Early warning systems augmented by bacterial genomics
Vitali Sintchenko A B C D E F and Nadine Holmes A D E GA Centre for Infectious Diseases and Microbiology – Public Health
B Institute of Clinical Pathology and Medical Research – Pathology West
C NSW Health Pathology and Westmead Hospital
D Marie Bashir Institute for Infectious Diseases and Biosecurity
E Sydney Medical School – Westmead, The University of Sydney
Tel: +61 2 9845 6255
Fax: + 61 2 9893 8659
F Email: vitali.sintchenko@sydney.edu.au
G Email: nadine.holmes@sydney.edu.au
Microbiology Australia 35(1) 44-48 https://doi.org/10.1071/MA14012
Published: 17 February 2014
The number of microbial threats – in the form of newly identified pathogens, infections crossing the species barrier to people, diseases adapting to new environments, transmissible drug-resistance genes and microbial agents appearing in more virulent forms – has multiplied to an unprecedented degree. The epidemiology of well-known infectious diseases has also been changing due to the globalisation of trade and in response to immunisation campaigns. This evolving epidemiology presents new challenges to countries’ healthcare systems, in terms of both understanding and monitoring of determinants of infections, as well as in terms of service provision and the implementation of appropriate prevention measures. In this article we discuss the concepts of early warning systems and genome sequencing for public health laboratory surveillance and outbreak detection and response. The added value of these new means of surveillance can be seen when clinical and public health laboratory data is harmonised, aggregated and shared.
Why early warning system are needed
Evidence suggests that traditional surveillance systems are vulnerable to the incomplete and delayed reporting of public health threats, highlighted by recent outbreaks. Many outbreaks have been characterised by delayed recognition and/or public health response. Such delays diminish the window of opportunity to mount effective response measures and are likely to be costly to society1. For example, it was estimated that a one-week delay in the implementation of control measures for SARS in Canada resulted in a 2.6-fold increase in the mean epidemic size and a four-week extension of the mean epidemic duration2. In addition, the rapid identification of outbreaks and the implementation of control measures have been crucial in limiting the impact of epidemics, both in preventing more casualties and in shortening the period during which the stringent control measures were needed3,4. Not surprisingly, researchers’ attention has been directed to the development and evaluation of early warning systems. The aspirational goal of these systems is to improve the timeliness and accuracy of biothreat detection using traditional and new sources of surveillance data. In contrast to historical passive surveillance instruments, early warning systems have capacity for the active monitoring of historical data and provide automated alerts when aberrations in disease trends are identified.
Prospective detection of communicable disease outbreaks
Public health professionals have been very good at identifying highly focal outbreaks where the majority of clinical cases occur in spatially and temporally tight clusters. Sophisticated surveillance and early warning systems are not needed to detect these outbreaks, as they stand out dramatically from the background of sporadic cases. Of course, outbreaks come in many shapes and sizes, and when clinical cases of infection are less concentrated in space or time, or have lower attack rates, they are less visible and harder to detect. Our public health colleagues expect that laboratory based surveillance systems could assist in the identification of these events.
The identification, control and prevention of outbreaks relies on interacting local, regional and national healthcare structures with complimentary roles. They collect and generate different types of data in relation to public health surveillance. It appears that prospective laboratory based surveillance offers the most specific surveillance signals. There is increasing evidence demonstrating the value of rapid molecular profiling as a means of assisting outbreak detection in hospital settings5,6. This can be of particular importance as the monitoring of mobile genetic elements, which spread antibiotic resistance genes, has become essential for successful infection control. In one prospective study, automated clonal alerts based on the real-time subtyping of hospital methicillin-resistant Staphylococcus aureus (MRSA) isolates and temporal-scan test statistics were 100% and 95.2% sensitive and specific in identifying outbreaks and more sensitive and timely than infection control nurses6.
To meet the increasing demands for accurate early warning systems microbiologists have applied advances in molecular microbiology in order to develop more rapid and easily standardisable methods of fingerprinting pathogens with epidemic potential. For example, multiple locus variable-number tandem-repeat analysis (MLVA) of Salmonella enterica serovar Typhimurium (STM) has been introduced by several jurisdictional enteric reference laboratories in Australia in order to improve the resolution of public health surveillance for human infections. Prospective MLVA typing of STM allowed the detection of community outbreaks, provided timely early warnings about possible STM clusters and demonstrated the sustained level of STM diversity. In addition, the monitoring of novel and persistent MLVA types offered a new benchmark for STM surveillance7.
The increasing complexity of bacterial typing methods and the interpretation of their results demand close collaborations between microbiologists and epidemiologists to ensure that alerts generated by early warning systems are sufficiently specific and sensitive and correspond well to epidemiological clusters. These collaborations are fostered through multidisciplinary networks. Such networks for subtype-based surveillance are being developed and adopted around the world for an increasing number of pathogens. The Enter-net system, coordinated out of the European Centre for Disease Prevention and Control, provides surveillance and early warning across the EU member states. The PulseNet model has been deployed in the USA and Canada for many years, and a memorandum of understanding permits each country to consult the other’s database of PFGE patterns. PulseNet International, with regional grouping for Latin America, Asia/Pacific, the Middle East and Europe, is now routinely exchanging patterns and information among a growing group of countries, paving the way toward a truly global network for subtype-based surveillance8. In parallel, the networks of national field epidemiology programs strive to develop uniform and collaborative investigative methods to match the growing laboratory capacity in prospective high-resolution subtyping. In Australia, a combined program for active surveillance and collaborative investigation, called OzFoodNet, now includes all of the states9. The appealing assumption is that new ways of networking and data sharing could enable the aggregation of clinical and public health data to support both clinical care and disease surveillance10.
Australian Biosecurity Intelligence Network (ABIN)
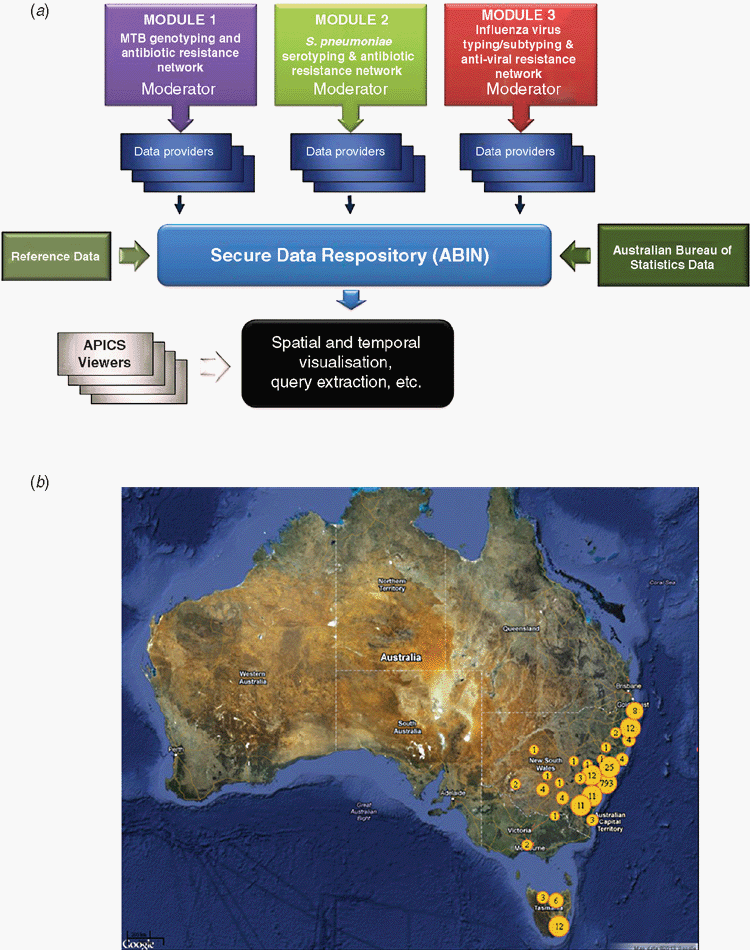
eResearch tools and open-source software for data mining and visualisation are gaining recognition and popularity among biomedical scientists. The Australian Pathogen Intelligence Community Space (APICS) (https://www.abin.org.au) aims to improve infectious disease surveillance by providing a secure online environment for collaborative research and the sharing of data, knowledge and resources between specialist pathology and microbiology providers, epidemiologists, public health professionals and multidisciplinary research teams. APICS developed data management and analysis tools for the secure uploading, trend/cluster analysis, and spatial/temporal visualisation of infectious disease data. Disease modules were built by integrating diverse pathogen information, including molecular and phenotypic subtyping and profiling information and de-identified clinical and geo-demographic patient data. Functionality was also created to incorporate external population and socioeconomic information from the Australian Bureau of Statistics. Three test disease modules were used to evaluate the utility of APICS. These modules were based on Mycobacterium tuberculosis genotyping and antibiotic resistance data, serotyping and antibiotic resistance monitoring of Streptococcus pneumoniae isolates associated with invasive pneumococcal disease and influenza sub-typing and antiviral resistance data. Output from these modules demonstrated the potential of APICS analytical and visualisation tools to link infectious disease cases with specific pathogen profiles, to distinguish epidemic strains from sporadic or emerging variants and to monitor spatio-temporal changes in pathogen subtypes and resistance profiles.
Figure 1 shows an overview of the APICS application structure and an APICS-generated map depicting the spatial clustering of pathogens isolated from patients residing in different postcodes across the eastern states of Australia. Wide-scale adoption of APICS has the potential to create data sharing and analysis networks for the prospective monitoring of pathogen transmission and changes in pathogen/disease patterns. Network information could also be used to pinpoint key areas for future infectious disease research and optimise disease prevention/intervention strategies.
Genomics enhanced public health laboratory surveillance
Recent advances in DNA sequencing technology have made whole-genome sequencing (WGS) of pathogens of public health significance, within a clinically relevant turn-around-time, both technically and economically feasible. DNA sequencing offers important advantages over other methods of pathogen characterisation. It provides a universal solution with high throughput, speed and quality. Sequencing is a standardised process regardless of the nature of microorganisms, and different species of pathogens could be processed simultaneously in a single sequencing run. This means that WGS could allow economies of scale at local or regional laboratories. Furthermore, a single WGS can often replace several traditional tests that reference laboratories will perform on a single isolate, while providing equivalent or superior-quality information. DNA sequences also represent an indisputable and possibly ‘future-proof’ data format amenable to exchange between jurisdictional laboratories and to comparison at the national and international levels. Finally, the potential utility of WGS for public health has been supported by the rapid growth of public databases with reference genomes.
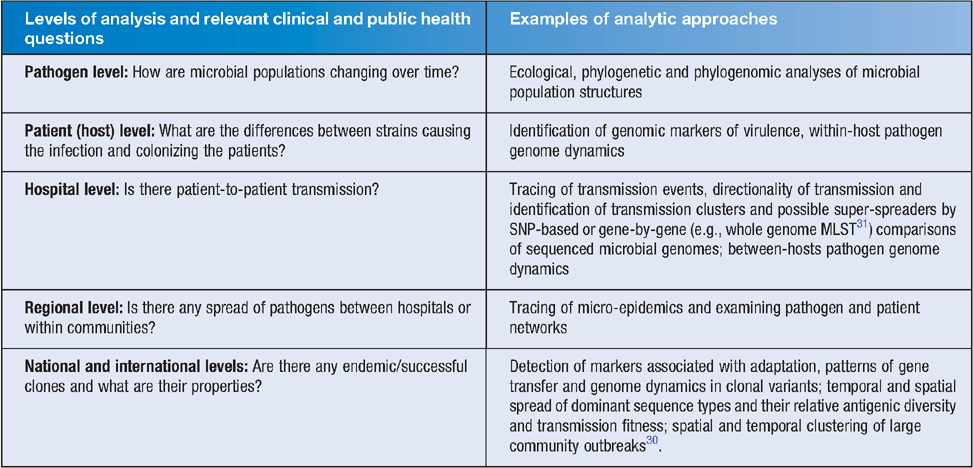
Table 1 highlights how WGS data can potentially enhance current investigations. WGS analysis has become the ultimate high-resolution alternative to bacterial subtyping. It has led to the identification of novel genomic targets for drugs and vaccines11, has provided a new impetus for the study of pathogen epidemiology and evolution12,13 and has been instrumental in deciphering the origin and trajectory of recent outbreaks14–16. Recent ground-breaking studies from The Netherlands17, Canada18, Germany19,20 and the UK21,22 have highlighted the potential utility of large-scale genome analysis for detecting and controlling outbreaks of communicable diseases. Emerging evidence suggests that WGS-based identification and characterisation of microbial pathogens improves monitoring for emerging clones or new pathogens and enhances the resolution of laboratory-based surveillance23. Specifically, this technology enhances the identification and tracing of outbreaks in community and hospital settings through the recognition of covert clusters as well as reconstruction of most probable transmission events within outbreaks. Recent proof-of-concept studies have demonstrated WGS superiority to current subtyping methods24,25. However, the vast majority of genomic data remains not medically actionable, at this time, as the WGS technology is still maturing and its cost-effectiveness for public health surveillance and laboratory workflow requires further assessment. The main added value of genomics-enabled surveillance appears to be in the improved accuracy of case clustering. Furthermore, WGS offers an unprecedented level of resolution for distinguishing the degrees of relatedness amongst bacterial isolates and compliments existing epidemiological tools by providing a means to reconstruct recent chains of transmission, identify sequential acquisition of strains and detect cryptic outbreaks that might otherwise go unnoticed. WGS is also effective at determining whether isolates within a potential cluster are unrelated, thereby saving time and resources on outbreak investigations.
|
New analytics for public health laboratory surveillance and early warning
The plummeting cost of sequencing has led to a radical shift in the bioscience capacity bottleneck from the generation of data to its analysis. The linkage of structured genomic data with the unstructured information captured in scientific literature has emerged as one of the most appealing domains for the ‘new analytics’. Genomic databases have grown exponentially and peer-reviewed publications made available through NCBI PubMed have grown at the rate of >4.8%, or more than two thousand new entries per day. Clearly the increased quantity of scientific and public health information is reaching the point where it cannot be sustained by current analytical practices. We argue that one possible solution could be software robots or ‘bots’26. They have proven their effectiveness in accessing resources across the Internet and analysing the results to gain knowledge, which is then applied to fulfil specific tasks. Recent ground-breaking projects such as RoboEarth© (www.roboearth.org), which has been building a parallel WWW for robots that can share information and learn from each other, are generating radically new data analysis techniques. Crucially, such robots could retrieve newly published genome sequences and integrate them with other data sources. This would represent a major scientific advance since no single library alone can index the entire science space. Web robots can continuously mine the knowledge space, searching for relationships following a well-validated ABC-principle, in which concepts A & C have no direct relationship but are connected via shared B intermediates. Examples of such relationships or events of public health significance are new ‘pathogen-gene’ associations, pathogen-gene-syndrome associations, or known associations in a new geographic location. However, these new analytics are unlikely to ever replace human judgement. Their role is to support the increasingly collaborative efforts of pathogen discovery and characterisation conducted by teams of microbiologists, clinicians, molecular biologists, public health professionals and statisticians27–30.
Conclusions
Robust and flexible surveillance methods, standardised and rapid pathogen subtyping networks, and collaborative epidemiological investigation strategies will be the marks of a successful approach to controlling communicable infections in the coming years. The development of the ‘smart analytics’ will transform the use of WGS for public health surveillance at regional, national and global levels. In particular, the simultaneous consideration of synergistic and dynamically evolving lines of evidence such as microbial genomics and population studies of infectious disease epidemiology will enhance the resolution of the detection and monitoring of infectious disease outbreaks. The exponential growth in available microbial genomics data opens a unique opportunity to start addressing the ‘big’ epidemiological and evolutionary questions, such as revealing the origins of novel infections, pathogen genome dynamics and patterns of gene transfer.
References
[1] Dato, V. et al. (2004) How outbreaks of infectious disease are detected: a review of surveillance systems and outbreaks. Public Health Rep. 119, 464–471.| How outbreaks of infectious disease are detected: a review of surveillance systems and outbreaks.Crossref | GoogleScholarGoogle Scholar | 15313109PubMed |
[2] Wallinga, J. and Teunis, P. (2004) Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 160, 509–516.
| Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures.Crossref | GoogleScholarGoogle Scholar | 15353409PubMed |
[3] Svoboda, T. et al. (2004) Public health measures to control the spread of the severe acute respiratory syndrome during the outbreak in Toronto. N. Engl. J. Med. 350, 2352–2361.
| Public health measures to control the spread of the severe acute respiratory syndrome during the outbreak in Toronto.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXks1Gjt7w%3D&md5=8a25303e6b8e3c52e6d4025af8a9c18aCAS | 15175437PubMed |
[4] Buckeridge, D.L. (2007) Outbreak detection through automated surveillance: a review of the determinants of detection. J. Biomed. Inform. 40, 370–379.
| Outbreak detection through automated surveillance: a review of the determinants of detection.Crossref | GoogleScholarGoogle Scholar | 17095301PubMed |
[5] Fournier, P.-E. et al. (2007) Bacterial genome sequencing and its use in infectious diseases. Lancet Infect. Dis. 7, 711–723.
| Bacterial genome sequencing and its use in infectious diseases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtlKhsbfN&md5=4979e5a7e90d304c58d0e36754c12144CAS | 17961857PubMed |
[6] Mellmann, A. et al. (2006) Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med. 3, e33.
| Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks.Crossref | GoogleScholarGoogle Scholar | 16396609PubMed |
[7] Sintchenko, V. et al. (2012) Improving resolution of public health surveillance for human Salmonella enterica serovar Typhimurium infection: 3 years of prospective multiple-locus variable-number tandem-repeat analysis (MLVA). BMC Infect. Dis. 12, 78.
| Improving resolution of public health surveillance for human Salmonella enterica serovar Typhimurium infection: 3 years of prospective multiple-locus variable-number tandem-repeat analysis (MLVA).Crossref | GoogleScholarGoogle Scholar | 22462487PubMed |
[8] Swaminathan, B. et al. (2006) Building PulseNet International: an interconnected system of laboratory networks to facilitate timely public health recognition and response to foodborne disease outbreaks and emerging foodborne diseases. Foodborne Pathog. Dis. 3, 36–50.
| Building PulseNet International: an interconnected system of laboratory networks to facilitate timely public health recognition and response to foodborne disease outbreaks and emerging foodborne diseases.Crossref | GoogleScholarGoogle Scholar | 16602978PubMed |
[9] Kirk, M.D. et al. (2010) Surveillance for outbreaks of gastroenteritis in long-term care facilities, Australia, 2002-2008. Clin. Infect. Dis. 51, 907–914.
| Surveillance for outbreaks of gastroenteritis in long-term care facilities, Australia, 2002-2008.Crossref | GoogleScholarGoogle Scholar | 20825308PubMed |
[10] Niesters, H.G. et al. (2013) Laboratory-based surveillance in the molecular era: the TYPENED model, a joint data-sharing platform for clinical and public health laboratories. Eurosurveillance 18, 20 387.
| 1:STN:280:DC%2BC3szktlKisQ%3D%3D&md5=799d444aeadb1c1e2f431c41c607a856CAS |
[11] Didelot, X. et al. (2012) Transforming clinical microbiology with bacterial genome sequencing. Nat. Rev. Genet. 13, 601–612.
| Transforming clinical microbiology with bacterial genome sequencing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFCrt7jE&md5=b3706a80c91e61d8387c2668d4497745CAS | 22868263PubMed |
[12] Ford, C.B. et al. (2011) Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat. Genet. 43, 482–486.
| Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXltFyqur0%3D&md5=c68e8cd0d2c19bf917ba1578dc2ae12dCAS | 21516081PubMed |
[13] Okoro, C.K. et al. (2012) High-resolution single nucleotide polymorphism analysis distinguishes recrudescence and reinfection in recurrent invasive nontyphoidal Salmonella typhimurium disease. Clin. Infect. Dis. 54, 955–963.
| High-resolution single nucleotide polymorphism analysis distinguishes recrudescence and reinfection in recurrent invasive nontyphoidal Salmonella typhimurium disease.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XktVGmtrs%3D&md5=8cfb49c0b37a485ced559fa46f6d636fCAS | 22318974PubMed |
[14] Snitkin, E.S. et al. (2012) Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumonia with whole-genome sequencing. Science Transl. Med. 4, 148ra116.
| Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumonia with whole-genome sequencing.Crossref | GoogleScholarGoogle Scholar |
[15] Jolley, K.A. et al. (2012) Resolution of a meningococcal disease outbreak from whole-genome sequence data with rapid web-based analysis methods. J. Clin. Microbiol. 50, 3046–3053.
| Resolution of a meningococcal disease outbreak from whole-genome sequence data with rapid web-based analysis methods.Crossref | GoogleScholarGoogle Scholar | 22785191PubMed |
[16] Walker, T.M. et al. (2013) Contact investigations for outbreaks of Mycobacterium tuberculosis: advances through whole genome sequencing. Clin. Microbiol. Infect. 19, 796–802.
| Contact investigations for outbreaks of Mycobacterium tuberculosis: advances through whole genome sequencing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtlWlurbI&md5=45a6e9278aba684881b51bcab00d016cCAS | 23432709PubMed |
[17] Schűrch, A.C. et al. (2010) High-resolution typing by integration of genome sequencing data in a large tuberculosis cluster. J. Clin. Microbiol. 48, 3403–3406.
| High-resolution typing by integration of genome sequencing data in a large tuberculosis cluster.Crossref | GoogleScholarGoogle Scholar |
[18] Gardy, J.L. et al. (2011) Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med. 364, 730–739.
| Whole-genome sequencing and social-network analysis of a tuberculosis outbreak.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXis1ymsb0%3D&md5=17f8737ecb384cf889db5a4e31e6e36fCAS | 21345102PubMed |
[19] Roetzer, A. et al. (2013) Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiology study. PLoS Med. 10, e1001387.
| Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiology study.Crossref | GoogleScholarGoogle Scholar | 23424287PubMed |
[20] Mellmann, A. et al. (2011) Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O124:H4 outbreak by rapid next generation sequencing technology. PLoS ONE 6, e22751.
| Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O124:H4 outbreak by rapid next generation sequencing technology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtVCnsr%2FP&md5=e629ed6bd6779f3f40ae78d5a51e49bfCAS | 21799941PubMed |
[21] Sherry, N.L. et al. (2013) Outbreak investigation using high-throughput genome sequencing within a diagnostic microbiology laboratory. J. Clin. Microbiol. 51, 1396–1401.
| Outbreak investigation using high-throughput genome sequencing within a diagnostic microbiology laboratory.Crossref | GoogleScholarGoogle Scholar | 23408689PubMed |
[22] Eyre, D.W. et al. (2012) A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2, e001124.
| A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance.Crossref | GoogleScholarGoogle Scholar | 22674929PubMed |
[23] Gilmour, M.W. et al. (2013) Public health genomics and the new molecular epidemiology of bacterial pathogens. Public Health Genomics 16, 25–30.
| Public health genomics and the new molecular epidemiology of bacterial pathogens.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3srhs1ynuw%3D%3D&md5=42fc1d80a4b9a6d67366ce60cca3154cCAS | 23548714PubMed |
[24] Köser, C.U. et al. (2012) Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 366, 2267–2275.
| Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak.Crossref | GoogleScholarGoogle Scholar | 22693998PubMed |
[25] Underwood, A.P. et al. (2013) Public health value of next-generation DNA sequencing of enterohaemorrhagic Escherichia coli from an outbreak. J. Clin. Microbiol. 51, 232–237.
| Public health value of next-generation DNA sequencing of enterohaemorrhagic Escherichia coli from an outbreak.Crossref | GoogleScholarGoogle Scholar | 23135946PubMed |
[26] Sintchenko, V. and Coiera, E. (2011) Translational web robots for pathogen genome analysis. Microbial Informatics and Experimentation 1, 10.
| Translational web robots for pathogen genome analysis.Crossref | GoogleScholarGoogle Scholar | 22587672PubMed |
[27] Okeke, I.N. and Wain, J. (2008) Postgenomic challenges for collaborative research in infectious diseases. Nat. Rev. Microbiol. 6, 858–864.
| Postgenomic challenges for collaborative research in infectious diseases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXht1CmtrrI&md5=7def4b661e75e514776de9cb3c855378CAS | 18711428PubMed |
[28] Sintchenko, V. et al. (2009) Towards bioinformatics assisted infectious disease control. BMC Bioinform. 10, S10.
| Towards bioinformatics assisted infectious disease control.Crossref | GoogleScholarGoogle Scholar |
[29] Lipkin, W.I. (2013) The changing face of pathogen discovery and surveillance. Nat. Rev. Microbiol. 11, 133–141.
| The changing face of pathogen discovery and surveillance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhvVymurvE&md5=f7a62e148983cd3adb210809f600caf8CAS | 23268232PubMed |
[30] Aarestrup, F.M. et al. (2012) Integrating genome-based informatics to modernize global disease monitoring, information sharing, and response. Emerg. Infect. Dis. 18, e1.
| Integrating genome-based informatics to modernize global disease monitoring, information sharing, and response.Crossref | GoogleScholarGoogle Scholar | 23092707PubMed |
[31] Maiden, M.C.J. et al. (2013) MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11, 728–736.
| MLST revisited: the gene-by-gene approach to bacterial genomics.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtlalu7nJ&md5=03bf50cdbfb5d694a97d049e9112b88aCAS |
Biographies
Dr Vitali Sintchenko FRCPA FACHI PhD MASM is a medical microbiologist and Director of the Centre for Infectious Diseases and Microbiology – Public Health in the Institute of Clinical Pathology and Medical Research-Pathology West at Westmead Hospital. He is an Associate Professor of Sydney Medical School and the Marie Bashir Institute for Infectious Diseases and Biosecurity, The University of Sydney. His major research interests are infectious disease informatics and genomics guided surveillance of bacteria with epidemic potential. Vitali is a NHMRC Career Development Fellow and his research is supported by NHMRC Centre for Research Excellence and Project Grants.
Dr Nadine Holmes (nee McCallum) PhD MASM is a Postdoctoral Research Fellow at the Marie Bashir Institute for Infectious Diseases and Biosecurity, the University of Sydney. Her research background is in bacterial genetics, focussing strongly on bacterial fitness, antibiotic resistance and virulence and molecular typing. Nadine is currently working on an NHMRC-funded project that investigates mechanisms of tuberculosis transmission, and has been involved in setting up an Ion Torrent PGM sequencing facility at the Centre for Infectious Diseases and Microbiology – Public Health and establishing a sequencing workflow for bacterial whole-genome sequencing.