Whole genome sequencing as a novel approach for characterising Neisseria meningitidis in Australia
Shakeel MowlaboccusSchool of Biomedical Sciences
The University of Western Australia
Perth, WA, Australia
Email: shakeel.mowlaboccus@uwa.edu.au
Microbiology Australia 38(3) 142-145 https://doi.org/10.1071/MA17052
Published: 22 August 2017
Neisseria meningitidis (meningococcus) is the causative agent of invasive meningococcal disease that manifests as life-threatening septicaemia and/or meningitis. This review provides a brief overview of the prevention of the disease and also highlights the importance of whole genome sequencing (WGS) in detecting outbreaks of meningococci in Australia. The use of WGS in identifying the emergence of a penicillin-resistant cluster of meningococci is Western Australia is used as an example for advocating the implementation of WGS on the routine surveillance in Australia.
Invasive meningococcal disease
Neisseria meningitidis, also known as the meningococcus (plural: meningococci), is a Gram-negative diplococcus that asymptomatically colonises the nasopharynx of approximately 10–30% of the adult population1. Occasionally, the bacterium crosses the epithelial layer and invades the host, causing invasive meningococcal disease (IMD) if the correct immune response is not elicited. IMD generally manifests as septicaemia and/or meningitis. Incidence of IMD follows a bimodal distribution with the first peak occurring in infants, due to lack of a mature immune system, and the second peak occurring in teenage years and early adulthood when the rate of transmission is highest due to lifestyle factors2. The onset of the disease is rapid and death may occur within 24 hours if the recommended antibiotics are not administered. Although IMD is rare, the mortality rate is approximately 10% and approximately 15% of survivors suffer from permanent sequelae such as limb loss and neurological disability3,4. Invasive strains of N. meningitidis express a polysaccharide capsule, which is used to classify the bacteria phenotypically into one of 12 serogroups – A, B, C, E, G, I, K, L, W, X, Y and Z5. IMD is predominantly caused by six serogroups (A, B, C, W, X and Y) and the distribution of these serogroups varies geographically and temporally6,7.
Prevention of IMD
IMD can be prevented through vaccination. Although there is no vaccine available against the most recent disease-causing serogroup X, conjugate polysaccharide vaccines targeting the capsule have been used to control endemic IMD caused by serogroups A, C, W and Y for the past decades8. However, the majority of IMD cases in developed countries, including Australia, has been caused by meningococcal serogroup B (MenB)9. Vaccine development against MenB has been hampered as the capsule polysaccharide elicits autoantibodies in humans. As of now, two multi-component vaccines against serogroup B disease have been developed, which target specific sub-capsular surface antigens expressed by N. meningitidis. These vaccines are Bexsero® (also known as 4CMenB)10 and Trumenba® (also known as rLP2086)11. Although both MenB vaccines have been licenced for use in the United States and the United Kingdom, Bexsero® is the only MenB vaccine available in Australia. Since the majority of cases (>65%) in Australia were caused by MenB post the implementation of the meningococcal C conjugate vaccine on the national immunisation programme (NIP) in 2003, inclusion of the Bexsero® vaccine on the NIP has been proposed to protect the Australian population from MenB infections. However, this focus has now shifted as there has been a switch in the predominant meningococcal serogroup in Australia where the majority of cases are currently caused by meningococcal serogroup W (MenW)9.
The MLST scheme
In addition to serogroup classification, meningococci can be classified genotypically into sequence types (STs) using multilocus sequence typing (MLST) of seven housekeeping genes12. The internal fragments of the seven genes (abcZ, adk, aroE, fumC, gdh, pdhC and pgm) are sequenced and an allele number is arbitrarily assigned to each different sequence for each locus. Each ST thus corresponds to a unique combination of seven integers. STs that are identical at four or more loci are grouped into the same clonal complex (cc), which is a direct measure of genetic lineage. Some clonal complexes are more commonly associated with disease than with carriage and are thus termed hypervirulent lineages. Examples of such lineages are cc1, cc5, cc11, cc32 and cc41/4413. In contrast to the MLST scheme, serogroup classification is not lineage specific and meningococci expressing different serogroup capsules may belong to the same cc. For instance, meningococci belonging to cc11 have been isolated as expressing either a serogroup B, serogroup C or serogroup W capsule14.
Characterising Neisseria meningitidis using whole genome sequencing
In Australia, diagnostic laboratories report the serogroup of all culturable meningococci from IMD cases to the National Neisseria Network. However, MLST profiles of meningococci are not reported as this action would require the amplification of the seven housekeeping loci by polymerase chain reaction (PCR) followed by sequencing of the fragments, which is not cost-effective. Although serogroup distribution is helpful for disease surveillance, this information is inadequate in regards to monitoring outbreaks and expansion of genetically related meningococci around the country. With the advent of next generation sequencing technologies, whole genome sequencing (WGS) has become relatively inexpensive and less time-consuming compared to conventional sequencing methods. In contrast to MLST profiling, which allows for lineage identification, WGS provides a better resolution and allows strain-to-strain differentiation. Furthermore, MLST profiles can be obtained from WGS data without the need for individual PCR on the MLST loci.
Genomes of N. meningitidis can be analysed on the freely accessible PubMLST website (https://pubmlst.org/neisseria) using the Bacterial Isolate Genome Sequence Database (BIGSdb) platform15. Once the WGS data are obtained from the sequencing platform (e.g. Illumina Miseq), the nucleotide sequences need to be assembled into contigs using an assembly software (e.g. SPAdes or VELVET) before using the BIGSdb Genome Comparator tool on the website. The core genome of the meningococcus has been characterised and is defined by 1605 core loci16. Comparison of meningococcal core genomes has previously been used to characterise N. meningitidis isolates causing outbreaks and epidemics14,17. With the collaboration of PathWest and under the supervision of A/Prof Charlene Kahler (UWA), I was able to implement one such analysis in Australia, as described below.
WGS reveals emerging cluster of penicillin-resistant N. meningitidis in Australia
In our recent study18, we used WGS as a tool to characterise the increasing number of invasive MenW isolates in Western Australia (WA). The first MenW case was recorded in 2013, which was followed by two cases in 2014 and three cases in 2015. In 2016, a significant increase in MenW incidence was detected and 13 such cases were reported then. We cultured the meningococci from all 19 cases, extracted and sequenced the genomic DNA. Analysis of the MLST loci revealed that all 19 MenW isolates belonged to the cc11 lineage although four different STs were identified. By aligning the nucleotide sequences of the core genome of each isolate, a phylogenetic tree was constructed to investigate similarities among the isolates (Figure 1). From the branching, we observed that the isolates fell into two main clusters, which we labelled as A and B. Cluster A comprised 8 isolates and Cluster B contained 10 isolates. One meningococcal strain (ExNm672), which was isolated from a traveller from Asia who had just arrived in WA, failed to cluster as its genome was significantly different from the WA isolates. Cluster A contained only ST-11 isolates whereas Cluster B contained all four STs identified in the collection. All meningococci in Cluster B were isolated in 2016. The spread of a single clone from Cluster B (ST-12351) in Kalgoorlie WA led to the one-off MenW vaccination program during Dec 2016–Apr 2017.
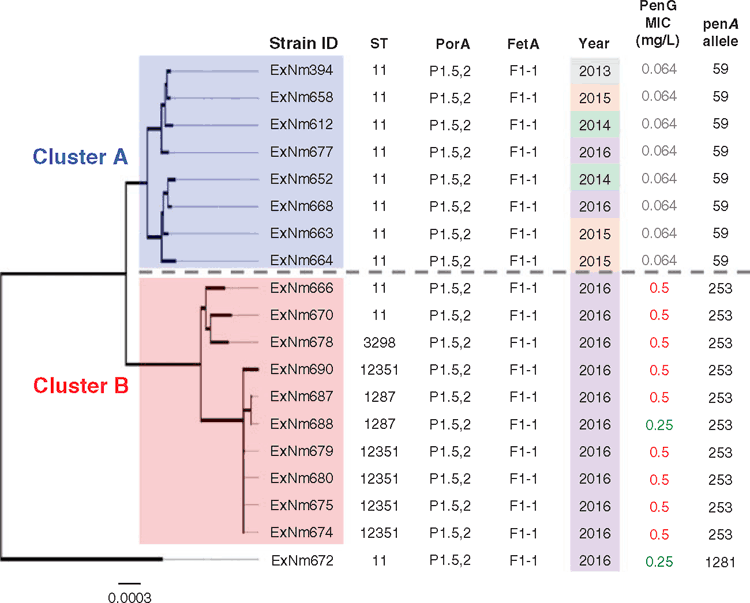
|
Interestingly, all isolates in Cluster A were sensitive to penicillin (MIC: ≤0.06 mg/L) whilst isolates in Cluster B showed reduced susceptibility to penicillin. The majority of isolates in Cluster B (9 out of 10) were resistant to penicillin (MIC: ≥0.5 mg/L). By comparing core genomes of the Cluster A isolates to the Cluster B isolates, we identified the penA locus that encodes a protein involved in peptidoglycan biosynthesis as the major contributor to this difference in penicillin susceptibility. Cluster A isolates harboured the penA_59 allele whereas Cluster B isolates harboured the penA_253 allele. These alleles differ at 101 nucleotides and the encoded peptides differ at 25 amino acid positions. Exchange of penA_59 to penA_253 in all Cluster A isolates resulted in reduced susceptibility to penicillin that confirmed the role played by this allele in conferring resistance to penicillin. This finding is of global concern because the penA_253 allele has been detected in at least five meningococcal isolates in Europe and although treatment with penicillin is still effective against penicillin-intermediate strains, low-dose treatment regimens may fail for cases involving meningococci with a penicillin MIC ≥ 0.5 mg/L19.
Conclusion and future direction
IMD is a debilitating disease with significant morbidity and mortality that can be controlled through vaccination. Although the meningococcal vaccine on the NIP in Australia protects only against meningococcal serogroup C, MenACWY and MenB vaccines are available on the private market. With the remarkable progress of next generation sequencing techniques, WGS has become the most cost-effective method for typing invasive meningococci for surveillance. By analysing WGS data of meningococci circulating in Western Australia, we detected the expansion of a MenW:cc11 clone in Kalgoorlie, which led to immediate vaccination of the community. Furthermore, we exploited WGS to identify the emergence of a penicillin-resistant clade of meningococci in WA that has prompted the screening of the penA_253 allele around the world as the establishment of this allele in the meningococcal population may have an impact on treatment regimens around the world. Implementing WGS analysis as a routine surveillance for monitoring meningococci in Australia will undoubtedly allow us to detect local outbreaks instantly, which will allow urgent actions such as emergency vaccination and will also help improve intervention strategies on a global level in the event where a cluster of antimicrobial resistant meningococci is detected.
Acknowledgements
The author acknowledges the support of the International Postgraduate Research Scholarship from The University of Western Australia. The author also thanks his supervisor Associate Professor Charlene Kahler for her excellent mentorship.
References
[1] Cartwright, K.A. et al. (1987) The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiol. Infect. 99, 591–601.| The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL1c7htF2kuw%3D%3D&md5=215387e8b8acd5d8eff76fc1b14d1ee6CAS |
[2] Gabutti, G. et al. (2015) Epidemiology of Neisseria meningitidis infections: case distribution by age and relevance of carriage. J. Prev. Med. Hyg. 56, E116–E120.
| 1:STN:280:DC%2BC28nisVGrtA%3D%3D&md5=4ad0cd7e86e3f04165cae654dbcab128CAS |
[3] Stephens, D.S. et al. (2007) Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369, 2196–2210.
| Epidemic meningitis, meningococcaemia, and Neisseria meningitidis.Crossref | GoogleScholarGoogle Scholar |
[4] Sadarangani, M. et al. (2015) Outcomes of invasive meningococcal disease in adults and children in Canada between 2002 and 2011: a prospective cohort study. Clin. Infect. Dis. 60, e27–e35.
| Outcomes of invasive meningococcal disease in adults and children in Canada between 2002 and 2011: a prospective cohort study.Crossref | GoogleScholarGoogle Scholar |
[5] Harrison, O.B. et al. (2013) Description and nomenclature of Neisseria meningitidis capsule locus. Emerg. Infect. Dis. 19, 566–573.
| Description and nomenclature of Neisseria meningitidis capsule locus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhslantLjK&md5=a0c000366fd40791826afc6a6b8adb34CAS |
[6] Jafri, R.Z. et al. (2013) Global epidemiology of invasive meningococcal disease. Popul. Health Metr. 11, 17.
| Global epidemiology of invasive meningococcal disease.Crossref | GoogleScholarGoogle Scholar |
[7] Mowlaboccus, S. et al. (2016) Temporal changes in BEXSERO(R) antigen sequence type associated with genetic lineages of Neisseria meningitidis over a 15-year period in Western Australia. PLoS One 11, e0158315.
| Temporal changes in BEXSERO(R) antigen sequence type associated with genetic lineages of Neisseria meningitidis over a 15-year period in Western Australia.Crossref | GoogleScholarGoogle Scholar |
[8] Ali, A. et al. (2014) Global practices of meningococcal vaccine use and impact on invasive disease. Pathog. Glob. Health 108, 11–20.
| Global practices of meningococcal vaccine use and impact on invasive disease.Crossref | GoogleScholarGoogle Scholar |
[9] Martin, N.V. et al. (2016) Rise in invasive serogroup W meningococcal disease in Australia 2013-2015. Commun. Dis. Intell. Q. Rep. 40, E454–E459.
[10] Serruto, D. et al. (2012) The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine 30, B87–B97.
| The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhs1OnurnF&md5=7e6b2fd808e19d8b2fb8fba47e9601caCAS |
[11] Gandhi, A. et al. (2016) Characteristics of a new meningococcal serogroup B vaccine, bivalent rLP2086 (MenB-FHbp; Trumenba(R)). Postgrad. Med. 128, 548–556.
| Characteristics of a new meningococcal serogroup B vaccine, bivalent rLP2086 (MenB-FHbp; Trumenba(R)).Crossref | GoogleScholarGoogle Scholar |
[12] Maiden, M.C. et al. (1998) Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95, 3140–3145.
| Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXitV2itLg%3D&md5=eaec59c7758a1a9cd7ce82dd6eeb4a28CAS |
[13] Caugant, D.A. and Maiden, M.C. (2009) Meningococcal carriage and disease--population biology and evolution. Vaccine 27, B64–B70.
| Meningococcal carriage and disease--population biology and evolution.Crossref | GoogleScholarGoogle Scholar |
[14] Lucidarme, J. et al. (2015) Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J. Infect. 71, 544–552.
| Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage.Crossref | GoogleScholarGoogle Scholar |
[15] Jolley, K.A. and Maiden, M.C. (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595.
| BIGSdb: scalable analysis of bacterial genome variation at the population level.Crossref | GoogleScholarGoogle Scholar |
[16] Bratcher, H.B. et al. (2014) A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics 15, 1138.
| A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes.Crossref | GoogleScholarGoogle Scholar |
[17] Kretz, C.B. et al. (2016) Whole-genome characterization of epidemic Neisseria meningitidis serogroup C and Resurgence of Serogroup W, Niger, 2015. Emerg. Infect. Dis. 22, 1762–1768.
| Whole-genome characterization of epidemic Neisseria meningitidis serogroup C and Resurgence of Serogroup W, Niger, 2015.Crossref | GoogleScholarGoogle Scholar |
[18] Mowlaboccus, S. et al. (2017) Clonal expansion of new penicillin-resistant clade of Neisseria meningitidis serogroup W clonal complex 11, Australia. Emerg. Infect. Dis. 23, 1364–1367.
| Clonal expansion of new penicillin-resistant clade of Neisseria meningitidis serogroup W clonal complex 11, Australia.Crossref | GoogleScholarGoogle Scholar |
[19] Turner, P.C. et al. (1990) Treatment failure in meningococcal meningitis. Lancet 335, 732–733.
| Treatment failure in meningococcal meningitis.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK3c7ptVWlug%3D%3D&md5=6b0dee1cb7e4ec4524139fa73b5ba58dCAS |
Biography
Shakeel (Shaxx) Mowlaboccus is a doctoral candidate and a sessional lecturer at The University of Western Australia, Perth, Australia. His primary research interests include the evolution and changing epidemiology of N. meningitidis and investigating the mechanism of antimicrobial resistance for this pathogen. Shaxx was awarded the ASM/BD Student Travel Award to present the data illustrated in this article at the Australian Society for Microbiology Annual Scientific Meeting 2017.


