The Australian Group on Antimicrobial Resistance
Geoffrey W. Coombs A B C * , Denise A. Daley B C , Jan M. Bell D , Shakeel Mowlaboccus A B , Jon R. Iredell E F G and on behalf of the Australian Group on Antimicrobial ResistanceA
B
C
D
E
F
G

Prof. Geoffrey Coombs is the Chair of Public Health at the School of Medical, Molecular and Forensic Sciences, Murdoch University, and a Clinical Scientist for PathWest Laboratory Medicine – WA at Fiona Stanley Hospital. At Murdoch University, he is the Chair of AGAR. |

Ms Denise Daley has a 40-year background in medical microbiology in NSW and WA, and currently works for PathWest Laboratory Medicine – WA at Fiona Stanley Hospital in Perth. Denise has been the AGAR Scientific Officer since 2011. |

Jan Bell is a Scientific Officer for the AGAR Gram-negative Surveillance Outcome Program (GnSOP). She is also a microbiology advisor to the Antimicrobial Use and Resistance in Australia (AURA), Infection Prevention Control (IPC) and Healthcare Associated Infection (HAI) team and Emerging Issues stream at the Australian Commission on Safety and Quality in Health Care. |

Dr Shakeel Mowlaboccus is an early-career researcher in the field of infectious diseases and AMR with over 10 years’ experience in microbiology, large-scale whole genome sequencing, bioinformatics analyses, and molecular cloning. He is the current International Young Ambassador representing Australia at the American Society for Microbiology and the Chair-elect of the Western Australia branch of the Australian Society for Microbiology. |

Jon Iredell is a physician and microbiologist based at the Westmead Institute for Medical Research, Westmead Hospital, The University of Sydney and is AMR lead at the Institute for Clinical Pathology and Medical Research, New South Wales Health Pathology. He is a past President of the Australian Society for Microbiology. |
Abstract
Antimicrobials are an integral component of healthcare delivery. When resistance to an antimicrobial emerges, it has a significant impact on an individual’s treatment and the community more broadly. In Australia, to monitor antimicrobial resistance in human health, the Australian Group on Antimicrobial Resistance (AGAR) performs three ongoing blood-based antimicrobial surveillance programs: the Australian Staphylococcus aureus Surveillance Outcome Program (ASSOP); the Australian Enterococcal Surveillance Outcome Program (AESOP); and the Gram-negative Surveillance Outcome Program (GnSOP). All data are available on the AGAR website and are included in the Australian Government Department of Health and Aged Care’s Antimicrobial Use and Resistance in Australia Surveillance System.
Keywords: AMR, antimicrobial resistance, E. coli, Enterococcus faecium, S. aureus, surveillance.
Introduction
The Australian Group on Antimicrobial Resistance (AGAR) is a unique collaboration of clinicians and scientists from private and public microbiology laboratories located across metropolitan and rural Australia. Coordinated by the Australian Society for Antimicrobials, the group conducts targeted surveillance of selected pathogens that cause bloodstream infections and collects demographic, outcome data, and data on antimicrobial resistance (AMR) rates. Funded by the Australian Government Department of Health and Aged Care, AGAR is a core component of the Antimicrobial Use and Resistance (AURA) Surveillance System,1 which was established by the Australian Commission on Safety and Quality in Health Care (ACSQHC) in 2014, and is coordinated by the Department of Health and Aged Care. AURA supports Australia’s efforts to prevent and contain AMR in human health and implementation of Australia’s National Antimicrobial Resistance Strategy – 2020 and Beyond.2
The AGAR, which was founded in 1985, initially involved 13 teaching hospitals and was sponsored by Eli Lilly Australia Pty Ltd. The group has subsequently grown, and now involves 33 laboratories servicing 56 institutions. This broadening of the group has meant that not only does the group have good information as to what is happening with major pathogens in the larger teaching hospitals (both adult and children) in each state and territory but can monitor what is happening with AMR rates in private hospitals.
The initial AMR surveillance program focused on resistance in Staphylococcus aureus and was known as the Staphylococcus Awareness Program (SAP). The first survey into enteric Gram-negatives began in 1992, and focused on Escherichia coli and Klebsiella spp. Later, Enterobacter spp. were added. With the emergence of vancomycin-resistant enterococci in Australia in the mid-1990s, Enterococcus spp. surveys commenced in 1995. Occasional surveys of other pathogens such as Streptococcus pneumoniae and Haemophilus influenzae have also been performed.
The AGAR surveillance programs
Since January 2013, the AGAR has focussed on three main surveillance programs examining blood culture isolates only:
Australian Staphylococcus aureus Sepsis Outcome Program (ASSOP);
Australian Enterococcal Sepsis Outcome Program (AESOP); and
Australian Enterobacteriaceae Sepsis Outcome Program (EnSOP).
The bacteria included in the AGAR surveillance programs are considered priority organisms for the surveillance of AMR in Australia.
In January 2015, Pseudomonas aeruginosa and Acinetobacter species were added to EnSOP forming the Gram-negative Sepsis Outcome Program (GnSOP). The term ‘Sepsis’ in the programs was changed in 2021 to ‘Surveillance’ to better reflect the AGAR’s surveillance of episodes of bacteraemia rather than sepsis. The AGAR contributing laboratories continuously provide minimum inhibitory concentration data and demographic data on isolates from all episodes of bacteraemia. This aligns closely with the approach taken by the European targeted-resistance surveillance system EARS-Net.3 Isolates are referred to the AGAR coordinating laboratories where whole genome sequencing is performed on all methicillin-resistant S. aureus (MRSA), Enterococcus faecium and selected GnSOP antimicrobial-resistant organisms.
The 2022 AGAR programs primary objectives
ASSOP
To determine the proportion of S. aureus bacteraemia (SAB) isolates demonstrating AMR, with particular emphasis on:
AESOP
To determine the proportion of E. faecalis and E. faecium bacteraemia isolates demonstrating AMR, with particular emphasis on:
GnSOP
To determine the proportion of Enterobacterales spp., P. aeruginosa and Acinetobacter species bacteraemia isolates with particular emphasis on:
co-resistance and multidrug resistance
emerging resistance to reserve agents such as carbapenems and colistin
the molecular basis of resistance to third generation cephalosporins, quinolones and carbapenems
The AGAR produces detailed annual reports on each program and publishes the programs’ key findings in the Communicable Diseases Intelligence journal.4–6 All reports are available on the AGAR website (see https://agargroup.org.au).
The ACSQHC also analyses data collected by the AGAR, which complements the two AMR surveillance programs developed by the Commission: the National Alert System for Critical Antimicrobial Resistances (CARAlert) and the Australian Passive AMR Surveillance (APAS). The data generated are used by AURA, which is Australia’s nationally coordinated surveillance system for gathering data on antimicrobial use and resistance in human health. The fifth AURA report on antimicrobial use and resistance in humans in Australia was released in 2023.7
The AGAR and APAS data are also submitted to the World Health Organization’s Global Antimicrobial Resistance and Use Surveillance System (GLASS) (see https://www.who.int/initiatives/glass).
In addition to the annual reports, the AGAR in collaboration with the ACSQHC produces an annual amalgam surveillance report.8
The 2022 AGAR amalgam report provides analyses of data on AMR associated with bloodstream infections across Australia in 2022.8 The report also includes considerations and implications for patient safety and delivery of care. Implications for health care identified by analyses of the 2022 AGAR data include:
A longitudinal trend of increasing resistance in gram-negative organisms.
Prevalence of extended spectrum β-lactamases (ESBLs) in hospital-onset E. coli infections and variations between states and territories.
Uncommon, but concerning, carbapenemase-producing gram-negative organisms.
Changing patterns of resistance in Enterococcus species.
Methicillin resistance in S. aureus.
Epidemiology of clinical manifestations of bacteraemia.
Variation across states and territories in patterns of resistance.
Variation between hospital and community settings in patterns of resistance.
Extended lengths of stay for patients with enterococcal and staphylococcal bacteraemia
Key AGAR findings in the 2022 surveillance outcome programs
ASSOP
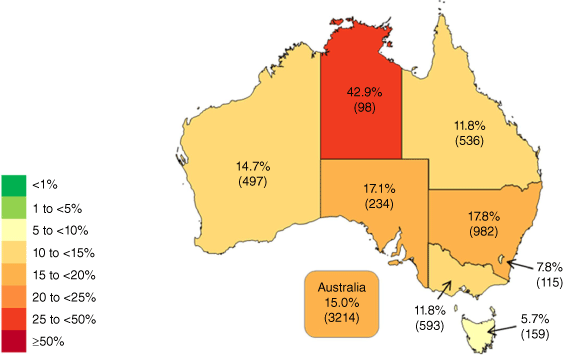
Between 1 January to 31 December, 3214 SAB episodes were reported, of which 77.5% were community onset. Of all episodes, 15.0% were due to methicillin-resistant isolates – ranging from 5.7% in Tasmania to 42.9% in the Northern Territory (Fig. 1)
The 30-day all-cause mortality was 17.5%. There was a significant difference in mortality between methicillin-susceptible S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA) (16.8 and 21.4% respectively, P = 0.0246); and between community-associated MRSA (CA-MRSA) and healthcare-associated MRSA (HA-MRSA) clones (19.9 and 34.5% respectively, P = 0.0166). There was no significant difference in mortality between hospital-onset (19.5%) and community-onset (17.0%) bacteraemia.
The 30-day all-cause mortality for S. aureus was significantly lower among children (<18 years) (5/219, 2.3%) compared to adults (459/2426, 18.9%) (P < 0.01).
The hospital length of stay was more than 30 days in 24.6% of patients (21.5% in MRSA; 25.2% in MSSA).
Resistance to non-β-lactam antimicrobials in MRSA have continued to decline overall, largely due to the substantial decline of the multi-resistant ST239-III clone.
CA-MRSA strains were the dominant cause of MRSA bacteraemia.
Two HA-MRSA clones were identified: ST22-IV (EMRSA-15) which was the dominant clone and ST239-III (Aus 2/3 EMRSA). No HA-MRSA isolates harboured the Panton–Valentine leukocidin (PVL)-associated genes.
Sixty-four CA-MRSA clones were identified; the dominant CA-MRSA clone was ST93-IV (Queensland clone). Overall, 42.8% of CA-MRSA isolates harboured PVL genes.
Percentage of Staphylococcus aureus from patients with bacteraemia with resistance, as defined by EUCAST, to methicillin in Australia (source: AGAR 20224).
AESOP
Between 1 January to 31 December 2022, 1535 episodes of bacteraemia were reported: the majority (92.8%) of enterococcal bacteraemia episodes were caused by Enterococcus faecalis or E. faecium.
The combined 30-day all-cause mortality for E. faecalis and E. faecium was 21.5%.
There was a significant difference in 30-day all-cause mortality between E. faecalis (17.2%) and E. faecium (26.9%) (P < 0.01), and between vancomycin-resistant and vancomycin-susceptible E. faecium episodes (34.4 and 19.7% respectively, P < 0.01).
The length of stay in hospital following enterococcal bacteraemia was more than 30 days for 22.3% of patients.
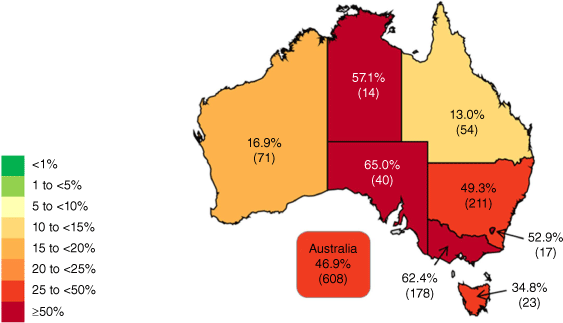
Of bacteraemia caused by E. faecium, 46.9% were phenotypically vancomycin-resistant – ranging from 16.9% in Western Australia to 65.0% in South Australia (Fig. 2).
Overall, 48.8% of E. faecium harboured vanA or vanB genes (vanA 13.7%; vanB 35.1%).
Percentage of Enterococcus faecium from patients with bacteraemia with resistance, as defined by EUCAST, to vancomycin in Australia (source: AGAR 20224).
GnSOP
From 1 January 2022 to 31 December 2022, 9739 episodes of gram-negative bacteraemia were reported, including Enterobacterales (90.1%), P. aeruginosa (8.6%) and Acinetobacter species (1.3%). Of the Enterobacterales, three genera – Escherichia (60.1%), Klebsiella (20.9%) and Enterobacter (5.7%) – contributed 86.7% of all Enterobacterales bacteraemias.
The all-cause 30-day mortality rate for gram-negative bacteraemia was 13.0% (12.5% for Enterobacterales, 18.4% for P. aeruginosa, and 13.0% for Acinetobacter species).
There was a significant difference in 30-day all-cause mortality between children and adults (3.7 v. 12.9%, P < 0.01) with Enterobacterales bacteraemia episodes.
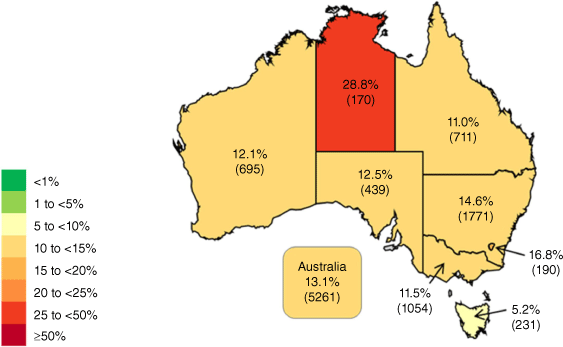
Overall, 13.1% of E. coli were resistant to third generation cephalosporins – ranging from 5.2% in Tasmania to 28.8% in the Northern Territory (Fig. 3).
Fluoroquinolone resistance in E. coli was 13.7% (up from 12.3% in 2021).
Fluoroquinolone resistance is commonly linked to cephalosporin resistance caused by ESBLs of the CTX-M type. A little over two-thirds (255/358, 71.2%) of E. coli that were ciprofloxacin-resistant and had confirmed ESBL β-lactamase gene(s) belonged to ST131 (n = 198, 55.3%) or ST1193 (n = 57, 15.9%).
Rates of carbapenemase-producing Enterobacterales (CPE) bacteraemic isolates remain low (0.3% overall, mostly carrying blaIMP-4). For Enterobacter cloacae complex the figure is higher at 2.1% overall.
Percentage of Escherichia coli from patients with bacteraemia with resistance, as defined by EUCAST, to third-generation cephalosporins (ceftriaxone or ceftazidime) in Australia (source: AGAR 20224).
International comparisons
Australia had relatively lower rates of resistance in 2022, compared to 2021 data available from the European Union (EU) and European Economic Area (EEA) countries reporting to EARS-Net9 or the WHO European Region countries (excluding EU and EEA countries) reporting to CAESAR.10 Australia ranks towards the middle in rates of resistance to methicillin in S. aureus compared to all European countries, and in the top quarter in rates of resistance to vancomycin in E. faecium. Australia also ranks towards the middle in rates of resistance to third-generation cephalosporins in E. coli, but in the bottom quarter in rates of resistance to fluoroquinolones.
Data availability
The data that support this study will be shared upon reasonable request to the corresponding author.
Declaration of funding
The Australian Group on Antimicrobial Resistance is funded by the Australian Government Department of Health and Aged Care.
Acknowledgements
AGAR gratefully acknowledges the Antimicrobial Resistance Laboratory, Microbial Genomics Reference Laboratory, CIDMLS, ICPMR, Westmead Hospital (Jenny Draper and Elena Martinez), the Westmead Institute for Medical Research (Sally Partridge and Alicia Fajardo Lubian) and the Antimicrobial Resistance and Infectious Diseases Research Laboratory, Murdoch University (Shakeel Mowlaboccus and Princy Shoby) for performing whole genome sequencing. Members of the AGAR from 2022 are: Australian Capital Territory: Peter Collignon and Susan Bradbury, Canberra Hospital; New South Wales: Alison Kesson and Andrew Jarrett, Children’s Hospital Westmead; Thomas Gottlieb and John Huynh, Concord Hospital; Gabrielle O’Kane and Nola Hitchick, Gosford Hospital; Hemalatha Varadhan and Bree Harris, John Hunter Hospital; Michael Maley and Helen Ziochos, Liverpool Hospital; James Branley and Linda Douglass, Nepean Hospital; Angela Wong, Royal North Shore Hospital; Sebastiaan van Hal and Thomas Le, Royal Prince Alfred Hospital; David Lorenz, St Vincent’s Hospital Sydney; Monica Lahra and Peter Huntington, Sydney Children’s Hospital and Prince of Wales Hospital; Jonathan Iredell and Elena Martinez, Westmead Hospital; Peter Newton and Melissa Hoddle, Wollongong Hospital; Northern Territory: James McLeod, Alice Springs Hospital; Rob Baird and Jann Hennessy, Royal Darwin Hospital; Queensland: Claire Heney and Narelle George, Pathology Queensland Central Laboratory, Royal Brisbane and Women’s Hospital, Queensland Children’s Hospital; Petra Derrington and Cheryl Curtis, Pathology Queensland Gold Coast University Hospital; Robert Horvath and Laura Martin, Pathology Queensland Prince Charles Hospital; Naomi Runnegar and Joel Douglas, Pathology Queensland Princess Alexandra Hospital; Jennifer Robson and Marianne Allen, Sullivan Nicolaides Pathology, Greenslopes Private Hospital and Mater Private Hospital Townsville; South Australia: Kelly Papanaoum and Xiao Ming Chen, SA Pathology, Flinders Medical Centre; Morgyn Warner and Kija Smith, SA Pathology, Royal Adelaide Hospital and Women’s and Children’s Hospital; Tasmania: Pankaja Kalukottege and Kathy Wilcox, Launceston General Hospital; Louise Cooley and David Jones, Royal Hobart Hospital; Victoria: Adam Jenney and Jacqueline Williams, Alfred Hospital; Marcel Leroi and Elizabeth Grabsch, Austin Health; Tony Korman, Despina Kotsanas and Kathryn Cisera, Dandenong Hospital, Monash Children’s Hospital, Monash Medical Centre; Katherine Bond and Rose Cotronei, Royal Melbourne Hospital; Andrew Daley and Gena Gonis, Royal Women’s and Children’s Hospital; Amy Crowe and Lisa Brenton, St Vincent’s Hospital; Western Australia: Shalinie Perera and Ian Meyer, Western Diagnostic Pathology, Joondalup Hospital; Denise Daley, PathWest Laboratory Medicine WA, Fiona Stanley Hospital; Christopher Blyth, PathWest Laboratory Medicine WA, Perth Children’s Hospital; Ronan Murray and Jacinta Bowman, PathWest Laboratory Medicine WA, Sir Charles Gairdner Hospital; Michael Leung, PathWest Laboratory Medicine WA, north-west regional WA; Owen Robinson and Geoffrey Coombs, PathWest Laboratory Medicine WA, Royal Perth Hospital; Sudha Pottumarthy-Boddu and Jacqueline Foster, Australian Clinical Laboratories, St John of God Hospital Murdoch. The ACSQHC produced the amalgam report. The Department of Health and Aged Care funds AGAR.
References
1 Australian Commission on Safety and Quality in Health Care (2022) Antimicrobial Use and Resistance in Australia (AURA). ACSQHC, Sydney, NSW, Australia. https://www.safetyandquality.gov.au/our-work/antimicrobial-resistance/antimicrobial-use-and-resistance-australia-aura
2 Department of Health and Aged Care and Department of Agriculture, Fisheries and Forestry (2020) Australia’s National Antimicrobial Resistance Strategy – 2020 and beyond. Date published 13 March 2020. Commonwealth of Australia. https://www.amr.gov.au/news/australias-national-antimicrobial-resistance-strategy-2020-and-beyond
3 European Centre for Disease Prevention and Control (2023) European Antimicrobial Resistance Surveillance Network (EARS-Net). European Union, Stockholm, Sweden. https://www.ecdc.europa.eu/en/about-us/networks/disease-networks-and-laboratory-networks/ears-net-data (last updated 17 November 2023)
4 Coombs GW et al. (2023) Australian Group on Antimicrobial Resistance (AGAR) Australian Staphylococcus aureus Surveillance Outcome Program (ASSOP) bloodstream infection annual report 2022. Commun Dis Intell 47, 1-19.
| Crossref | Google Scholar | PubMed |
5 Coombs GW et al. (2023) Australian Group on Antimicrobial Resistance (AGAR) Australian Enterococcal Surveillance Outcome Program (AESOP) bloodstream infection annual report 2022. Commun Dis Intell 47, 1-11.
| Crossref | Google Scholar | PubMed |
6 Bell JM et al. (2023) Australian Group on Antimicrobial Resistance (AGAR) Australian Gram-negative Surveillance Outcome Program (GnSOP) Bloodstream Infection Annual Report 2022. Commun Dis Intell 47, 1-13.
| Crossref | Google Scholar | PubMed |
7 Australian Commission on Safety and Quality in Health Care (2023) AURA 2023: Fifth Australian report on antimicrobial use and resistance in human health – report. ACSQHC, Sydney, NSW, Australia. https://www.safetyandquality.gov.au/publications-and-resources/resource-library/aura-2023-fifth-australian-report-antimicrobial-use-and-resistance-human-health-report
8 Australian Commission on Safety and Quality in Health Care (2023) Surveillance outcome programs bloodstream infections: 2022 report. D23-53608. November 2023. ACSQHC, Sydney, NSW, Australia. https://www.safetyandquality.gov.au/sites/default/files/2023-12/AGAR%20Surveillance%20Outcome%20Programs%202022%20Report.pdf
9 European Centre for Disease Prevention and Control (2022) Antimicrobial resistance in the EU/EEA (EARS-Net) – annual epidemiological report for 2021. European Union, Stockholm, Sweden. https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2021
10 European Centre for Disease Prevention and Control (2023) Antimicrobial resistance surveillance in Europe 2023–2021 data. Surveillance report. European Union, Stockholm, Sweden. https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2023-2021-data
 Prof. Geoffrey Coombs is the Chair of Public Health at the School of Medical, Molecular and Forensic Sciences, Murdoch University, and a Clinical Scientist for PathWest Laboratory Medicine – WA at Fiona Stanley Hospital. At Murdoch University, he is the Chair of AGAR. |
 Ms Denise Daley has a 40-year background in medical microbiology in NSW and WA, and currently works for PathWest Laboratory Medicine – WA at Fiona Stanley Hospital in Perth. Denise has been the AGAR Scientific Officer since 2011. |
 Jan Bell is a Scientific Officer for the AGAR Gram-negative Surveillance Outcome Program (GnSOP). She is also a microbiology advisor to the Antimicrobial Use and Resistance in Australia (AURA), Infection Prevention Control (IPC) and Healthcare Associated Infection (HAI) team and Emerging Issues stream at the Australian Commission on Safety and Quality in Health Care. |
 Dr Shakeel Mowlaboccus is an early-career researcher in the field of infectious diseases and AMR with over 10 years’ experience in microbiology, large-scale whole genome sequencing, bioinformatics analyses, and molecular cloning. He is the current International Young Ambassador representing Australia at the American Society for Microbiology and the Chair-elect of the Western Australia branch of the Australian Society for Microbiology. |
 Jon Iredell is a physician and microbiologist based at the Westmead Institute for Medical Research, Westmead Hospital, The University of Sydney and is AMR lead at the Institute for Clinical Pathology and Medical Research, New South Wales Health Pathology. He is a past President of the Australian Society for Microbiology. |


