The impact of antimicrobial resistance on induction, transmission and treatment of Clostridium difficile infection
Stacey Hong A B , Daniel R Knight A and Thomas V Riley A B C D EA Medical, Molecular and Forensic Sciences, Murdoch University, Murdoch, WA 6105, Australia
B School of Biomedical Sciences, The University of Western Australia, Queen Elizabeth II Medical Centre, Nedlands, WA 6009, Australia
C School of Medical and Health Sciences, Edith Cowan University, Joondalup, WA 6027, Australia
D Department of Microbiology, PathWest Laboratory Medicine, Queen Elizabeth II Medical Centre, Nedlands, WA 6009, Australia
E Tel: +61 8 6457 3690, Email: thomas.riley@uwa.edu.au
Microbiology Australia 40(2) 77-81 https://doi.org/10.1071/MA19022
Published: 18 April 2019
Clostridium difficile infection (CDI) of the gastrointestinal (GI) tract is a potentially life-threatening disease that has surpassed multi-drug-resistant Staphylococcus aureus as the commonest antimicrobial-resistant organism associated with healthcare1. This obligate anaerobic spore-forming Gram-positive bacillus colonises the GI tract and its numbers increase after disruption of the commensal GI microbiota often induced by exposure to antimicrobial agents2. Paradoxically, the disease that may follow its outgrowth necessitates further antimicrobial treatment. Already a major challenge to infection prevention and control strategies, there are indications that C. difficile is developing further resistance to currently used antimicrobial agents.
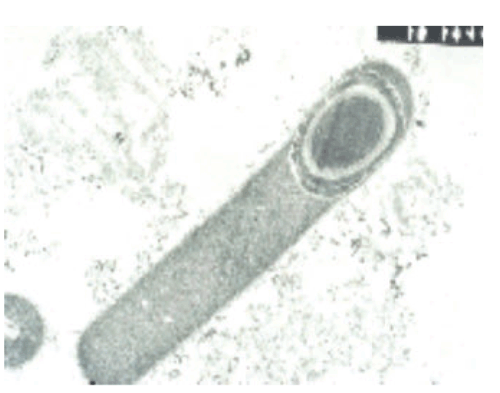
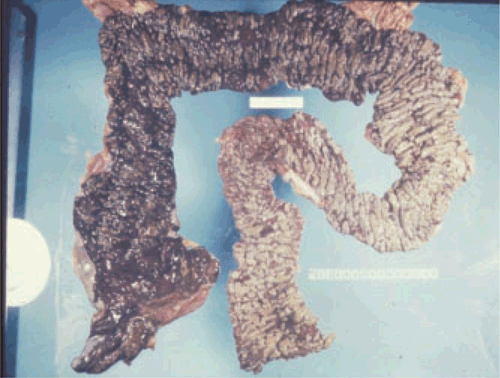
C. difficile (Figure 1) produces toxins that cause a wide spectrum of disease, ranging from mild to self-limiting diarrhoea through to severe complications including pseudomembranous colitis (PMC) (Figure 2), toxic megacolon and death2. Historically, clindamycin and third-generation cephalosporins have been recognised for their propensity to promote CDI3; however, almost all antimicrobial agents have been implicated. In Australia, treatment with metronidazole and vancomycin remains the preferred options as first-line therapy drugs for mild-to-moderate and severe cases of CDI, respectively4. Resistance to multiple antimicrobials represents a selective advantage for the emergence of new C. difficile strains.
|
|
The prevalence of CDI outbreaks has risen since the early 2000s associated with the appearance in multiple countries of an epidemic strain of C. difficile, PCR ribotype (RT) 0272. This strain displays high levels of fluoroquinolone (FQ) resistance not seen previously and infection has been associated with increased morbidity and mortality leading to description of the strain as ‘hyper-virulent’2. The increased virulence may be due to enhanced production of toxins A and B in vitro, in addition to the presence of binary toxin. Despite improved clinical management strategies for CDI, healthcare costs for treating CDI remain high. C. difficile is now recognised by the Centers for Disease Control and Prevention as the most urgent public health threat in the USA, causing more than 453 000 infections per year with approximately 29 300 deaths and $4.8 billion in excess medical costs per year5.
The emergence and rapid transmission of C. difficile strains resistant to multiple antimicrobials is now a significant problem worldwide. C. difficile has evolved multiple mechanisms for antimicrobial resistance (AMR) including chromosomal mutations and acquired mobile genetic elements (MGE), causing alterations in the targets of antimicrobials and/or in metabolic pathways. Rates of AMR in C. difficile vary considerably between studies, related to geography and antimicrobial policy. In this short review, important classes of antimicrobial agents will be discussed in relation to their role in inducing and treating CDI.
Antimicrobials that incite CDI
β-Lactams
Broad spectrum β-lactam antimicrobials such as aminopenicillins and cephalosporins are known for their propensity to cause CDI3. Cephalosporin resistance is a hallmark of C. difficile; data extrapolated from previous studies revealed that the majority of C. difficile isolates tested were resistant (95% second generation; 38% third generation) and there is usually significant clustering around the breakpoint6. Thus, C. difficile is often described as ‘intrinsically resistant’ to cephalosporins, mediated by class D β-lactamases and a β-lactam inducing penicillin binding protein encoded on cdd and blaR genes, respectively7,8.
Macrolide-lincosamide-streptogramin B (MLSB)
The macrolide-lincosamide-streptogramin (MLS) B class of antimicrobials contains structurally different but functionally similar drugs, that all bind to the 50S ribosomal subunit9. Clindamycin was the first antimicrobial agent linked to PMC in the mid-1970s2. Although the use of clindamycin has declined since its close association with CDI was first described, clindamycin resistance remains common10. In a recent review by Spigaglia et al., 51.1% of C. difficile clinical isolates in 46 papers published between 2012 and 2017 were resistant to clindamycin6. Resistant strains exhibit varying minimal inhibitory concentrations (MICs) ranging from 16 to ≥265 mg/L between RTs and often according to the geographical location6.
In C. difficile, resistance to the MLSB family is usually conferred by erythromycin methylase B, encoded by ermB, which prevents drug binding by methylation of bacterial 23S rRNA. The ermB gene is often located on MGEs such as Tn5398 and Tn6194, with multiple genetic organisations with no clear RT association11. However, clindamycin-resistant ermB negative and clindamycin-susceptible ermB positive strains of C. difficile have been reported12. Thus, alternative mechanisms of resistance may be present in C. difficile.
Erythromycin is a macrolide that inhibits protein synthesis. Macrolides alone do not have a strong association with CDI; however, the true risks are likely underestimated given that macrolides are often co-administered with other antimicrobial agents13. Similar to clindamycin, erythromycin resistance may be mediated through the expression of ermB, as well as ermFS, mutation in 23S rDNA and a possible cme efflux pump11,14,15.
FQs
FQ resistance is well documented in C. difficile owing to the FQ-resistant epidemic strain of C. difficile RT027. FQs such as ciprofloxacin and moxifloxacin target DNA gyrase (gryA and gryB) and prevent the synthesis of enzymes responsible for supercoiling bacterial DNA. Resistance to FQs in C. difficile is invariably due to alterations in drug target structure via nucleotide substitutions of gyrA and/or gyrB within the quinolone-resistance determining region of DNA gyrase subunits. The selective pressure for resistance was likely due to heavy reliance on FQs in a clinical environment, particularly with levofloxacin being one of the most commonly prescribed antimicrobials in North America during the late 1990s and early 2000s16. More recently, Freeman et al. reported 35.8% of 2694 of C. difficile isolates collected over 3 years were resistant to moxifloxacin, in multiple RTs but particularly in RTs 001, 018, 356, 017 and 19810. The detection of FQ resistance is now an important epidemiological target for the identification of epidemic C. difficile strains.
Antimicrobials for CDI treatment
Metronidazole
Metronidazole remains the primary drug of choice in the treatment of mild-to-moderate CDI in Australia although in the USA metronidazole is no longer recommended for first-line treatment17. The key mode of action is direct DNA damage following reduction of its nitro group once inside the bacterium18. Since metronidazole has been a mainstay of therapy for CDI for over 30 years, reduced efficacy has been reported. In a prospective observational study of 207 CDI patients, only 50% were successfully treated, and 22% continued to experience symptomatic CDI despite ≥10 days of treatment. Furthermore, 28% of patients experienced symptomatic recurrence within 90 days19. Publications from France20, Spain21 and elsewhere have reported clinical isolates with reduced susceptibility to metronidazole, however, observations of metronidazole resistance in vitro are scarce. A recent pan-European survey of antimicrobial susceptibility in 2694 C. difficile isolates over 3 years reported 0.2% were resistant to metronidazole, mostly RT027 and its close relative RT19810. In addition, resistance to metronidazole appears to be heterogeneous within a population and MICs were largely dependent on the antimicrobial susceptibility method21. More important, C. difficile strains isolated from patients who failed metronidazole therapy appeared to have similar MICs to those isolated from successfully treated cases. Thus, reduced efficacy of metronidazole in the clinical setting was unlikely to be attributed to decreased susceptibility and more likely linked to other bacterial and host factors that remain to be elucidated22. Currently, oral metronidazole remains the preferred antimicrobial agent for mild CDI in Europe due to low costs4.
Vancomycin
Vancomycin is the first-line treatment for moderate to severe CDI4. Antimicrobial activity is achieved through inhibiting the biosynthesis of the bacterial cell wall peptidoglycan. Unlike metronidazole, vancomycin is poorly absorbed in the gastrointestinal tract after oral administration leading to high concentrations in the gut and rapid suppression of CDI23. The mechanism for vancomycin resistance in C. difficile is still unclear but is possibly linked to the possession of a vanG homolog (vanGCd). The homolog is inducible by vancomycin in vitro but does not promote vancomycin resistance in C. difficile24. Susceptibility to vancomycin remained high with Freeman et al. reporting 98.6% of isolates susceptible10. However, there have been sporadic reports of elevated MICs ≥4-8 mg/L from a previous pan-European survey25, although the underlying mechanism of resistance was not determined. There has been a report of a cryptic vanB2 gene cassette (Tn1549-like) in a C. difficile strain isolated from an Australian calf26. Notably, vancomycin is now the preferred treatment over metronidazole for an initial episode of CDI as recommended by the Infectious Diseases Society of America and Society for Healthcare Epidemiology of America in 201817.
Rifamycins
The rifamycins achieve selective toxicity in bacteria by targeting bacterial DNA-dependent RNA polymerase27. Both rifampicin and rifaximin have been used to treat CDI due to very low MICs and they are often used as adjunctive post-vancomycin therapy for recurrent CDI17. One potential concern about the use of rifaximin is the rapid development of resistance during treatment27. Resistance is likely conferred by RpoB amino acid substitutions that appear to be independently derived rather than disseminated from specific resistant clones27. Thus, prolonged use of rifamycins is not recommended.
Fidaxomicin
Fidaxomicin is a macrocyclic narrow spectrum bactericidal agent that targets bacterial RNA polymerase28. It is the first drug to be licensed and recommended for CDI treatment for adults in over 25 years28 and it is currently the recommended treatment for an initial episode of CDI in the USA along with vancomycin17. Fidaxomicin offers lower recurrence rates in CDI patients compared to vancomycin and metronidazole29, and reduced susceptibility to fidaxomicin is very rare17. It has minimal impact on the native gut microbiota, sparing the Bacteroides group, and achieves high concentrations in the gut and faeces. The use of fidaxomicin has been limited due to its high cost.
AMR in C. difficile and One Health
C. difficile colonises the gastrointestinal tracts of all animals during the neonatal period, multiplies and is excreted, but cannot/does not compete well when other bacterial species start to colonise. CDI should always have been considered a zoonosis, either direct or indirect. The One Health concept is a worldwide strategy for interdisciplinary collaboration and communication in all aspects of healthcare for humans, animals and the environment30. In recent years, 70% of emerging or re-emerging infections have been vector-borne or zoonoses – animal diseases transmissible to humans31. Adult humans treated with antimicrobials fool C. difficile into thinking it is colonising a neonatal gut. In the 1980s and 90s, there was an expansion of CDI in hospitals driven by cephalosporins to which C. difficile is intrinsically resistant (see section on β-lactams). Since 1990 in North America, cephalosporins have been licensed for use in food animals. There has been amplification of C. difficile in food animals, with subsequent contamination of meat, and vegetables grown in soil containing animal faeces32. Animal strains of C. difficile are now infecting humans causing a rise in the incidence of community-acquired CDI33. In Australia, tetracyclines are used commonly in food animals34 and C. difficile RT014 human and porcine strains isolated in Australia demonstrated tetracycline resistance8.
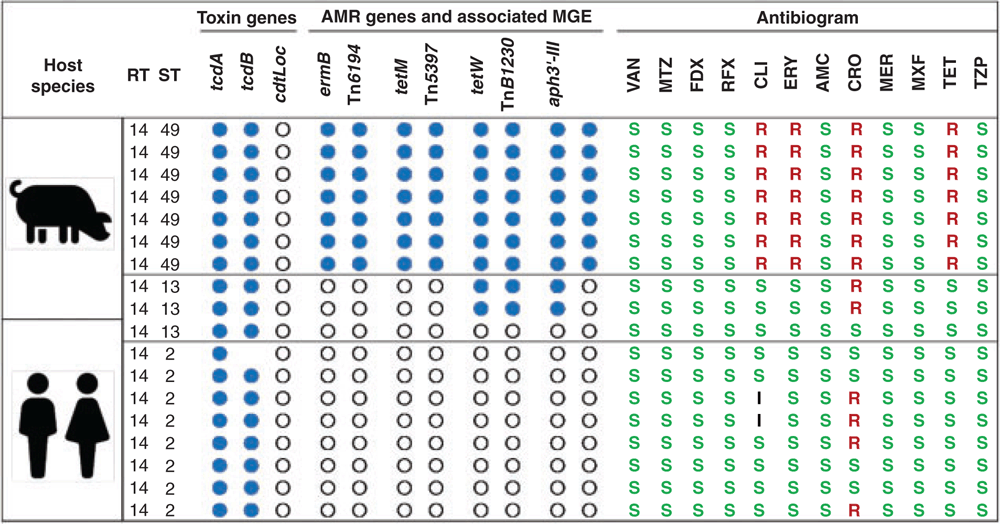
The tetracycline family of antimicrobials has broad activity against Gram-positive and Gram-negative bacteria, including anaerobes, and tetracycline resistance occurs frequently in many organisms35. Whilst tetracycline is not usually associated with the induction of CDI, nor used for treatment, tetracycline resistance has been reported in C. difficile and is transferable between strains36. Resistance in C. difficile is mostly mediated by tet genes carried on transposable elements related to Tn916 that encodes the TetM cytoplasmic protein that protect ribosomes from tetracycline binding35. Figure 3 is a comparative analysis of AMR genotype and phenotype in C. difficile RT014 strains isolated from pigs and humans in Australia in 2013 (Knight et al. unpublished), and clearly shows macrolide and tetracycline resistance genes concentrated in porcine isolates.
Conclusion
Antimicrobial agents are an intrinsic part of CDI; in its induction, transmission and treatment. The widespread emergence of hyper-virulent strains around the world has highlighted the importance of AMR in facilitating the spread of epidemic C. difficile clones. There are worrying trends of increasing resistance to the current therapies used to treat this infection. The contribution of antimicrobial use in production animals and how this contributes to the emergence and spread of new strains of C. difficile is underappreciated. A One Health approach could alleviate pressures towards the development of further AMR in this significant healthcare pathogen.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This research did not receive any specific funding.
References
[1] Miller, B.A. et al. (2011) Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect. Control Hosp. Epidemiol. 32, 387–390.| Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals.Crossref | GoogleScholarGoogle Scholar | 21460491PubMed |
[2] Freeman, J. et al. (2010) The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23, 529–549.
| The changing epidemiology of Clostridium difficile infections.Crossref | GoogleScholarGoogle Scholar | 20610822PubMed |
[3] Slimings, C. and Riley, T.V. (2014) Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J. Antimicrob. Chemother. 69, 881–891.
| Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis.Crossref | GoogleScholarGoogle Scholar | 24324224PubMed |
[4] Debast, S.B. et al. (2014) European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 20, 1–26.
| European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection.Crossref | GoogleScholarGoogle Scholar | 24118601PubMed |
[5] Lessa, F.C. et al. (2015) Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 825–834.
| Burden of Clostridium difficile infection in the United States.Crossref | GoogleScholarGoogle Scholar | 25714160PubMed |
[6] Spigaglia, P. et al. (2018) Antibiotic resistances of Clostridium difficile. Adv. Exp. Med. Biol. 1050, 137–159.
| Antibiotic resistances of Clostridium difficile.Crossref | GoogleScholarGoogle Scholar | 29383668PubMed |
[7] Toth, M. et al. (2018) Intrinsic class D β-lactamases of Clostridium difficile. MBio 9, e01803-18.
| 30563905PubMed |
[8] Knight, D.R. et al. (2017) Genome analysis of Clostridium difficile PCR ribotype 014 lineage in Australian pigs and humans reveals a diverse genetic repertoire and signatures of long-range interspecies transmission. Front. Microbiol. 7, 2138.
| 28123380PubMed |
[9] Tenson, T. et al. (2003) The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330, 1005–1014.
| The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome.Crossref | GoogleScholarGoogle Scholar | 12860123PubMed |
[10] Freeman, J. et al. (2018) The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011-2014. Clin. Microbiol. Infect. 24, 724–731.
| The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011-2014.Crossref | GoogleScholarGoogle Scholar | 29066403PubMed |
[11] Spigaglia, P. et al. (2011) Multidrug resistance in European Clostridium difficile clinical isolates. J. Antimicrob. Chemother. 66, 2227–2234.
| Multidrug resistance in European Clostridium difficile clinical isolates.Crossref | GoogleScholarGoogle Scholar | 21771851PubMed |
[12] Tang-Feldman, Y.J. et al. (2005) Prevalence of the ermB gene in Clostridium difficile strains isolated at a university teaching hospital from 1987 through 1998. Clin. Infect. Dis. 40, 1537–1540.
| Prevalence of the ermB gene in Clostridium difficile strains isolated at a university teaching hospital from 1987 through 1998.Crossref | GoogleScholarGoogle Scholar | 15844079PubMed |
[13] Baines, S.D. and Wilcox, M.H. (2015) Antimicrobial resistance and reduced susceptibility in Clostridium difficile: potential consequences for induction, treatment, and recurrence of C. difficile Infection. Antibiotics (Basel) 4, 267–298.
| Antimicrobial resistance and reduced susceptibility in Clostridium difficile: potential consequences for induction, treatment, and recurrence of C. difficile Infection.Crossref | GoogleScholarGoogle Scholar | 27025625PubMed |
[14] Schmidt, C. et al. (2007) Antimicrobial phenotypes and molecular basis in clinical strains of Clostridium difficile. Diagn. Microbiol. Infect. Dis. 59, 1–5.
| Antimicrobial phenotypes and molecular basis in clinical strains of Clostridium difficile.Crossref | GoogleScholarGoogle Scholar | 17509804PubMed |
[15] Lebel, S. et al. (2004) The cme gene of Clostridium difficile confers multidrug resistance in Enterococcus faecalis. FEMS Microbiol. Lett. 238, 93–100.
| 15336408PubMed |
[16] Linder, J.A. et al. (2005) Fluoroquinolone prescribing in the United States: 1995 to 2002. Am. J. Med. 118, 259–268.
| Fluoroquinolone prescribing in the United States: 1995 to 2002.Crossref | GoogleScholarGoogle Scholar | 15745724PubMed |
[17] McDonald, L.C. et al. (2018) Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 66, e1–e48.
| Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA).Crossref | GoogleScholarGoogle Scholar | 29462280PubMed |
[18] Kaihovaara, P. et al. (1998) Flavodoxin-dependent pyruvate oxidation, acetate production and metronidazole reduction by Helicobacter pylori. J. Antimicrob. Chemother. 41, 171–177.
| Flavodoxin-dependent pyruvate oxidation, acetate production and metronidazole reduction by Helicobacter pylori.Crossref | GoogleScholarGoogle Scholar | 9533458PubMed |
[19] Musher, D.M. et al. (2005) Relatively poor outcome after treatment of Clostridium difficile colitis with metronidazole. Clin. Infect. Dis. 40, 1586–1590.
| Relatively poor outcome after treatment of Clostridium difficile colitis with metronidazole.Crossref | GoogleScholarGoogle Scholar | 15889354PubMed |
[20] Barbut, F. et al. (1999) Antimicrobial susceptibilities and serogroups of clinical strains of Clostridium difficile isolated in France in 1991 and 1997. Antimicrob. Agents Chemother. 43, 2607–2611.
| Antimicrobial susceptibilities and serogroups of clinical strains of Clostridium difficile isolated in France in 1991 and 1997.Crossref | GoogleScholarGoogle Scholar | 10543736PubMed |
[21] Peláez, T. et al. (2008) Metronidazole resistance in Clostridium difficile is heterogeneous. J. Clin. Microbiol. 46, 3028–3032.
| Metronidazole resistance in Clostridium difficile is heterogeneous.Crossref | GoogleScholarGoogle Scholar | 18650353PubMed |
[22] Johnson, S. et al. (2000) Metronidazole resistance in Clostridium difficile. Clin. Infect. Dis. 31, 625–626.
| Metronidazole resistance in Clostridium difficile.Crossref | GoogleScholarGoogle Scholar | 10987742PubMed |
[23] Al-Nassir, W.N. et al. (2008) Comparison of clinical and microbiological response to treatment of Clostridium difficile-associated disease with metronidazole and vancomycin. Clin. Infect. Dis. 47, 56–62.
| Comparison of clinical and microbiological response to treatment of Clostridium difficile-associated disease with metronidazole and vancomycin.Crossref | GoogleScholarGoogle Scholar | 18491964PubMed |
[24] Ammam, F. et al. (2013) The functional vanGCd cluster of Clostridium difficile does not confer vancomycin resistance. Mol. Microbiol. 89, 612–625.
| The functional vanGCd cluster of Clostridium difficile does not confer vancomycin resistance.Crossref | GoogleScholarGoogle Scholar | 23782343PubMed |
[25] Freeman, J. et al.. (2015) Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin. Microbiol. Infect. 21, 248.e9–248.e16.
[26] Knight, D.R. et al. (2016) A phenotypically silent vanB2 operon carried on a Tn1549-like element in Clostridium difficile. mSphere 1, e00177-16.
| 27536735PubMed |
[27] O’Connor, J.R. et al. (2008) Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob. Agents Chemother. 52, 2813–2817.
| Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile.Crossref | GoogleScholarGoogle Scholar | 18559647PubMed |
[28] Venugopal, A.A. and Johnson, S. (2012) Fidaxomicin: a novel macrocyclic antibiotic approved for treatment of Clostridium difficile infection. Clin. Infect. Dis. 54, 568–574.
| Fidaxomicin: a novel macrocyclic antibiotic approved for treatment of Clostridium difficile infection.Crossref | GoogleScholarGoogle Scholar | 22156854PubMed |
[29] Cornely, O.A. et al. (2014) Clinical efficacy of fidaxomicin compared with vancomycin and metronidazole in Clostridium difficile infections: a meta-analysis and indirect treatment comparison. J. Antimicrob. Chemother. 69, 2892–2900.
| Clinical efficacy of fidaxomicin compared with vancomycin and metronidazole in Clostridium difficile infections: a meta-analysis and indirect treatment comparison.Crossref | GoogleScholarGoogle Scholar | 25074856PubMed |
[30] Järhult, J.D. (2015) One Health: a doctor’s perspective. Vet. Rec. 176, 351–353.
| One Health: a doctor’s perspective.Crossref | GoogleScholarGoogle Scholar | 25837947PubMed |
[31] Blancou, J. et al. (2005) Emerging or re-emerging bacterial zoonoses: factors of emergence, surveillance and control. Vet. Res. 36, 507–522.
| Emerging or re-emerging bacterial zoonoses: factors of emergence, surveillance and control.Crossref | GoogleScholarGoogle Scholar | 15845237PubMed |
[32] Lim, S.C. et al. (2018) Antimicrobial susceptibility of Clostridium difficile isolated from food and environmental sources in Western Australia. Int. J. Antimicrob. Agents 52, 411–415.
| Antimicrobial susceptibility of Clostridium difficile isolated from food and environmental sources in Western Australia.Crossref | GoogleScholarGoogle Scholar | 29802886PubMed |
[33] Slimings, C. et al. (2014) Increasing incidence of Clostridium difficile infection, Australia, 2011-2012. Med. J. Aust. 200, 272–276.
| Increasing incidence of Clostridium difficile infection, Australia, 2011-2012.Crossref | GoogleScholarGoogle Scholar | 24641152PubMed |
[34] Jordan, D. et al. (2009) Antimicrobial use in the Australian pig industry: results of a national survey. Aust. Vet. J. 87, 222–229.
| Antimicrobial use in the Australian pig industry: results of a national survey.Crossref | GoogleScholarGoogle Scholar | 19489779PubMed |
[35] Chopra, I. and Roberts, M. (2001) Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260.
| Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance.Crossref | GoogleScholarGoogle Scholar | 11381101PubMed |
[36] Jasni, A.S. et al. (2010) Demonstration of conjugative transposon (Tn5397)-mediated horizontal gene transfer between Clostridium difficile and Enterococcus faecalis. Antimicrob. Agents Chemother. 54, 4924–4926.
| Demonstration of conjugative transposon (Tn5397)-mediated horizontal gene transfer between Clostridium difficile and Enterococcus faecalis.Crossref | GoogleScholarGoogle Scholar | 20713671PubMed |
Biographies
Stacey Hong is a final year PhD candidate at The University of Western Australia. Her research focuses on the descriptive epidemiology, microbiology and comparative genomics of an emerging strain of Clostridium difficile in Australia.
Daniel Knight is an NHMRC Early Career Fellow at Murdoch University, Western Australia. His research focus is evolutionary and One Health aspects of Clostridium difficile infection.
Tom Riley holds positions in various universities in Western Australia, as well as in Pathwest Laboratory Medicine. He has had a long-standing interest in healthcare-related infections, particularly the diagnosis, pathogenesis and epidemiology of Clostridium difficile infection, in both humans and animals.