Intestinal Spirochaetes and Brachyspiral colitis
David J HampsonSchool of Veterinary and Life Sciences
Murdoch University
Western Australia
Tel: +61 8 9360 2287
Fax: +61 8 9319 4144
Email: d.hampson@murdoch.edu.au
Microbiology Australia 34(1) 34-37 https://doi.org/10.1071/MA13011
Published: 20 March 2013
The “intestinal spirochaetes” are a group of anaerobic bacteria assigned to various species in the genus Brachyspira. They inhabit the large intestines of birds and animals – but also may be found in human beings. These bacteria first came to prominence in the early 1970s when a spirochaete named Treponema hyodysenteriae (now Brachyspira hyodysenteriae) was shown to be the agent of swine dysentery, a colonic infection of pigs that is endemic in many countries and is of considerable economic significance. Since the initial description, related spirochaetes have been identified and characterised and various name changes have occurred – finally resulting in the genus Brachyspira and its seven officially recognised species. Many different hosts are colonised with the various Brachyspira species, but disease is mainly reported in pigs and in adult chickens. Humans are colonised with the zoonotic Brachyspira pilosicoli and Brachyspira aalborgi. Reduced susceptibility to various antimicrobials is now starting to represent a major problem for effective control of Brachyspiral colitis in pigs and other species, and consequently attention is focusing on the development of new vaccines. The Brachyspira species have specialised growth requirements, and different species can take from three days to three weeks to form a thin film of visible growth on selective isolation plates. Genetic manipulation of individual strains remains difficult, and this has limited understanding of gene function and disease pathogenesis. Recently whole genomic sequencing projects have started to reveal much that was previously unknown about these specialised bacteria.
Taxonomy and disease association
Currently the genus Brachyspira includes seven officially named species and a variety of unofficially proposed members. Some species have well-established pathogenic potential in certain hosts, causing forms of Brachyspira colitis, whilst others are considered to be largely commensal (see Table 1). The Brachyspira are genetically distinct from other spirochaetes, and the close similarities between some of the species in their 16S rRNA sequences suggest that speciation in the genus has occurred relatively recently and rapidly. Apart from the strength of haemolysis, there are few clear phenotypic differences between the species, and indeed the boundaries between some of the named species are indistinct both genetically and phenotypically.
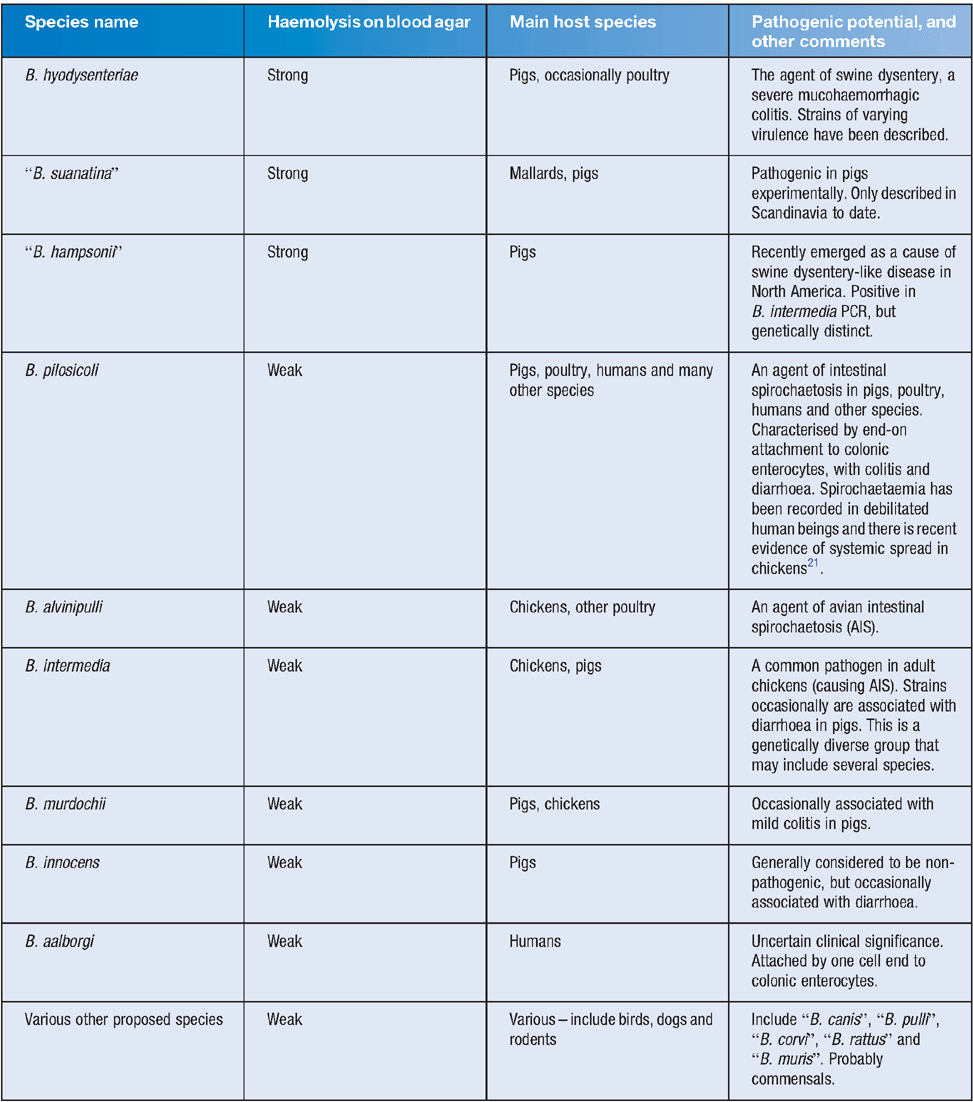
|
Population structures, evolution and genetic variation
For some species, such as B. hyodysenteriae, studies using multilocus sequence typing have provided clear evidence of the population structure being clonal1,2, while for B. pilosicoli the population appears to be recombinant3. In the case of B. hyodysenteriae, the adaptation to a lifestyle in the large intestine appears to have included acquisition of various genes from Escherichia coli and Clostridium species, especially those encoding proteins associated with transport and metabolism4. These are likely to have been acquired in the densely populated, complex and specialised environment of the large intestine.
The existence of extensive genetic rearrangements can be observed within and between Brachyspira species, with sequence drift also generating diversity. The variation and fluidity of the genomes can be seen in the case of B. pilosicoli, where three sequenced strains had genome sizes of ~2,765, 2.890 and 2.596 Mb respectively5, with genome rearrangements that largely correlated with the positions of mobile genetic elements. Novel bacteriophages were detected, as they were in a previous genomic study on B. intermedia6. These bacteriophages, that have themselves undergone extensive gene remodelling, are involved in intra- and inter-species horizontal gene transfer, and are likely to be a major force in the evolution of the Brachyspira species. In addition, novel genetic information may be acquired through the activity of prophage-like gene transfer agents that are present in the genome of different Brachyspira species7,8. Evidence for rapid genetic change can be seen at the farm level, where, for example, “microevolution” of B. hyodysenteriae strains resulting in changed DNA profiles has been recorded over relatively short time periods9,10.
Pathogenesis
The basis of virulence in the various Brachyspira species is still imperfectly understood. In order for pathogenic Brachyspira species to induce disease it is essential for them to colonise the large intestine and to grow to large numbers. Their anaerobic metabolism and use of substrates has been fine tuned to allow them to thrive in the milieu of the large intestine. There are complex physical and chemical interactions that occur between components of the diet and the normal colonic microbiota: these profoundly influence the environment, and it has become clear that the resultant conditions can affect colonisation by the spirochaetes11.
As part of the colonisation process Brachyspira cells must move through the mucus overlying the epithelium of the large intestine. The corkscrew-like motility of B. hyodysenteriae has long been known to be an important virulence attribute, allowing it to penetrate the mucus. In the case of B. pilosicoli, this spirochaete shows increased motility under viscous conditions12, including mucin concentrations equivalent to those found in the colon13. In addition to their motility, the cells of different Brachyspira species demonstrate a chemotactic attraction to colonic mucin. Comparison of the genome sequences of B. hyodysenteriae and B. pilosicoli has shown that B. pilosicoli has fewer methyl-accepting chemotaxis genes than B. hyodysenteriae, and completely lacks mcpC genes; hence these species are predicted to have different chemotactic responses, and this in turn may help to explain their different host ranges and colonisation sites in the large intestine14. Experimentally, although cells of both B. hyodysenteriae and B. pilosicoli are attracted to and enter mucin solutions, this was reduced at mucin concentrations above 6% for B. hyodysenteriae but not for B. pilosicoli13, again providing a possible explanation for their different colonisation sites.
A likely virulence determinant in B. hyodysenteriae is the strong haemolytic activity of this spirochaete. This is supported by the fact that two other recently described strongly haemolytic species are also pathogenic15,16 (see Table 1). Currently eight genes encoding proteins with predicted haemolytic activity have been described in B. hyodysenteriae4,14, but their respective roles have not all been confirmed experimentally.
A recent in vitro study using Caco-2 cell monolayers has provided some insights into how B. pilosicoli interacts with colonic enterocytes to cause disease17, and similar detailed studies are required with B. hyodysenteriae and other species. In that study17 the Caco-2 cell junctions were shown to be the initial targets of the characteristic end-on attachment by B. pilosicoli. Colonised monolayers then demonstrated a time-dependent series of changes, including accumulation of actin at the cell junctions, loss of tight junction integrity and condensation and fragmentation of nuclear material consistent with the occurrence of apoptosis. The colonised monolayers demonstrated a significant up-regulation of interleukin-1ß (IL-1ß) and IL-8 expression. These cytokines/chemokines are likely to be responsible for attracting inflammatory cells to the colonisation site, and causing localised colitis. Potential mechanisms for inducing such cellular damage include the biological activity of lipooligosaccharides and/or the action of membrane proteases.
Sequencing of the genome of B. hyodysenteriae strain WA1 resulted in the identification of a previously unrecognised plasmid that contained 31 genes, including six rfbA-D genes that were predicted to be involved with rhamnose biosynthesis, and hence LOS structure, as well as others associated with glycosylation4. Subsequently avirulent strain A1 was shown to lack the plasmid, and when an Australian field isolate lacking the plasmid was selected and used experimentally to infect pigs, significantly fewer became colonised and developed dysentery compared to the pigs infected with a control strain that contained the plasmid18. The results support the likelihood that plasmid-encoded genes of B. hyodysenteriae are involved in colonisation and/or in disease expression.
Recombinant vaccines
Recently recombinant protein vaccines have received attention as potential vaccine components for Brachyspira species: for example, vaccination with recombinant outer-membrane lipoprotein Bhlp29.7 from B. hyodysenteriae provided a 50% reduction in the incidence of disease compared to unvaccinated controls following experimental infection19. The availability of genome sequences has provided the opportunity to broaden this approach through the application of “reverse vaccinology”, where scores of such predicted proteins can be identified from the genome sequence, screened and tested as vaccine candidates. This approach has been used successfully with B. hyodysenteriae20, and it is anticipated that a new generation of commercial vaccines based on this approach will become available in the next few years.
Summary
With the recent availability of Brachyspira genome sequences and new technologies better insights into the growth requirements and pathogenic mechanisms of Brachyspira species are emerging. This information is of direct benefit for control of the infections, since, for example, information about growth and colonisation requirements derived from metabolic reconstructions of the spirochaetes can help to predict what changes in the colonic environment are likely to reduce their growth4,14. Further detailed studies are needed to determine how the colonic microbiota is influenced by different dietary substrates, and how this impacts on colonisation by Brachyspira species. The sequence data has also allowed the use of a reverse vaccinology approach to vaccine development.
References
[1] La, T. et al. (2009) Multilocus sequence typing as a tool for studying the molecular epidemiology and population structure of Brachyspira hyodysenteriae. Vet. Microbiol. 138, 330–338.| Multilocus sequence typing as a tool for studying the molecular epidemiology and population structure of Brachyspira hyodysenteriae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtV2ksLjP&md5=c31b153ec8137974a6c2fac0761dacccCAS |
[2] Osorio, J. et al. (2012) Dissemination of clonal groups of Brachyspira hyodysenteriae amongst pig farms in Spain, and their relationships to isolates from other countries. PLoS ONE 7, e39082.
| Dissemination of clonal groups of Brachyspira hyodysenteriae amongst pig farms in Spain, and their relationships to isolates from other countries.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XptlSnsLs%3D&md5=5cf4c1f9b7e17ac872e4165dd132722fCAS |
[3] Trott, D.J. et al. (1998) Population genetic analysis of Serpulina pilosicoli and its molecular epidemiology in villages in the Eastern Highlands of Papua New Guinea. Int. J. Syst. Bacteriol. 48, 659–668.
| Population genetic analysis of Serpulina pilosicoli and its molecular epidemiology in villages in the Eastern Highlands of Papua New Guinea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXmt1Sgs7Y%3D&md5=20f30131b40fe70c287dbed6a5a0f10aCAS |
[4] Bellgard, M.I. et al. (2009) Genome sequence of the pathogenic intestinal spirochete Brachyspira hyodysenteriae reveals adaptations to its lifestyle in the porcine large intestine. PLoS ONE 4, e4641.
| Genome sequence of the pathogenic intestinal spirochete Brachyspira hyodysenteriae reveals adaptations to its lifestyle in the porcine large intestine.Crossref | GoogleScholarGoogle Scholar |
[5] Mappley, L.J. et al. (2012) Comparative genomics of Brachyspira pilosicoli strains: extensive genome rearrangements and reductions, and correlation of genetic compliment with phenotypic diversity. BMC Genomics 13, 454.
| Comparative genomics of Brachyspira pilosicoli strains: extensive genome rearrangements and reductions, and correlation of genetic compliment with phenotypic diversity.Crossref | GoogleScholarGoogle Scholar |
[6] Håfström, T. et al. (2011) Complete genome sequence of Brachyspira intermedia reveals unique genomic features in Brachyspira species and phage-mediated horizontal gene transfer. BMC Genomics 12, 395.
| Complete genome sequence of Brachyspira intermedia reveals unique genomic features in Brachyspira species and phage-mediated horizontal gene transfer.Crossref | GoogleScholarGoogle Scholar |
[7] Matson, E.G. et al. (2005) Identification of genes of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae. J. Bacteriol. 187, 5885–5892.
| Identification of genes of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXps1yrsrY%3D&md5=827465f6c51a86e434bad4997333da64CAS |
[8] Motro, Y. et al. (2009) Identification of genes associated with prophage-like gene transfer agents in the pathogenic intestinal spirochaetes Brachyspira hyodysenteriae, Brachyspira pilosicoli and Brachyspira intermedia. Vet. Microbiol. 134, 340–345.
| Identification of genes associated with prophage-like gene transfer agents in the pathogenic intestinal spirochaetes Brachyspira hyodysenteriae, Brachyspira pilosicoli and Brachyspira intermedia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtlOqs7s%3D&md5=59e080cda94e1a26c2d3111f5b9f34bcCAS |
[9] Atyeo, R.F. et al. (1999) Analysis of Serpulina hyodysenteriae strain variation and its molecular epidemiology using pulsed-field gel electrophoresis. Epidemiol. Infect. 123, 133–138.
| Analysis of Serpulina hyodysenteriae strain variation and its molecular epidemiology using pulsed-field gel electrophoresis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXmtFOkurg%3D&md5=000e3bf405be58eecdda5e5765796c30CAS |
[10] Hidalgo, A. et al. (2010) Multiple-locus variable-number tandem-repeat analysis of the swine dysentery pathogen, Brachyspira hyodysenteriae. J. Clin. Microbiol. 48, 2859–2865.
| Multiple-locus variable-number tandem-repeat analysis of the swine dysentery pathogen, Brachyspira hyodysenteriae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXht1Omu7vK&md5=0a4b5b286aea6711389cb36d3729fd20CAS |
[11] Hansen, C.F. et al. (2010) Diets containing inulin but not lupins help to prevent swine dysentery in experimentally challenged pigs. J. Anim. Sci. 88, 3327–3336.
| Diets containing inulin but not lupins help to prevent swine dysentery in experimentally challenged pigs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtF2rtbzJ&md5=de6af66166a089410725024190317d38CAS |
[12] Nakamura, S. et al. (2006) Improvement in motion efficiency of the spirochete Brachyspira pilosicoli in viscous environments. Biophys. J. 90, 3019–3026.
| Improvement in motion efficiency of the spirochete Brachyspira pilosicoli in viscous environments.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XjsFequ7o%3D&md5=f646cdd4593286c5aec202557f449b55CAS |
[13] Naresh, R. et al. (2010) Attraction of Brachyspira pilosicoli to mucin. Microbiology 156, 191–197.
| Attraction of Brachyspira pilosicoli to mucin.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhvV2isbY%3D&md5=fc4e670134ebb5ba70962ec5835074faCAS |
[14] Wanchanthuek, P. et al. (2010) The complete genome sequence of the pathogenic intestinal spirochete Brachyspira pilosicoli and comparison with other Brachyspira genomes. PLoS ONE 5, e11455.
| The complete genome sequence of the pathogenic intestinal spirochete Brachyspira pilosicoli and comparison with other Brachyspira genomes.Crossref | GoogleScholarGoogle Scholar |
[15] Råsbäck, T. et al. (2007) A novel enteropathogenic, strongly haemolytic spirochaete isolated from pig and mallard, provisionally designated ’Brachyspira suanatina’ sp. nov. Environ. Microbiol. 9, 983–991.
| A novel enteropathogenic, strongly haemolytic spirochaete isolated from pig and mallard, provisionally designated ’Brachyspira suanatina’ sp. nov.Crossref | GoogleScholarGoogle Scholar |
[16] Chander, Y. et al. (2012) Phenotypic and molecular characterization of a novel strongly hemolytic Brachyspira species, provisionally designated "Brachyspira hampsonii". J. Vet. Diagn. Invest. 24, 903–910.
[17] Naresh, R. et al. (2009) The intestinal spirochete Brachyspira pilosicoli attaches to cultured Caco-2 cells and induces pathological changes. PLoS ONE 4, e8352.
| The intestinal spirochete Brachyspira pilosicoli attaches to cultured Caco-2 cells and induces pathological changes.Crossref | GoogleScholarGoogle Scholar |
[18] La, T. et al. (2011) Evidence that the 36kb plasmid of Brachyspira hyodysenteriae contributes to virulence. Vet. Microbiol. 153, 150–155.
| Evidence that the 36kb plasmid of Brachyspira hyodysenteriae contributes to virulence.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtFKiur%2FK&md5=da0e11fa2cea7956c8029690b66b4521CAS |
[19] La, T. et al. (2004) Protection of pigs from swine dysentery by vaccination with recombinant BmpB, a 29.7 kDa outer-membrane lipoprotein of Brachyspira hyodysenteriae. Vet. Microbiol. 102, 97–109.
| Protection of pigs from swine dysentery by vaccination with recombinant BmpB, a 29.7 kDa outer-membrane lipoprotein of Brachyspira hyodysenteriae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXmt1WhsLs%3D&md5=6b272bc33767d53e850298d92078debdCAS |
[20] Song, Y. et al. (2009) A reverse vaccinology approach to swine dysentery vaccine development. Vet. Microbiol. 137, 111–119.
| A reverse vaccinology approach to swine dysentery vaccine development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXltlChtLY%3D&md5=53e7283a54b0924651577ed6c9af5786CAS |
[21] Mappley, L.J. et al. (2013) Evidence for systemic spread of the potentially zoonotic intestinal spirochaete Brachyspira pilosicoli in experimentally challenged laying chickens. J. Med. Microbiol. 62, 297–302.
Biography
David Hampson BVetMed, PhD, DSc, FASM, FAAM, FRCPath, FRCVS is Professor of Veterinary Microbiology and Dean of the School of Veterinary and Life Sciences at Murdoch University in Perth. He qualified as a veterinarian from the Royal Veterinary College in London in 1979, and has a PhD from the University of Bristol and a DSc from the University of London. He has worked as an academic at Murdoch University since 1986.


