Monitoring the viable grapevine microbiome to enhance the quality of wild wines
Brady L. Welsh A * , Raphael Eisenhofer A B , Susan E. P. Bastian C and Stephen P. Kidd A D EA School of Biological Sciences, The University of Adelaide, North Terrace Campus, Adelaide, SA 5005, Australia.
B Center for Evolutionary Hologenomics, GLOBE Institute, University of Copenhagen, DK-1353 Copenhagen, Denmark.
C School of Agriculture, Food & Wine, Waite Research Institute, The University of Adelaide, Waite Campus, PMB 1, Glen Osmond, SA 5064, Australia.
D Research Centre of Infectious Disease, The University of Adelaide, North Terrace Campus, Adelaide, SA 5005, Australia.
E Australian Centre for Antimicrobial Resistance Ecology (ACARE), The University of Adelaide, North Terrace Campus, Adelaide, SA 5005, Australia.

Brady Welsh is a student at The University of Adelaide undertaking a Doctor of Philosophy (PhD) in the field of grapevine microbiology investigating the interactions between the living grapevine microbiome and common vineyard fungicides. |

Dr Raphael Eisenhofer is a postdoctoral researcher at the Globe Institute, University of Copenhagen, Denmark, and an adjunct assistant lecturer at the University of Adelaide. His research focus is studying the microbiomes of native Australian mammals, though he applies his metagenomic expertise to diverse study systems. |

Assoc. Prof. Susan Bastian is a qualified winemaker and has been a wine educator and researcher at the University of Adelaide for more than two decades. Broadly, her research group examines the whole wine value chain, specifically grape and wine quality; viticultural and winemaking production impacts, including the influence of wine yeasts, on wine sensory and chemical composition; and wine consumer preference. |

Dr Stephen Kidd is a research group leader at the University of Adelaide. His group studies the bacterial response to environmental stress, specifically the adaptation over long periods of time and the impact on the functionality of the microorganisms. This has extended to the responses of microbial communities, such as the grapevine microbiome. |
Microbiology Australia 44(1) 13-17 https://doi.org/10.1071/MA23004
Submitted: 17 January 2023 Accepted: 26 February 2023 Published: 9 March 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Grapevines that are used for winemaking host a diverse range of microorganisms that make up their microbiome. The microbes that inhabit the grapevine have been used by winemakers to produce wine for centuries, although modern wine producers often rely on inoculated microorganisms such as Saccharomyces cerevisiae. In the Australian wine industry, there is a movement towards returning to the utilisation of the microbiome for wine fermentation. With the recent increase in the understanding of the role of the grapevine microbiome in grapevine health, fermentation and subsequent wine sensory traits, the microbial world offers a new level of complexity that can be harnessed for winemaking. In order to develop and maintain a desired vineyard micro-biodiversity, extensive microbial monitoring is required. Here we discuss the utilisation of a viability selection dye in order to distinguish between microorganisms that are live and associated with the host, and relic signals generated from non-living sources.
Keywords: fermentation, metagenomics, micro-biodiversity, microbiome, microbiota, wild, wine.
Fermentation and the grapevine microbiome
The process of producing wine from grape juice was discovered to be the result of microbial organisms in the nineteenth century by Louis Pasteur. With advancements in biochemistry and microbiology, the understanding behind this complex fermentation process has been investigated extensively. As such, winemaking is one of the oldest human utilisations of microorganisms through the fine-tuned control of these complex fermentation reactions. The fermentation process is mostly completed during ‘primary fermentation’, typically by inoculated yeasts, wherein most of the alcohol content is produced by the alcoholic fermentation of glucose and fructose into ethanol. This is followed by ‘secondary fermentation’, which leads to microbial stability and advance the wine’s sensory profile (its flavour and aroma) through processes such as malolactic fermentation.1 Two categories of microorganisms, fungi (predominantly yeasts) and bacteria, are recognised as the driving force of the primary and secondary fermentation processes, as well as the more recently recognised ‘spontaneous fermentation’ process performed by wild yeasts and bacteria, which originate from the plant’s microbiome.2–6
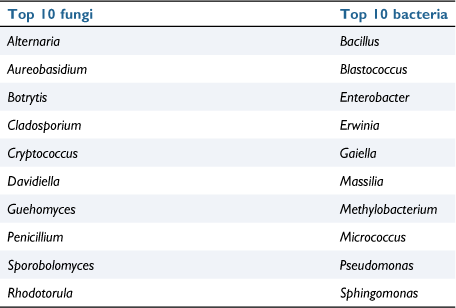
The grapevine microbiome, similar to other, complex microbiomes such as the human gut microbiome, is separated into distinct compartments with each hosting its own diversity of microbes. The main subdivisions of the grapevine microbiome are the rhizosphere microbiome (the area surrounding the roots), the endosphere microbiome (the area within the plant’s tissues) and the phyllosphere microbiome (the surface of the aerial portion of the plant). Of these, the rhizosphere has been the main focus since its coinage7; however, current literature is beginning to recognise the importance of the phyllosphere and endosphere microbiomes because of their role in vine health as well as wine fermentation.8 Both phyllospheric and endospheric microorganisms inhabit the grape berry tissues, collectively known as the carposphere microbiome, and as such act as diverse pools of fungal and bacterial taxa that can be utilised as wild inoculations to be utilised during spontaneous fermentation9,10 (Table 1).
|
The movement towards wild wines
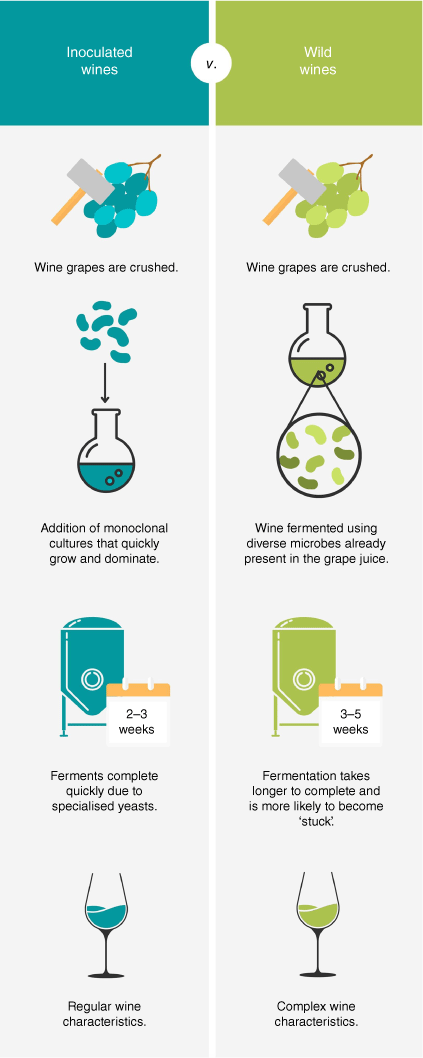
Currently, within the Australian wine industry, a movement is underway with some wineries adopting traditional practises for winemaking through the production of ‘wild wines’. Wild wines are fermented exclusively through spontaneous fermentation, using wild microorganisms derived from the grapevine microbiome to ferment wines to completion as opposed to exogenously inoculated Saccharomyces cerevisiae cultures (Fig. 1). By utilising the grapevine microbiome for wine fermentation, winemakers are capable of producing complex wines that have unique and diverse sensory traits.6,11 Furthermore, many methods of maintaining optimal micro-biodiversity in vineyards, such as bio-dynamic and low-input approaches, align with the Australian wine industry’s promotion of ‘organic’ and pesticide-free products for health-conscious target consumers and more sustainable viticultural practises.12
|
Although wild wines are becoming favourable, there is a multitude of challenges that must be overcome to successfully complete fermentation and produce wines with palatable characteristics. Two methods of producing wild wines currently exist. The first utilises vine-borne microbial isolates that can be inoculated into wine ferments to drive and support the complex fermentation reactions.13–15 Although this method does not use specialised microorganisms such as S. cerevisiae, it also does not completely depend on the endogenous grapevine microbiota, and thus does not require the involved viticultural processes, such as monitoring, which come along with wild ferments. The second, and more-traditional method, utilises the vineyard’s own micro-biodiversity, which is cultivated on the grapevine that makes its way into the wine ferment during the crushing process. This can produce complex sensory traits that vary between seasons and regions depending on a variety of different factors (natural, human-derived and environmental).
As with most organisms, a healthy grapevine microbiome is one that holds a richness and diversity specific to its host organism,16 with the opposite typically being an indicator of a disease state within the host vine (e.g. one pathogenic taxon dominating the niche).16–18 As the health of a plant can be dictated by that state of its microbiome, as well as the outcomes of wild fermentation, suitable micro-biodiversities must be maintained within vineyards for effective wine production. Although the occurrence of each taxon is largely host-dependent,10,18 these microbial communities are also affected by various stressors from the environment that shape the structure of the grapevine microbiome. The frequency of each taxon is also dependent on season, geographic location, water availability, UV exposure and human intervention.16 Controlled cultivation of these specific communities is crucial to the host plant’s development; however, for wild winemaking, effective monitoring of the micro-biodiversity is also of great importance in order to encourage the development of beneficial species and hinder the growth of unwanted microorganisms such as those related to wine spoilage.
Dead or alive? Monitoring the viable grapevine microbiome
Metagenomic monitoring methods, including many of the PCR-based quantifications and DNA sequencing technologies that are typically applied to microbiome research, must be utilised to properly ensure vineyards are maintaining a healthy micro-biodiversity. The introduction of these methods would allow for incredibly efficient characterisation of the microbial richness and diversity present on a vineyard’s grapevines. Understanding which microorganisms are present in the grapevine microbiomes of a vineyard will allow grape growers to understand what involvement is necessary to cultivate or preserve a healthy microbiota. However, one of the most significant downsides to current microbiome analyses is the inability to discern which organisms were ‘alive’, at the time of sampling.19,20 Relic DNA makes up all DNA from non-living sources, such as dead cells, compromised cells or environmental DNA (eDNA),21 and can be just as easily amplified and sequenced as DNA extracted from viable cells. This indiscriminate nature of PCR can provide false positives to vineyard biodiversity monitoring, which, with the wrong intervention, could lead to undesired impacts on the wild microorganisms and downstream outcomes.
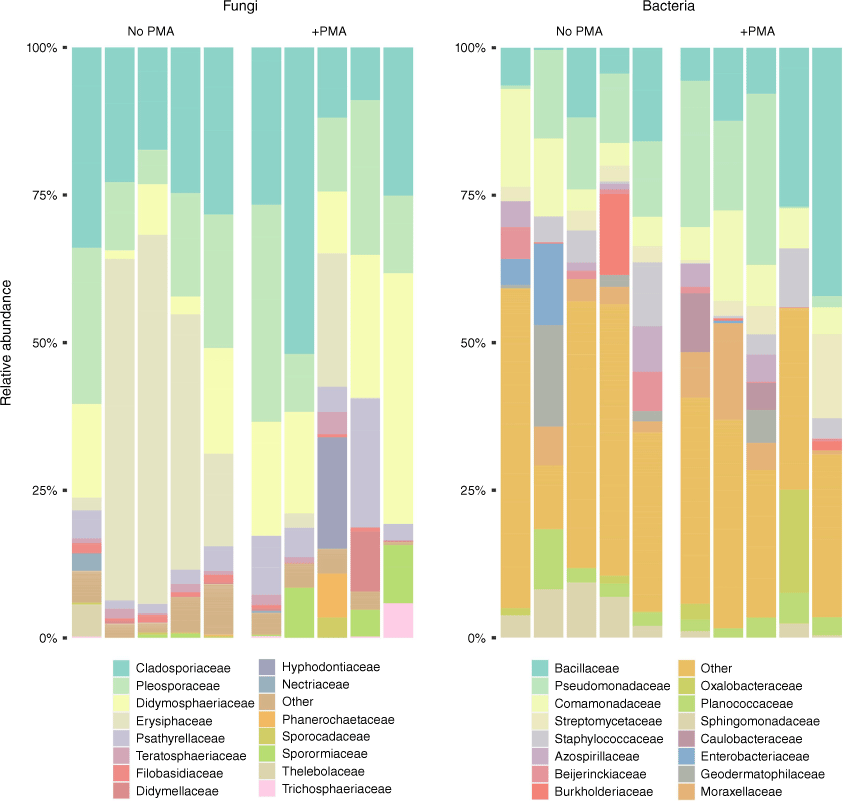
Discrimination between live and dead cells is an important milestone that must be overcome to allow grape growers the ability to determine which taxa are present and associated with their host vines. To combat this, we have utilised the viability selection dye, propidium monoazide (PMA),20–23 which is capable of covalently binding to DNA when exposed to light. The binding of the PMA dye to DNA prevents it from being amplified by PCR, and thus, cannot be observed. However, the dye cannot cross the intact membrane of live cells, allowing it to only bind to relic DNA from compromised cells or environmental sources. We tested this dye using leaf phyllosphere microbiome samples collected from Vitis vinifera ‘Syrah’ (Shiraz) grapevines and were able to gain sequencing data representative of the living phyllosphere microbiome.
Our PMA-Seq results utilised both 16S and ITS rRNA gene amplicon sequencing to observe the bacterial and fungal communities respectively. Sequencing was conducted on two sets of grape leaf swab samples; both sets were prepared identically with one being treated with PMA (representative of the viable communities) and the other left untreated (representative of the total DNA within the sample) prior to DNA extraction and amplification (B. Welsh, unpubl. data). DNA sequencing of these samples demonstrated the PMA dye’s ability to effectively remove relic DNA from grapevine microbiome samples without sacrificing the quality of the data (Fig. 2). For example, from the untreated sequencing data, we observed a high relative abundance of the fungal family Erysiphaceae, a taxon responsible for powdery mildew (a common grapevine infection); however, this abundance was lower in the samples treated with PMA. This result suggests that the observed Erysiphaceae taxa were, in fact, false positives, and the DNA responsible for those sequences likely came from non-viable sources. In a vineyard setting, if these metagenomic monitoring practices were taking place without the use of PMA viability selection, these results would have suggested a disease instance within the vines, resulting in fungicidal sprays that could affect the established structure of the grapevine microbiome. Responses to false positives could be catastrophic to the micro-biodiversity of vineyards resulting in unfavourable sensory characteristics in wild wines.
Conclusions
With the growing movement towards wild wines within the wine industry, new viticultural and oenological practices will be required. To produce grapevines with sufficient micro-biodiversity for the fermentation of wine, metagenomic analytical techniques must be adopted for sufficient monitoring of the grapevine microbiome. With the inclusion of metagenomic monitoring in vineyards, considerations must be taken to ensure these microbial communities are observed with extreme scrutiny to account for potential false positives such as those demonstrated in our results. By specifically monitoring the living microbiome, the wine industry can begin to discover and adopt practices for cultivating microbial communities which produce healthier grapevines for more sustainable practices, as well as higher quality wild wines for the current growing market.
Data availability
The data that support this study will be shared upon reasonable request to the corresponding author.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
This work was supported by Wine Australia, with levies from Australia’s grape growers and winemakers and matching funds from the Australian Government.
References
[1] Styger, G et al. (2011) Wine flavor and aroma. J Ind Microbiol Biotechnol 38, 1145–1159.| Wine flavor and aroma.Crossref | GoogleScholarGoogle Scholar |
[2] Piao, H et al. (2015) Insights into the bacterial community and its temporal succession during the fermentation of wine grapes. Front Microbiol 6, 809.
| Insights into the bacterial community and its temporal succession during the fermentation of wine grapes.Crossref | GoogleScholarGoogle Scholar |
[3] Gutiérrez, AR et al. (1999) Ecology of spontaneous fermentation in one winery during 5 consecutive years. Lett Appl Microbiol 29, 411–415.
| Ecology of spontaneous fermentation in one winery during 5 consecutive years.Crossref | GoogleScholarGoogle Scholar |
[4] Beneduce, L et al. (2004) Molecular characterization of lactic acid populations associated with wine spoilage. J Basic Microbiol 44, 10–16.
| Molecular characterization of lactic acid populations associated with wine spoilage.Crossref | GoogleScholarGoogle Scholar |
[5] Clemente-Jimenez, JM et al. (2004) Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiol 21, 149–155.
| Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must.Crossref | GoogleScholarGoogle Scholar |
[6] Combina, M et al. (2005) Dynamics of indigenous yeast populations during spontaneous fermentation of wines from Mendoza, Argentina. Int J Food Microbiol 99, 237–243.
| Dynamics of indigenous yeast populations during spontaneous fermentation of wines from Mendoza, Argentina.Crossref | GoogleScholarGoogle Scholar |
[7] Hiltner, L (1904) Uber nevere Erfahrungen und Probleme auf dem Gebiet der Boden Bakteriologie und unter besonderer Beurchsichtigung der Grundungung und Broche. Arbeit Deut Landw Ges Berlin 98, 59–78.
[8] Gouka, L et al. (2022) Ecology and functional potential of phyllosphere yeasts. Trends Plant Sci 27, 1109–1123.
| Ecology and functional potential of phyllosphere yeasts.Crossref | GoogleScholarGoogle Scholar |
[9] Martins, G et al. (2013) Characterization of epiphytic bacterial communities from grapes, leaves, bark and soil of grapevine plants grown, and their relations. PLoS One 8, e73013.
| Characterization of epiphytic bacterial communities from grapes, leaves, bark and soil of grapevine plants grown, and their relations.Crossref | GoogleScholarGoogle Scholar |
[10] Bettenfeld, P et al. (2022) The microbiota of the grapevine holobiont: a key component of plant health. J Adv Res 40, 1–15.
| The microbiota of the grapevine holobiont: a key component of plant health.Crossref | GoogleScholarGoogle Scholar |
[11] König, H and Claus, H (2018) A future place for Saccharomyces mixtures and hybrids in wine making. Fermentation 4, 67.
| A future place for Saccharomyces mixtures and hybrids in wine making.Crossref | GoogleScholarGoogle Scholar |
[12] Sustainable Winegrowing Australia (2022) Annual Operating Plan: 1 July 2022–30 June 2023. https://sustainablewinegrowing.com.au/wp-content/uploads/2022/09/SWA_AOP_2022-23_W.pdf
[13] Alperstein, L et al. (2020) Yeast bioprospecting versus synthetic biology—which is better for innovative beverage fermentation? Appl Microbiol Biotechnol 104, 1939–1953.
| Yeast bioprospecting versus synthetic biology—which is better for innovative beverage fermentation?Crossref | GoogleScholarGoogle Scholar |
[14] Hranilovic, A et al. (2021) Impact of Lachancea thermotolerans on chemical composition and sensory profiles of merlot wines. Food Chem 349, 129015.
| Impact of Lachancea thermotolerans on chemical composition and sensory profiles of merlot wines.Crossref | GoogleScholarGoogle Scholar |
[15] Hranilovic, A et al. (2022) Impact of Lachancea thermotolerans on chemical composition and sensory profiles of viognier wines. J Fungi 8, 474.
| Impact of Lachancea thermotolerans on chemical composition and sensory profiles of viognier wines.Crossref | GoogleScholarGoogle Scholar |
[16] Bashir, I et al. (2022) Phyllosphere microbiome: diversity and functions. Microbiol Res 254, 126888.
| Phyllosphere microbiome: diversity and functions.Crossref | GoogleScholarGoogle Scholar |
[17] Redford, AJ et al. (2010) The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol 12, 2885–2893.
| The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves.Crossref | GoogleScholarGoogle Scholar |
[18] Gong, T and Xin, X-F (2021) Phyllosphere microbiota: community dynamics and its interaction with plant hosts. J Integr Plant Biol 63, 297–304.
| Phyllosphere microbiota: community dynamics and its interaction with plant hosts.Crossref | GoogleScholarGoogle Scholar |
[19] Carini, P et al. (2016) Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat Microbiol 2, 16242.
| Relic DNA is abundant in soil and obscures estimates of soil microbial diversity.Crossref | GoogleScholarGoogle Scholar |
[20] Emerson, JB et al. (2017) Schrödinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 5, 86.
| Schrödinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems.Crossref | GoogleScholarGoogle Scholar |
[21] Wang, Y et al. (2021) Whole microbial community viability is not quantitatively reflected by propidium monoazide sequencing approach. Microbiome 9, 17.
| Whole microbial community viability is not quantitatively reflected by propidium monoazide sequencing approach.Crossref | GoogleScholarGoogle Scholar |
[22] Nocker, A et al. (2006) Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods 67, 310–320.
| Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs dead bacteria by selective removal of DNA from dead cells.Crossref | GoogleScholarGoogle Scholar |
[23] Baymiev, AK et al. (2020) Modern approaches to differentiation of live and dead bacteria using selective amplification of nucleic acids. Microbiology 89, 13–27.
| Modern approaches to differentiation of live and dead bacteria using selective amplification of nucleic acids.Crossref | GoogleScholarGoogle Scholar |