The movement of humans and the spread of Salmonella into existing and pristine ecosystems
John B Iveson A , S Donald Bradshaw B and David W Smith A C DA Department of Microbiology, PathWest Laboratory Medicine WA, Hospital Avenue, Nedlands, WA 6009, Australia
B Faculty of Science, University of Western Australia, Nedlands, WA 6009, Australia
C Faculty of Medicine and Health Sciences, University of Western Australia, Nedlands, WA 6009, Australia
D Email: david.smith@health.wa.gov.au
Microbiology Australia 38(4) 201-203 https://doi.org/10.1071/MA17070
Published: 2 November 2017
The spread of infectious diseases by the international and national movement of people, animals, insects and products has a documented history dating back several centuries1. The role of human movements has been fundamental to this, and has increased as global travel has risen in amount and speed. This has been exemplified by international epidemics of influenza, antimicrobial resistant bacteria, SARS coronavirus, dengue, chikungunya virus, Zika viruses and many others. Foodborne pathogens have also regularly come to our attention for their ability to move internationally, and outbreaks of salmonellosis due to importation of contaminated foods are well described2,3. An extensive collection of non-typhoidal Salmonella and related species isolated from human, food, animal and environmental sources has been accumulated within Western Australia (WA) since the mid-20th century, and has proven an important historical source of information about the role of humans in the dissemination of microorganisms across and within diverse ecosystems4–6. It is clear that the movement of microorganisms into and out of Australia is by no means a new phenomenon, and that humans have been important contributors to that spread. These are important markers of our impact on established and pristine ecosystems.
It had been postulated that increases in human visitors to the Antarctic might be responsible for the introduction of new Salmonella species into the sub-Antarctic wildlife7. The WA collection provided an opportunity to look in more detail at the impact of humans and their accompanying animals arising from the establishment of the various Antarctic research bases and the increasing tourist traffic5. Estimates from the International Association of Antarctica Tour Operators showed a rise in tourist numbers into the Antarctic waters from 13 475 ship passengers in 2000–01 to 44 402 in 2016–178. Most tourist activities currently take place off-shore, i.e. there is limited time on the land. On-land activities are nearly all carried out by the smaller number of staff in the research stations, who have introduced dogs and other domesticated animals5,9.
Iveson et al.5 surveyed penguins and seals to determine whether Salmonella enterica infections in Antarctic and sub-Antarctic wildlife were likely native serovars (i.e. antigenically unique and therefore likely to be part of the pre-existing local enzootic Salmonella population) or exotic (i.e. antigenically similar to serovars already know to be present elsewhere). They found that 10/12 (83%) of the S. enterica serovars isolated from the penguins and seals matched those found commonly within Australia, including in humans and domesticated animals. This dominance of exotic serovars, and the low frequency of any serovars likely to have been native to the Antarctic ecosystem, indicated extensive contamination of the Antarctic and sub-Antarctic environments by introduced Salmonella serovars. Some of these exotic serovars were found in Adélie penguins in Commonwealth Bay about 250 km from any recent human presence, suggesting that there had been contamination of the Antarctic environment from human sewage. It was postulated that spread to Commonwealth Bay may have occurred via contamination of krill swarms that the penguins feed on, or spread via carrier Antarctic birds5,10.
A further analysis of this data set was undertaken to look for evidence of introduction of exotic Salmonella serovars into the Western Australian ecosystem from overseas, as had been described for the islands of Java, Bali, and the Wallacea region following European colonisation in the 17th century6. The risk to the WA ecosystem dates back to European settlement of the southern temperate regions of WA, which began in 1827, and the tropical Kimberley in 1890. In more recent times, there has also been the potential for introduction by the increasing numbers of travellers returning from areas with relatively high rates of food- and waterborne salmonellosis, such as Bali.
A comprehensive analysis of isolates collected over a 50-year period from 1950 to 2000 supported the potential for introduction of exotic bacterial organisms, with over 90 different Salmonella serovars being detected in humans arriving from Asian, African and European countries. That included more than 600 cases reported in travellers returning from Bali or other areas of southern Indonesia.
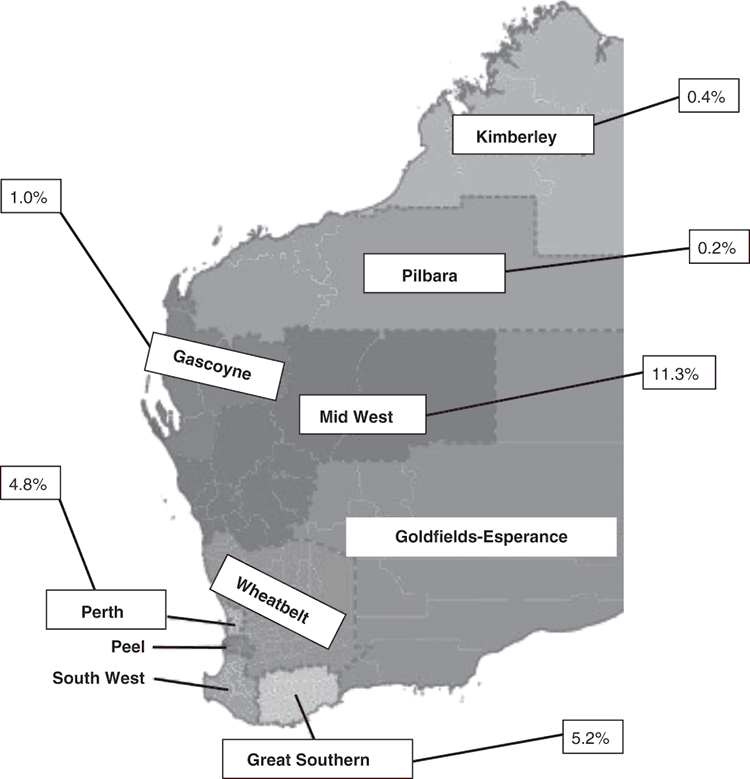
The data from this collection also provided evidence that, once introduced into Australia, these exotic Salmonella serovars then spread into other pristine environments within Australia. Exotic serovars were found in wildlife populations even in remote unpopulated areas of northern WA, such as the Mitchell Plateau in the Kimberley, though in much smaller numbers than found in wildlife from areas with larger human populations in the south-west of the state (Figure 1). Furthermore, the Salmonella serovars from the areas with human contact were dominated by exotic serovars, while those from remote sites were predominantly native serovars. This suggested a path of transmission from human populations in more heavily populated areas, to local wildlife in those areas, then further transmission in animals such as migratory birds and marsupials, in humans, or in both.
|
One particular strain, S. enterica serotype Enteriditis phage type 4, had previously been found in increasing numbers in sub-Antarctic fur seals7. In WA, it was found in travellers returning to WA from Indonesia, and was also found in the samples from the Adélie penguins in the Australian Antarctic territory5,6. This phage type was known to have emerged and spread from Europe in the 1980s, and importation in returned travellers has been well documented11. The WA data suggests a potential role for the Western Australian ecosystem as a ‘staging post’ for the onward spread of organisms by human movement into Antarctica.
The data obtained from this collection has shown the historical and ongoing impact of human colonisation and other travel on the spread of important microorganisms into Australia, within Australia and, probably, onwards from there. That has had a significant impact on human, animal and environmental ecosystems in settled and pristine locations.
These observations clearly demonstrate the value of collections of organisms for better understanding disease ecology and for ongoing surveillance, even when restricted to traditional typing methods like phage-typing and serotyping. It is unfortunate that this historical collection is no longer available for further analysis using contemporary techniques such as whole genome sequencing, which has shown its additional value in identifying sources and mapping the transmission pathways of different Salmonella strains12,13.
Acknowledgements
The authors acknowledge the staff of the Enteric Laboratory of the Western Australia State Health Laboratory Service, which was subsequently incorporated into PathCentre and then into PathWest.
References
[1] Institute of Medicine (2006) The impact of globalization on infectious disease emergence and control: exploring the consequences and opportunities: workshop summary. Washington, DC: The National Academic Press.[2] OzFoodNet Working Group (2005) Reported foodborne illness and gastroenteritis in Australia: annual report of the OzFoodNet network, 2004. Commun. Dis. Intell. Q. Rep. 29, 165–192.
[3] Unicomb, L.E. et al. (2005) Sesame seed products contaminated with Salmonella: three outbreaks associated with tahini. Epidemiol. Infect. 133, 1065–1072.
| Sesame seed products contaminated with Salmonella: three outbreaks associated with tahini.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD2MrpsF2qtQ%3D%3D&md5=7b4e2d39aee8cd933bc143457b8bbfb4CAS |
[4] Taux, R.V. (1999) Salmonella Enteritidis: the continuing public health challenge XI–XIII. In Salmonella enterica serovar Enteritidis in humans and animals: epidemiology, pathogenesis and control (Saeed, A.M., ed.) pp. 3–18, Iowa State University Press.
[5] Iveson, J.B. et al. (2009) Salmonella infections in Antarctic fauna and island populations of wildlife exposed to human activities in coastal areas of Australia. Epidemiol. Infect. 137, 858–870.
| Salmonella infections in Antarctic fauna and island populations of wildlife exposed to human activities in coastal areas of Australia.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1M3ptlegtg%3D%3D&md5=239d7bc144fcda0d8926ec5fcfbc17bbCAS |
[6] Iveson, J.B. et al. (2014) Human migration is important in the international spread of exotic Salmonella serovars in animal and human populations. Epidemiol. Infect. 142, 2281–2296.
| Human migration is important in the international spread of exotic Salmonella serovars in animal and human populations.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC2c3mtVSisg%3D%3D&md5=11ef384dc8630c1ee3324588c161747cCAS |
[7] Palmgren, H. et al. (2000) Salmonella in sub-Antarctica: low heterogeneity in Salmonella serotypes in South Georgian seals and birds. Epidemiol. Infect. 125, 257–262.
| Salmonella in sub-Antarctica: low heterogeneity in Salmonella serotypes in South Georgian seals and birds.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3M%2FmsVOisg%3D%3D&md5=72315ab008720b46de715c4d91ebce0fCAS |
[8] The International Association of Antarctica Tour Operators (2017) Tourism statistics. https://iaato.org/tourism-statistics (accessed 24 September 2017).
[9] Australian Antarctic Division (2017) Human impacts in Antarctica. http://www.antarctica.gov.au/environment/human-impacts-in-antarctica (accessed 24 September 2017).
[10] Nicol, S. et al. (2000) Products derived from krill. In Krill Biology, Ecology and Fisheries (Everson, I., ed.) pp. 262–283, Blackwell Science.
[11] Nygård, K. et al. (2004) Emergence of new Salmonella Enteriditis phage types in Europe? Surveillance of infections in returning travellers. BMC Med. 2, 32.
| Emergence of new Salmonella Enteriditis phage types in Europe? Surveillance of infections in returning travellers.Crossref | GoogleScholarGoogle Scholar |
[12] Yachison, C.A. et al. (2017) The validation and implications of using whole genome sequencing as a replacement for traditional serotyping for a national Salmonella reference laboratory. Front. Microbiol. 8, 1044.
| The validation and implications of using whole genome sequencing as a replacement for traditional serotyping for a national Salmonella reference laboratory.Crossref | GoogleScholarGoogle Scholar |
[13] Phillips, A. et al. (2016) Whole genome sequencing of Salmonella Typhimurium illuminates distinct outbreaks caused by an endemic multi-locus variable number tandem repeat analysis type in Australia. BMC Microbiol. 16, 211.
| Whole genome sequencing of Salmonella Typhimurium illuminates distinct outbreaks caused by an endemic multi-locus variable number tandem repeat analysis type in Australia.Crossref | GoogleScholarGoogle Scholar |
Biographies
John Iveson was a senior scientist in the enteric section of the State Health Laboratory Service in Western Australia for over 50 years. The collection of organisms in the Salmonella typing laboratory was a lifetime endeavour and achievement for him, and his work with it continued following his retirement, up until a few days before he passed away in August, 2016. He remained a keen competitive cyclist throughout his life.
Don Bradshaw is an Emeritus Professor and Senior Honorary Research Fellow in the School of Biological Sciences at the University of Western Australia, where he was Chair of Zoology from 1969 to 2005. He has had a wide interest in research related to fish, frogs, reptiles and marsupials, producing three books and over 250 scientific papers. His current activities are directed to trying to conserve the amazing biodiversity in the southwest of WA.
David Smith is a Clinical Microbiologist at PathWest Laboratory Medicine WA, and a Clinical Professor in the Faculty of Medicine and Heath Sciences at the University of Western Australia. His research interests include arboviruses, respiratory viruses, emerging infections and laboratory-based epidemiology.


