recently released a 5-year pathogen genomics strategy to prepare for and respond to infectious disease threats to public health.7 In Australia, the Communicable Diseases Genomics Network (see https://www.cdgn.org.au/) was established in 2015 with the aim to foster collaboration and consistency in approach to enhance the implementation of pathogen genomics in public health. The AusTrakka platform was deployed early in the COVID-19 pandemic as the platform to facilitate trans-jurisdictional consistency in genomic data sharing and reporting in public health surveillance nationally (see https://www.cdgn.org.au/austrakka) (Fig. 2).8
AusTrakka and its interface with public health laboratories in genomic epidemiology. Collaboration in data sharing, analytics and visualisation with public health officials proved invaluable in pandemic-related epidemiology; this model is tenable to expansion by inclusion of One Health stakeholders from broader human, animal and environmental domains.
The true utility and capacity of these genomic initiatives was demonstrated during the SARS-CoV-2 pandemic and the need for a One Health approach to genomics was highlighted by the broad utilisation of sequencing to inform public health decision making, establishment of global wastewater epidemiology consortiums, and veterinary testing to describe spillback from humans into domestic and wild animals. The increased capacity for sequencing, epidemiological interrogation of data and cross-sectoral collaboration critical to the pandemic response can be further leveraged to address AMR from a One Health perspective.
Currently, One Health genomic epidemiological studies typically fall within an academic research environment rather than cross-jurisdictional, government-led activities. The Australian Pathogen Genomics Program (AusPathoGen, see https://www.auspathogen.org.au/), which utilises the AusTrakka platform, represents a multidisciplinary approach to national public health genomics, comprising leading academics and public health laboratory experts from 12 academic institutions across Australia. With cross-disciplinary expertise in antimicrobial resistance, bioinformatics, epidemiology, evaluation and implementation science, health economics, infectious diseases, genomic modelling, medical microbiology and phylodynamics – the One Health E. coli project represents a comprehensive genomics approach to One Health. This project has brought together experts spanning human health, veterinary medicine, food-safety, water quality and wildlife monitoring to work toward a national, One Health genomic surveillance network to provide insights into human, animal and environmental health (see https://www.auspathogen.org.au/studies/e-coli). Other notable examples of national One Health frameworks that incorporate genomics to varied extents include Canada and Sweden (see https://www.slu.se/en/Collaborative-Centres-and-Projects/one-health-sweden/). The Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS), coordinated by the Public Health Agency of Canada, brings together the National Microbiology Laboratory, federal, provincial and private industry partners to analyse trends in antimicrobial use and AMR for select bacteria from humans, animals, and retail meat across the country with increasing utilisation of genomics.9
Building the foundation for integrated AMR surveillance through collaborative genomic epidemiology
Antimicrobial resistance is a highly complex issue requiring large-scale, cross-sectoral collaboration. Nation-scale integrated genomics ecosystems spanning systematic sampling, metagenomic and genomic sequencing of microbes spanning human, animal and environmental sectors (and subsequent analytics and information dissemination) are required to improving AMR-related outcomes in human and animal health, industry and the environment. Critical in this regard are efforts to collaboratively: (i) develop, maintain and leverage global ‘omics’ databases; (ii) promote equitable and robust ‘omics’ analytics and visualisation capacity; (iii) build democratised data analytics infrastructure that simultaneously empowers and protects stakeholders with diverse data-related needs.
Global databases are critical infrastructure herein, including (i) retrospective metagenomic and genomic datasets; (ii) ‘complete’ sequences of plasmids and other complex mobile genetic elements (MGEs) such as genomic islands, and (iii) ARGs and AMR-associated loci (e.g. insertion sequences). Mining of these databases enables identification of emerging hyper resistant lineages as well as other organisms, MGEs, ARGs and sequences of concern that can be targets of ongoing surveillance efforts. Stakeholder engagement, balancing the demands on data access and privacy, and respecting industry sensitivities lie at the health of the development of a successful national surveillance program. Concerns of stakeholders, as it relates to antimicrobial resistance (and adjacent challenges, particularly infectious disease, sanitisation, and food safety), differ greatly. Collaboration therefore ensures common good for these diverse, contributing individuals and organisations.
Analytics and visualisation of AMR-centric ‘omics’ data should be undertaken using innovative and open-source pipelines that optimise for reproducibility, portability and analytical robustness. Standardisation of bioinformatic analysis platforms and the sharing of these workflows by open-source code repositories aligns with One Health themes of collaboration, equity and empowerment of AMR stakeholders. This is particularly effective when combined with capacity building programs run in less well-resourced regions such as parts of South-east Asia (e.g. Centre for Pathogen Genomics10), Latin America (e.g. Capacity building for bioinformatics in Latin America,11 see https://www.cabana.online) and Africa (e.g. African Biogenome Project, see https://africanbiogenome.org). Establishing foundational sequencing infrastructure is crucial for equitable access to essential genome sequencing technologies. This requires obtaining both short-read and long-read sequencing platforms, enabling comprehensive AMR metagenomic and genomic analyses and enhancing local capabilities to contribute significantly to global AMR surveillance. The need for capacity building, innovative data analytics and visualisation methods, gold-standard bioinformatic tools, computational infrastructure, artificial intelligence, and cyber-security will be crucial for the management and monitoring of AMR.
Generation of actionable insights relevant to such stakeholders, by a centralised analytics teams or by analysts associated with individual sectors or contributing organisations, can then be used to inform intervention strategies that aim to improve outcomes to a given sector or organisation. Leveraging data contributed by other sectors likely maximises the likelihood of generating such actionable insights within any given sector. To this end, strict and tiered access allowing stakeholders to determine which of their data are utilised, and by whom, can encourage deposition of samples or sequences to a centralised analytics platform that may otherwise be prevented by misaligned incentive structures (e.g. those relating to data sensitivity, which are common in industry and healthcare). For instance, Nebula Genomics and LunaDNA employ blockchain technology12 to provide secure, decentralised platforms for storing and accessing genomic data. This ensures that data are accessible only to authorised users, thereby maintaining privacy. Data, including ‘-omics’ data and metadata, must be stored in a highly secure environment to ensure all stakeholders are protected from unwanted consequences of collaboration, promoting their continual engagement. Permissioned and needs-based access can then be provided to stakeholders to utilise this analytics platform. Governance of such a system may prove challenging, however AMR surveillance and management is a complex problem warranting elegant, if initially challenging, solutions.
Such stakeholder engagement rewards contribution to surveillance efforts; such contributions (theoretically) may be financial in nature, by submission of samples or isolates for metagenomic and genomic analyses, or contribution of labour or other resources. In this way, synergistic and mutually beneficial relationships can iteratively reinforce the development of such an AMR surveillance system and enhance the prosperity borne of it.
Conclusion
A One Health genomics approach that encompasses all disciplines and sectors is critical in understanding and mitigating the impacts of AMR. Leadership, effective and considerate cross-sectoral communication, and elegant data-driven solutions that leverage cutting edge analytics tools represent the best approach to tackle the AMR crisis.
Declaration of funding
This work was supported by the Medical Research Future Fund-supported AusPathoGen Program (FSPGN000049), the Australian Centre for Genomic Epidemiological Microbiology (AusGEM), a collaborative research initiative between the New South Wales Department of Primary Industries and the University of Technology—Sydney, and the Cooperative Research Centre for Solving Antimicrobial resistance in Agribusiness, Food and Environments (CRC SAAFE). B. P. Howden is supported by a National Health and Medical Research Council (NHMRC) Fellowship (GNT1196103).
Acknowledgements
The authors thank members of the AusTrakka team for assistance with preparation of Fig. 2.
References
1 Djordjevic SP et al. (2024) Genomic surveillance for antimicrobial resistance — a One Health perspective. Nat Rev Genet 25, 142-157.
| Crossref | Google Scholar | PubMed |
2 Sakagianni A et al. (2023) Using machine learning to predict antimicrobial resistance – a literature review. Antibiotics 12, 452.
| Crossref | Google Scholar | PubMed |
3 Anahtar MN et al. (2021) Applications of machine learning to the problem of antimicrobial resistance: an emerging model for translational research. J Clin Microbiol 59, e01260-20.
| Crossref | Google Scholar | PubMed |
4 Lane CR et al. (2021) Search and contain: impact of an integrated genomic and epidemiological surveillance and response program for control of carbapenemase-producing Enterobacterales. Clin Infect Dis 73, 3912-3920.
| Crossref | Google Scholar | PubMed |
5 Gorrie CL et al. (2021) Key parameters for genomics-based real-time detection and tracking of multidrug-resistant bacteria: a systematic analysis. Lancet Microbe 2, 575-583.
| Crossref | Google Scholar | PubMed |
6 Timme RE et al. (2019) Utilizing the public GenomeTrakr database for foodborne pathogen traceback. Methods Mol Biol 1918, 201-212 [Erratum in Methods Mol Biol. 1918, C1.].
| Crossref | Google Scholar | PubMed |
7 Allard MW et al. (2016) Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol 54, 1975-1983.
| Crossref | Google Scholar | PubMed |
8 Hoang T et al. (2022) AusTrakka: fast-tracking nationalized genomics surveillance in response to the COVID-19 pandemic. Nat Commun 13, 865.
| Crossref | Google Scholar | PubMed |
9 Public Health Agency of Canada (2024) The Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS): 2022 executive summary: key and integrated findings. Cat.: HP40-351/2022E-PDF. Public Health Agency of Canada, Ottawa, ON, Canada. https://www.canada.ca/content/dam/phac-aspc/documents/services/publications/drugs-health-products-canadian-integrated-program-antimicrobial-resistance-surveillance-2022-executive-summary/canadian-integrated-program-antimicrobial-resistance-surveillance-2022-executive-summary-en.pdf
10 The University of Melbourne (2023) Centre for Pathogen Genomics: building and supporting collaborative opportunities for research, genomics-enhanced infectious disease surveillance, and capacity building and training. https://biomedicalsciences.unimelb.edu.au/departments/microbiology-Immunology/research/our-research-centres-and-programs/centre-for-pathogen-genomics (accessed 14 March 2024)
11 Stroe O (2022) Building bioinformatics capacity in Latin America. EMBLetc 2022(99), 16 November 2022. https://www.embl.org/news/embletc/issue-99/building-bioinformatics-capacity-in-latin-america/.
| Google Scholar |
12 Crow D (2019) A new wave of genomics for all. Cell 177(1), 5-7.
| Crossref | Google Scholar | PubMed |
 Dr Veronica Jarocki is a CRC SAAFE Foundational Postdoctoral Fellow based at the Australian Institute of Microbiology and Infection (AIMI) at the University of Technology—Sydney. Her research interests include genomic AMR surveillance in wastewater, humans and animals and characterising the mechanisms behind AMR dissemination between sources. She has a background in subunit vaccine design and development against animal pathogens. |
 Dr Max Cummins is a researcher at AIMI at the University of Technology—Sydney, whose research interests include microbial genomics, antimicrobial resistance, bioinformatics, big data, AI and data visualisation. Max co-leads the Australian Pathogen Genomics Program’s One Health theme, utilising bioinformatic tools to generate meaningful and informative insights into bacterial populations found across humans, animals, food and the environment. |
 Dr Celeste Donato is a postdoctoral researcher at the Doherty Institute and her research interests include pathogen genomics, molecular and genomic epidemiology with expertise in viral enteric pathogens. Celeste co-leads the One Health and Candida auris projects within the Australian Pathogen Genomics program. |
 Benjamin Howden is Infectious Diseases Physician and Public Health Microbiologist. He is Professor and Director of the Microbiological Diagnostic Unit Public Health Laboratory at The University of Melbourne (Doherty Institute). His research interests include genomics for understanding and responding bacterial pathogens of public health significance and antimicrobial resistance. He is lead of the Australian Pathogen Genomics Program (AusPathoGen). |
 Steven Djordjevic is a Distinguished Professor of Infectious Disease in the Australian Institute for Microbiology & Infection at the University of Technology—Sydney. His research interests are in antimicrobial resistance and pathogen evolution through a One Health lens. His laboratory conducts genomic epidemiological analyses of a broad range of Gram-negative bacterial pathogens, and the mobile elements they carry, from hospital and veterinary clinical environments, food production animals, urban and native wildlife, wastewater and natural water systems. |