Uropathogenic Escherichia coli biofilms
Nguyen Thi Khanh Nhu A B C * , Chitra Ravi A B C and Mark A. Schembri A B C *A Institute for Molecular Bioscience (IMB), The University of Queensland, Brisbane, Qld, Australia.
B School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Qld, Australia.
C Australian Infectious Diseases Research Centre, The University of Queensland, Brisbane, Qld, Australia.

Dr Nguyen Thi Khanh Nhu is a postdoctoral researcher in the IMB Centre for Superbug Solutions at The University of Queensland (UQ). She attained her BSc (Biotechnology) and MSc (Microbiology) at the University of Natural Sciences, Vietnam, and then worked as a research assistant at the Oxford University Clinical Research Unit in Vietnam, where she investigated molecular mechanisms of virulence and antibiotic resistance in a range of enteric pathogens. Dr Nhu completed her PhD on uropathogenic E. coli at UQ and now applies genomic and molecular methods to understand how uropathogenic E. coli cause human infection. |

Chitra Ravi is a PhD student in the School of Chemistry & Molecular Biosciences, and the IMB Centre for Superbug Solutions, at The University of Queensland (UQ). She attained a Diploma (Molecular Biotechnology) at Nanyang Polytechnic, Singapore, and her BSc(Hons) (Microbiology) at UQ. Her PhD research investigates the regulation and molecular mechanisms of cellulose biosynthesis in uropathogenic E. coli, and the role of cellulose as a key component of the extracellular matrix in uropathogenic E. coli biofilms. |

Prof. Mark Schembri is the Director of the IMB Centre for Superbug Solutions at The University of Queensland. His expertise lies in molecular microbiology and bacterial pathogenesis, and he investigates how antibiotic resistant uropathogenic E. coli cause urinary tract and bloodstream infections, diseases of major significance to human health. |
Microbiology Australia 44(2) 109-112 https://doi.org/10.1071/MA23030
Submitted: 13 April 2023 Accepted: 8 May 2023 Published: 24 May 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Urinary tract infection (UTI) is one of the most common infectious diseases, with a global annual incidence of ~175 million cases. Uropathogenic Escherichia coli (UPEC) is the major cause of UTI (>80%) and increasingly associated with rising antibiotic resistance. UPEC form biofilms during infection of the urinary tract, either on the luminal surface of the bladder, intracellularly within bladder superficial epithelial cells, or on the surface of indwelling catheters. This lifestyle of sessile growth promotes enhanced resistance, persistence and increased rates of recurrent UTI. UPEC employ a range of virulence factors to form biofilms, including fimbrial adhesins for attachment and autotransporters to promote cell-to-cell aggregation. In addition, UPEC biofilms are encased in an extracellular matrix comprised of proteins such as curli amyloid fibres and polysaccharides such as cellulose, which together form a hydrating glue that provides structural support for the biofilm and protects its component cells. Here, we describe the key features of UPEC biofilms and their importance for UPEC pathogenesis of the urinary tract.
Keywords: fimbrial adhesins, UPEC biofilms, UPEC pathogenesis, urinary tract infections, uropathogenic Escherichia coli.
Urinary tract infections
Urinary tract infection (UTI) involves infection of the bladder (cystitis) and kidney (pyelonephritis) and can lead to life-threatening sepsis. Approximately 25% of women who suffer UTI will experience a recurrence within 6 months of the initial infection,1 either with the same strain or a new organism.2 Overall, recurrent UTI is associated with increasing antibiotic resistance, treatment failure, decreased quality of life and mounting pressure on our healthcare system. Catheter-associated UTIs (CAUTIs) are also a major problem, particularly in the hospital setting. In Australia, it is estimated that the economic burden of antibiotic-resistant UTIs could mount to A$1.6 billion per annum by 2030 if nothing is done to halt increasing rates of infection.3
Uropathogenic Escherichia coli
UTI is caused by a diverse range of pathogens, the most common being uropathogenic Escherichia coli (UPEC), which is responsible for >80% of all infections.4 UPEC possess multiple virulence factors that enable infection of the urinary tract and promote disease pathogenesis. These include: (i) adhesive organelles that mediate attachment to uroepithelial cells in the bladder (e.g. type 1 fimbriae) and the kidney (e.g. P fimbriae) and facilitate colonisation5,6; (ii) autotransporter proteins (e.g. Ag43) that mediate aggregation7,8; (iii) iron acquisition systems involving siderophores and heme-binding proteins that scavenge iron from the host to enable survival in the iron-poor urinary tract9; (iv) toxins such as cytotoxic necrotising factor-1 and hemolysin that damage host cells, thereby enabling penetration into deeper tissue layers10; (v) surface polysaccharides such as the capsule and O-antigen that provide protection against soluble and cellular mediators of host innate immunity11; and (vi) flagella that promote motility, enabling ascension from the bladder into the kidneys.12 UPEC also exhibit high rates of antibiotic resistance, including resistance to last line carbapenems and polymyxins, and frequently carry conjugative plasmids that facilitate the rapid dissemination of antibiotic resistance genes.13
UPEC biofilm formation
UPEC can exist in extracellular and intracellular niches in the urinary tract, both of which are associated with the formation of biofilms. UPEC form biofilms on the luminal surface of the bladder and on the surface of catheters. In addition, UPEC can form biofilm-associated intracellular bacterial communities (IBCs) within superficial bladder epithelial cells, where they establish tightly packed aggregates comprising 104–105 cells enclosed in an extracellular matrix.14 The pathway of IBC formation has been described extensively in mice,14 and evidence for IBC formation in human patients has also been reported.15 The formation of biofilms by UPEC during infection enables the establishment of a physical barrier that protects against host innate immune defences (e.g. neutrophils and antimicrobial peptides such as cathelicidins) and antibiotics.16 In laboratory mice, UPEC intracellular growth within bladder epithelial cells results in the persistence of infection despite treatment with antibiotics with different target specificities and from different classes.17 The close proximity of bacterial cells within biofilms also contributes to the exchange of DNA, including antimicrobial resistance plasmids. Overall, the impact of biofilm formation by UPEC is associated with increased resistance, persistence and recurrent infections.
Mechanisms of UPEC biofilm formation
UPEC biofilm formation involves several stages of progression: (i) adhesion, (ii) aggregation and early development of the biofilm structure, (iii) biofilm expansion and maturation, and (iv) dispersal of cells from the biofilm (Fig. 1). Initial attachment requires the expression of fimbriae such as type 1 fimbriae that mediate specific binding to mannosylated bladder superficial epithelial cells by a tip-located FimH adhesin, and subsequent invasion and the formation of IBCs.14 UPEC also produce other fimbriae that contribute to adhesion by interaction with different receptors, including P, F1C, F9, Afa, and type 3 fimbriae.18–20 After attachment, UPEC proliferation leads to the formation of aggregates or clusters by cell-to-cell interactions. One important UPEC surface factor that drives aggregation and biofilm development is the autotransporter protein Ag43,8 but other autotransporters also promote similar phenotypes.21,22 A key process that occurs during UPEC biofilm maturation is the production of an extracellular matrix, a glue composed of protein and polysaccharide that functions as an external support to maintain the structural integrity of the biofilm. The primary building blocks of the UPEC biofilm extracellular matrix are curli and cellulose. Curli are extracellular amyloid fibres that form connections between cells and facilitate interactions with UPEC-produced cellulose.23 Other polysaccharides such as β-1,6-N-acetyl-D-glucosamine (PGA) and colanic acid also function as extracellular matrix components that shape the architecture of the developing biofilm. The last stage of UPEC biofilm maturation involves detachment, leading to dispersal of cells from the biofilm and the capacity to seed new biofilms at distal sites.
Schematic diagram of biofilm formation and dispersal. Biofilm developmental process involving: (i) surface attachment of uropathogenic Escherichia coli (UPEC), (ii) cell aggregation and early development, (iii) expansion and maturation, and (iv) dispersal that can result in the formation of new biofilms at distal sites. Diguanylate cyclases (DGC) synthesise c-di-GMP from two GTP molecules, increasing levels of c-di-GMP that promotes biofilm formation. In the reverse step, c-di-GMP is degraded into two GMP molecules by phosphodiesterases (PDE), reducing c-di-GMP levels and promoting motility and dispersal from biofilms. Image was created with Biorender.
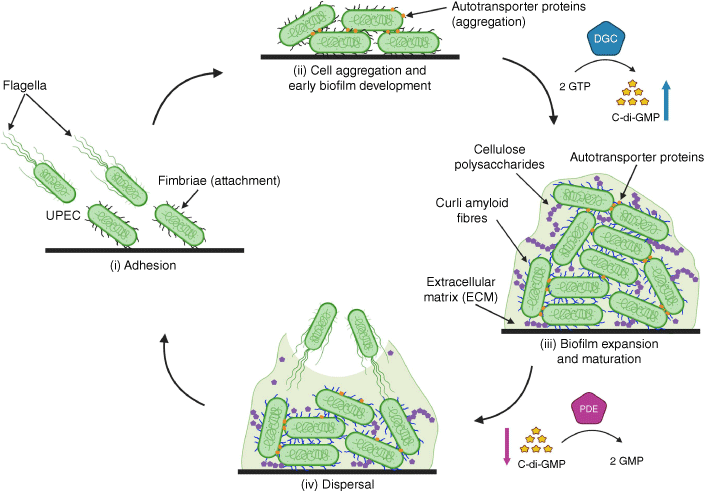
A molecular mechanism that triggers the transition of UPEC from a sessile (attached) state to a planktonic (free-swimming) state is the production of the second messenger molecule bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP).24 High UPEC intracellular concentrations of c-di-GMP, generated by diguanylate cyclase enzymes that synthesise c-di-GMP, promote the production of adherence, aggregation and extracellular matrix components that drive biofilm formation. By contrast, low intracellular concentrations of c-di-GMP, achieved by the activation of phosphodiesterase enzymes that degrade c-di-GMP, promote the production of flagella and UPEC dispersal from the biofilm.
UPEC biofilm extracellular matrix components – curli
Curli are extracellular amyloid fibres that contribute to UPEC biofilm formation by interacting with host cell matrix proteins and enhancing adherence, aggregation and colonisation of the urinary tract.25 The genes involved in curli production (referred to as curli-specific genes, csg) are found at the same location on the chromosome and arranged as two divergent operons; csgBAC and csgDEFG. The csgBAC operon encodes the major subunit protein CsgA, the minor subunit and nucleator protein CsgB, and the CsgC periplasmic chaperone. The csgDEFG operon encodes the positive regulator CsgD (which regulates both operons), the auxiliary secretion factors CsgE and CsgF, and the CsgG pore that facilitates secretion of curli subunit proteins across the outer membrane.25
Curli biosynthesis is regulated by a complex network involving environmental and stress-sensing mechanisms that are relayed through the curli regulator CsgD. These involve the stationary phase sigma factor RpoS, the two-component regulatory systems OmpR-EnvZ and CpxA-R, the Rcs signal transduction system, integration host factor (IHF), the histone-like nucleoid-associated protein H-NS, small non-coding RNAs and c-di-GMP. Most of our knowledge on curli regulation and biosynthesis comes from studies on commensal E. coli K-12 strains. We recently devised a high-resolution genetic screen employing saturated transposon mutagenesis coupled with phenotypic detection using the curli-specific dye Congo Red to define the genes involved in curli biogenesis.23 Transposon mutants with impaired curli production, identified by their inability to bind Congo Red, were characterised en masse using transposon-directed insertion site sequencing (TraDIS). In addition to the known genes mentioned above, our method identified novel genes and pathways involved in curli production, including purine biosynthesis, lipopolysaccharide (LPS) biogenesis, stress and stationary phase regulation, metabolism, sodium transport and septum formation. Excitingly, we also discovered a new curli repressor that we named rcpA (i.e. repressor of curli production A). Overexpression of rcpA reduced the transcription of genes encoding the curli regulator CsgD and the curli major subunit CsgA. The rcpA gene encodes a protein of 93 amino acids with two predicted transmembrane helices, a cytoplasmic C-terminal domain, and a high hydrophobic amino acid ratio, all of which suggest that RcpA may localise to the inner membrane and function by sensing and responding to environmental signals. Ongoing work in our lab is aimed at understanding the function of RcpA and elucidating the molecular mechanisms by which other factors identified in our screen affect curli production.
UPEC biofilm extracellular matrix components – cellulose
Cellulose is a linear exopolysaccharide polymer comprising β-1,4-linked glucosyl residues. The genes responsible for UPEC cellulose synthesis are encoded on two divergent operons; bcsRQABZC encoding proteins involved in synthesis and secretion and bcsEFG encoding the machinery required for cellulose modification. Similar to curli, cellulose biosynthesis is controlled by a complex regulatory network, mostly through CsgD. Therefore, cellulose is often co-produced with curli, explaining why both of these molecules make up the primary components of the biofilm extracellular matrix. The second messenger molecule c-di-GMP is also a critical checkpoint for cellulose production, as it functions as a direct activator of the BcsA synthase that transfers glucosyl residues from UDP-glucose onto the growing β-D-1,4-glucan chain in cellulose biogenesis. In UPEC, cellulose is modified by the addition of phosphoethanolamine, mediated by the BcsG transferase enzyme,26 although the functional effect of this modification remains to be properly understood. Current work in our lab is directed at defining the complete set of genes involved in UPEC cellulose production, and understanding the intersection of pathways involved in cellulose and curli synthesis and degradation.
Conclusion
The capacity to form biofilms plays an integral part in UPEC disease pathogenesis. We now have a good understanding of the virulence components that promote UPEC biofilm formation, including mechanisms of attachment, cell aggregation and extracellular matrix production. Future work will involve investigating the complex genetic networks that control these processes, with the goal to develop new therapeutics to inhibit and disrupt UPEC biofilms.
References
[1] Flores-Mireles, AL et al. (2015) Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13, 269–284.| Urinary tract infections: epidemiology, mechanisms of infection and treatment options.Crossref | GoogleScholarGoogle Scholar |
[2] Chen, Z et al. (2018) The urinary microbiome in patients with refractory urge incontinence and recurrent urinary tract infection. Int Urogynecol J 29, 1775–1782.
| The urinary microbiome in patients with refractory urge incontinence and recurrent urinary tract infection.Crossref | GoogleScholarGoogle Scholar |
[3] University of Technology Sydney (2020) Outbreak – a One Health antimicrobial resistance economic perspective. https://outbreakproject.com.au/wp-content/uploads/2020/12/OUTBREAK_REPORT_2020_economics_ERRATUM.pdf
[4] Foxman, B (2010) The epidemiology of urinary tract infection. Nat Rev Urol 7, 653–660.
| The epidemiology of urinary tract infection.Crossref | GoogleScholarGoogle Scholar |
[5] Connell, I et al. (1996) Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA 93, 9827–9832.
| Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract.Crossref | GoogleScholarGoogle Scholar |
[6] Johnson, JR et al. (1998) papG Alleles of Escherichia coli strains causing first-episode or recurrent acute cystitis in adult women. J Infect Dis 177, 97–101.
| papG Alleles of Escherichia coli strains causing first-episode or recurrent acute cystitis in adult women.Crossref | GoogleScholarGoogle Scholar |
[7] Ulett, GC et al. (2007) Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun 75, 3233–3244.
| Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract.Crossref | GoogleScholarGoogle Scholar |
[8] Heras, B et al. (2014) The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc Natl Acad Sci USA 111, 457–462.
| The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping.Crossref | GoogleScholarGoogle Scholar |
[9] Watts, RE et al. (2012) Contribution of siderophore systems to growth and urinary tract colonization of asymptomatic bacteriuria Escherichia coli. Infect Immun 80, 333–344.
| Contribution of siderophore systems to growth and urinary tract colonization of asymptomatic bacteriuria Escherichia coli.Crossref | GoogleScholarGoogle Scholar |
[10] Nhu, NTK et al. (2019) Complex multilevel control of hemolysin production by uropathogenic Escherichia coli. mBio 10, e02248-19.
| Complex multilevel control of hemolysin production by uropathogenic Escherichia coli.Crossref | GoogleScholarGoogle Scholar |
[11] Sarkar, S et al. (2014) Role of capsule and O antigen in the virulence of uropathogenic Escherichia coli. PLoS One 9, e94786.
| Role of capsule and O antigen in the virulence of uropathogenic Escherichia coli.Crossref | GoogleScholarGoogle Scholar |
[12] Lane, MC et al. (2007) Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci USA 104, 16669–16674.
| Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract.Crossref | GoogleScholarGoogle Scholar |
[13] Hancock, SJ et al. (2020) Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB. Nat Microbiol 5, 1340–1348.
| Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.Crossref | GoogleScholarGoogle Scholar |
[14] Anderson, GG et al. (2003) Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301, 105–107.
| Intracellular bacterial biofilm-like pods in urinary tract infections.Crossref | GoogleScholarGoogle Scholar |
[15] Rosen, DA et al. (2007) Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med 4, e329.
| Detection of intracellular bacterial communities in human urinary tract infection.Crossref | GoogleScholarGoogle Scholar |
[16] Kai-Larsen, Y et al. (2010) Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog 6, e10010.
| Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37.Crossref | GoogleScholarGoogle Scholar |
[17] Blango, MG and Mulvey, MA (2010) Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob Agents Chemother 54, 1855–1863.
| Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics.Crossref | GoogleScholarGoogle Scholar |
[18] Hancock, SJ et al. (2022) Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli. PLoS Pathog 18, e1010582.
| Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.Crossref | GoogleScholarGoogle Scholar |
[19] Alvarez-Fraga, L et al. (2022) Differential Afa/Dr fimbriae expression in the multidrug-resistant Escherichia coli ST131 clone. mBio 13, e03519-21.
| Differential Afa/Dr fimbriae expression in the multidrug-resistant Escherichia coli ST131 clone.Crossref | GoogleScholarGoogle Scholar |
[20] Ong, C-LY et al. (2008) Identification of type 3 fimbriae in uropathogenic Escherichia coli reveals a role in biofilm formation. J Bacteriol 190, 1054–1063.
| Identification of type 3 fimbriae in uropathogenic Escherichia coli reveals a role in biofilm formation.Crossref | GoogleScholarGoogle Scholar |
[21] Allsopp, LP et al. (2012) Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect Immun 80, 321–332.
| Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073.Crossref | GoogleScholarGoogle Scholar |
[22] Allsopp, LP et al. (2010) UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect Immun 78, 1659–1669.
| UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073.Crossref | GoogleScholarGoogle Scholar |
[23] Nhu, NTK et al. (2018) Discovery of new genes involved in curli production by a uropathogenic Escherichia coli strain from the highly virulent O45:K1:H7 lineage. mBio 9, e01462-18.
| Discovery of new genes involved in curli production by a uropathogenic Escherichia coli strain from the highly virulent O45:K1:H7 lineage.Crossref | GoogleScholarGoogle Scholar |
[24] Valentini, M and Filloux, A (2016) Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem 291, 12547–12555.
| Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria.Crossref | GoogleScholarGoogle Scholar |
[25] Barnhart, MM and Chapman, MR (2006) Curli biogenesis and function. Annu Rev Microbiol 60, 131–147.
| Curli biogenesis and function.Crossref | GoogleScholarGoogle Scholar |
[26] Thongsomboon, W et al. (2018) Phosphoethanolamine cellulose: a naturally produced chemically modified cellulose. Science 359, 334–338.
| Phosphoethanolamine cellulose: a naturally produced chemically modified cellulose.Crossref | GoogleScholarGoogle Scholar |


