Biofilms of Campylobacter concisus: a potential survival mechanism in the oral cavity
Taghrid Istivan A * and Mohsina Huq A BA School of Science, STEM College, RMIT University, Melbourne, Vic., Australia.
B Department of Medical Laboratories, College of Applied Medical Sciences, Qassim University, Buraydah, Saudi Arabia.

Assoc. Prof. Taghrid Istivan is an academic in the School of Science at RMIT University, as an educator, and research leader. She has a PhD in molecular microbiology and MSc in microbial genetics. Her research on pathogens and therapies focuses on microbial colonisation and virulence of bacterial pathogens, in addition to developing novel peptide therapeutics to fight antimicrobial resistance and cancer. She has published more than 40 peer-reviewed scientific papers and book chapters in these fields. Her recent research on drug delivery systems is protected by a patent on controlled released biomaterials for the delivery of therapeutics. |

Dr Mohsina Huq completed her MSc and PhD in Biotechnology at RMIT University, on research projects focussed on the detection and pathogenesis of the oral bacterium Campylobacter concisus, including biofilm formation and biofilm related genes. She has published several peer-reviewed research papers and a book chapter on the detection and virulence of this bacterium. She is currently an Assistant Professor at Qassim University, in Saudi Arabia. |
Microbiology Australia 44(2) 100-103 https://doi.org/10.1071/MA23028
Submitted: 14 April 2023 Accepted: 8 May 2023 Published: 24 May 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Campylobacter concisus, a member of the human’s oral microflora, is a Gram-negative, fastidious, microaerophilic bacterium. However, it is debatable whether it should be recognised as a commensal of the human oral cavity, or an opportunistic pathogen as it has been linked to oral and gastrointestinal infections. But there is no doubt that its biofilm-forming capacity has enhanced its survival mechanism whether as a commensal or a pathogen. Hence, through our investigation to assess C. concisus biofilms, we believe that its survival strategy in the oral cavity is enhanced by being protected in the biofilm environment with other oral microbes. Our hypothesis is supported by the findings that oral isolates of this bacterium possess a significantly higher biofilm forming capability than those isolated from the gastrointestinal tract.
Keywords: biofilm formation, Campylobacter concisus, extracellular polymeric substances, hydrogen-requiring campylobacters, luxS gene.
Association of Campylobacter concisus with the gastrointestinal tract
Campylobacter concisus is a Gram-negative, microaerophilic and hydrogen-requiring fastidious bacterium, which is a motile, spiral or curved-shaped rod.1 C. concisus was first recognised and named as a member of the microflora of the human oral cavity in the early 1980s by Tanner et al.2 Since then, it has been isolated from gingivitis, periodontitis, foot ulcers, gastritis, and from intestinal biopsies of patients with inflammatory bowel disease (IBD).3 It is likely that the human gastrointestinal tract is the sole reservoir for C. concisus, as it has no known primary animal host. But it is debatable whether it should be recognised as commensal of the human oral cavity or an opportunistic pathogen. Macuch and Tanner4 suggested that this bacterium colonises the oral cavity and is an opportunistic oral pathogen under certain medical conditions. The detection rate of C. concisus in permanent teeth was significantly higher than that of indeciduous teeth (P < 0.001).5,6 The prevalence of C. concisus in the healthy human oral cavity was also confirmed by PCR7,8 and by cultivation and culture-independent molecular methods such as 16S rRNA sequencing.9 The association of C. concisus with human periodontal diseases was well known since 1981,2,6,10 as antibody levels against C. concisus were found to be higher in periodontally diseased subjects compared to the healthy controls.11 Later on, it was reported to be associated with gingivitis and periodontal sites12 and was isolated from enlarged lesions of gingivitis.13 The association of C. concisus with periodontitis was also supported by significantly higher isolation rates when gingival crevicular fluid of patients was positive for aspartate aminotransferase (AST) compared to patients negative for AST in gingival crevicular fluid.14
Biofilm formation by Campylobacter spp.
Different Campylobacter spp., including C. jejuni and C. concisus are known to form biofilms,15–17 which are microbial communities enveloped within an extracellular polymeric matrix that provides protection from physical, environmental and biological stresses, to persist in a diverse range of ecological niches.18 In 2009, 14 Campylobacter species, both microaerophilic and hydrogen-requiring microaerophilic were tested on different surfaces such as glass, stainless steel, and polystyrene plastic for biofilm-forming ability. Of the eight microaerophilic Campylobacter species tested, C. jejuni, C. coli, C. lari, C. upsaliensis, C. sputorum, C. hyointestinalis, C. helveticus and C. fetus, only C. jejuni strain 81–176 reliably produced a visible biofilm on multiple surfaces. However, all six strains of the hydrogen-requiring microaerophilic Campylobacter species including C. rectus, C. showae, C. mucosalis, C. concisus, C. curvus and C. gracilis reliably produced visible biofilms on multiple surfaces.17 Furthermore, Sampathkumar et al.19 studied the transcriptional and translational expression profiles of C. jejuni 11168 biofilms. The proteomic analysis showed higher levels of expression of proteins involved in the motility complex, including the flagellins (FlaA, FlaB), the filament cap (FliD), the basal body (FlgG, FlgG2), and the chemotactic protein (CheA). Flagella and quorum-sensing (QS) are known to play important roles in biofilm formation. The inactivation of flaAB (flagella subunits) and luxS (responsible for QS) resulted in reduced biofilm formation. Hence, it was suggested that both flab and luxS are required for biofilm formation in C. jejuni.16 Since then, many researchers investigated the role of luxS and demonstrated that this gene is involved in a variety of physiologic pathways, motility, autoagglutination, cytolethal distending toxin (CDT) expression, flagellar expression, oxidative stress, and animal colonisation in C. jejuni.16,20–22
Biofilm formation by hydrogen-requiring campylobacters
In following studies, C. concisus and C. rectus were detected in four of seven patients (57%) with Barrett’s oesophagus (BO)23 when micro-colonies were detected in the form of mucosal biofilm in biopsy samples taken from those patients. This study was supported by the research conducted by Blackett et al.24, on oesophageal biofilms in patients with BO and gastro-oesophageal reflux disease (GORD), where C. concisus was the dominant species, therefore the authors suggested the emergence of C. concisus as the dominant species in the refluxed oesophagus. Moreover, C. concisus strains isolated from IBD patients, gastroenteritis and healthy individuals were reported to be capable of forming biofilms on glass coverslips.25 C. concisus has recently been reported to have been isolated from subgingival microbiota of individuals with HIV26 and from biofilms of the dental plaque.27 Hence, biofilm formation by this bacterium being a fastidious oral cavity coloniser, is likely to be an important requirement for it to survive in this environment. However, to date, there has been no thorough investigation on C. concisus biofilm formation.
Investigation of C. concisus biofilms formed by oral and clinical strains
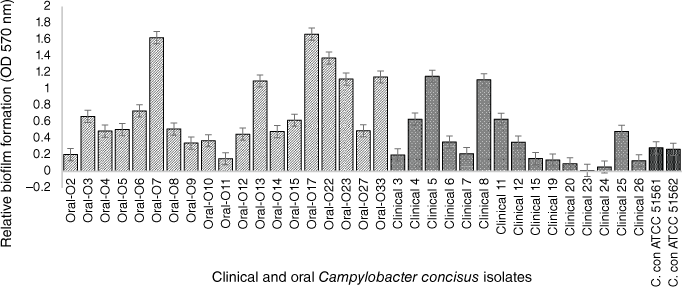
In the past few years, our research team focussed on the evaluation and characterisation of C. concisus biofilms. To assess the biofilm forming capability of strains isolated from different gastrointestinal tract sections, we tested 19 oral isolates collected from saliva samples of healthy volunteers at RMIT University, Melbourne, in addition to 14 intestinal clinical isolates previously collected using the Cape Town Protocol28 from children suffering from mild to severe bloody diarrhoea at the Royal Children’s Hospital, Melbourne.29 Two C. concisus reference strains (ATCC 51561 and ATCC 51562) were also characterised in our research. All intestinal and oral C. concisus isolates were screened for biofilm formation by the modified quantitative crystal violet assay.16 Among the 14 intestinal isolates, only 11 produced biofilms at low to moderate levels, whereas all tested oral isolates formed higher levels of biofilms (Fig. 1), indicating that the oral isolates are significantly more prolific biofilm producers than the clinical isolates (P < 0.05) including the two reference strains that also have a faecal origin. A possible explanation for forming more biofilm by the oral isolates could be an advantageous trait within the oral cavity to escape from toxic oxygen and other adverse conditions. In other studies, C. jejuni NCTC 11168 has been shown to develop biofilm more rapidly under environmental and food-chain-relevant aerobic conditions (20% O2) than under microaerobic conditions (5% O2, 10% CO2).30
Crystal violet, biofim quantitative assay of 34 C. concisus strains. The oral strains were isolated from saliva of healthy persons and the intestinal strains were isolated from patients with gastritis. The oral isolates exhibited significant biofilm forming potential than clinical isolates (by Mann–Whitney test; P = 0.0354). C. concisus ATCC 51561 and 51562 were used as control strains. The results represent the mean values and standard errors of three independent experiments. Each experiment was performed three times independently and in biological–technical triplicates.
Microscopic observation of C. concisus biofilms
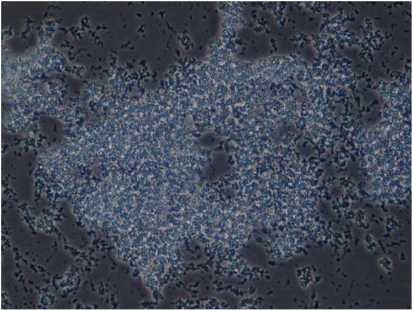
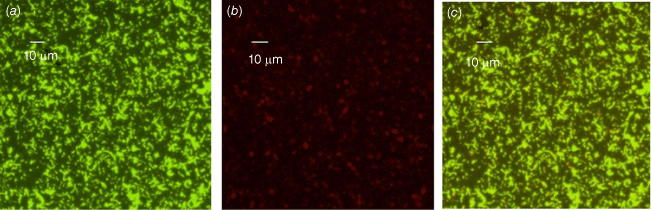
We also conducted phenotypic characterisation of biofilms formed by selected oral and intestinal C. concisus strains, which included phase contrast microscopy, and confocal laser scanning microscopy (CLSM). Different morphological stages of biofilm formation (attachment, maturation and dispersion) were observed by phase contrast microscopy. Cell clusters were observed to undergo alterations in their structure due to the dispersion of bacterial cells from their interior within 96 h (Fig. 2). These bacterial cells were observed to swim away from the inner portions of the cell cluster. The ability of bacteria to swim freely within the void spaces as observed by microscopy indicated the absence of dense polymers or other gel-like material in the void space. Completely developed biofilms were observed by CLSM after 96 h, with a mixture of dead and live bacteria within the biofilm’s structure (Fig. 3).
Biofilm complex produced by a C. concisus oral isolate observed at 400× magnification by phase contrast microscopy showing the bacterial cells embedded within an extracellular material and cells evacuated interior portions of cell clusters, forming void spaces.
Biofilms of a C. concisus oral isolate on glass coverslip observed by confocal laser scanning microscopy (CLSM) after 96 h of incubation showing a mixture of live and dead cells (a) stained with SYTO-9 (bright green for live cells), (b) stained with PI (red or orange, or loss of bright green for dead cells), (c) combined image of SYTO-9 and PI.
In conclusion, we have shown that the ability of C. concisus to form biofilms is correlated with the source where it was collected from. Oral isolates comparatively produced higher biofilm levels in vitro than gut isolates, as only a few intestinal isolates could form good biofilms. This could be considered as a survival mechanism for the oral colonising bacterium in its normal habitat. Furthermore, C. concisus biofilms were observed to be complex matrices composed of live and dead cells embedded in an extracellular polymeric substance (EPS). Hence, expression of biofilm related genes in these oral and clinical isolates has currently been undertaken by our research team. Further studies to evaluate the environmental effects on biofilm formation and the role of chemotaxis related genes in this mechanism are also in progress.
Data availability
Data related to this research topic are available from Dr Mohsina Huq’s PhD thesis, ‘Molecular characterization of biofilm production and whole genome sequencing of selected Campylobacter concisus oral and clinical strains’, which is available through RMIT’s Open Access Research Repository).31
References
[1] Garrity GM et al. (2005) Genus I. Campylobacter. In Bergey’s Manual of Systematic Bacteriology, 2nd edn. Springer, New York, NY, USA.[2] Tanner, ACR et al. (1981) Wolinella gen. nov., Wolinella succinogenes (Vibrio succinogenes Wolin et al.) comb. nov., and description of Bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp. nov., and Eikenella corrodens from humans with periodontal disease. Int J Syst Bacteriol 31, 432–45.
| Wolinella gen. nov., Wolinella succinogenes (Vibrio succinogenes Wolin et al.) comb. nov., and description of Bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp. nov., and Eikenella corrodens from humans with periodontal disease.Crossref | GoogleScholarGoogle Scholar |
[3] Kaakoush, NO et al. (2015) Global epidemiology of Campylobacter infection. Clin Microbiol Rev 28, 687–720.
| Global epidemiology of Campylobacter infection.Crossref | GoogleScholarGoogle Scholar |
[4] Macuch, PJ and Tanner, ACR (2000) Campylobacter species in health, gingivitis, and periodontitis. J Dent Res 79, 785–92.
| Campylobacter species in health, gingivitis, and periodontitis.Crossref | GoogleScholarGoogle Scholar |
[5] Kamma, JJ et al. (2000) Profile of subgingival microbiota in children with primary dentition. J Periodontal Res 35, 33–41.
| Profile of subgingival microbiota in children with primary dentition.Crossref | GoogleScholarGoogle Scholar |
[6] Kamma, JJ et al. (2000) Profile of subgingival microbiota in children with mixed dentition. Oral Microbiol Immunol 15, 103–11.
| Profile of subgingival microbiota in children with mixed dentition.Crossref | GoogleScholarGoogle Scholar |
[7] Petersen, RF et al. (2007) A PCR-DGGE method for detection and identification of Campylobacter, Helicobacter, Arcobacter and related epsilobacteria and its application to saliva samples from humans and domestic pets. J Appl Microbiol 103, 2601–15.
| A PCR-DGGE method for detection and identification of Campylobacter, Helicobacter, Arcobacter and related epsilobacteria and its application to saliva samples from humans and domestic pets.Crossref | GoogleScholarGoogle Scholar |
[8] Zhang, L et al. (2010) Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J Clin Microbiol 48, 2965–7.
| Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease.Crossref | GoogleScholarGoogle Scholar |
[9] Dewhirst, FE et al. (2010) The human oral microbiome. J Bacteriol 192, 5002–17.
| The human oral microbiome.Crossref | GoogleScholarGoogle Scholar |
[10] Tanner, ACR et al. (1987) Wolinella recta, Campylobacter concisus, Bacteroides gracilis, and Eikeneila corrodens from periodontal lesions. J Periodontal Res 22, 327–30.
| Wolinella recta, Campylobacter concisus, Bacteroides gracilis, and Eikeneila corrodens from periodontal lesions.Crossref | GoogleScholarGoogle Scholar |
[11] Taubman, MA et al. (1992) Longitudinal monitoring of humoral antibody in subjects with destructive periodontal diseases. J Periodontal Res 27, 511–21.
| Longitudinal monitoring of humoral antibody in subjects with destructive periodontal diseases.Crossref | GoogleScholarGoogle Scholar |
[12] Tanner, A et al. (1998) Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol 25, 85–98.
| Microbiota of health, gingivitis, and initial periodontitis.Crossref | GoogleScholarGoogle Scholar |
[13] Kamma, JJ et al. (1998) Subgingival microflora and treatment in prepubertal periodontitis associated with chronic idiopathic neutropenia. J Clin Periodontol 25, 759–65.
| Subgingival microflora and treatment in prepubertal periodontitis associated with chronic idiopathic neutropenia.Crossref | GoogleScholarGoogle Scholar |
[14] Kamma, JJ et al. (2001) Association of early onset periodontitis microbiota with aspartate aminotransferase activity in gingival crevicular fluid. J Clin Periodontol 28, 1096–105.
| Association of early onset periodontitis microbiota with aspartate aminotransferase activity in gingival crevicular fluid.Crossref | GoogleScholarGoogle Scholar |
[15] Joshua, GWP et al. (2006) Biofilm formation in Campylobacter jejuni. Microbiology 152, 387–96.
| Biofilm formation in Campylobacter jejuni.Crossref | GoogleScholarGoogle Scholar |
[16] Reeser, RJ et al. (2007) Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl Environ Microbiol 73, 1908–13.
| Characterization of Campylobacter jejuni biofilms under defined growth conditions.Crossref | GoogleScholarGoogle Scholar |
[17] Gunther IV, NW and Chen, C-Y (2009) The biofilm forming potential of bacterial species in the genus Campylobacter. Food Microbiol 26, 44–51.
| The biofilm forming potential of bacterial species in the genus Campylobacter.Crossref | GoogleScholarGoogle Scholar |
[18] Costerton, JW et al. (1999) Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–22.
| Bacterial biofilms: a common cause of persistent infections.Crossref | GoogleScholarGoogle Scholar |
[19] Sampathkumar, B et al. (2006) Transcriptional and translational expression patterns associated with immobilized growth of Campylobacter jejuni. Microbiology 152, 567–77.
| Transcriptional and translational expression patterns associated with immobilized growth of Campylobacter jejuni.Crossref | GoogleScholarGoogle Scholar |
[20] Jeon, B et al. (2005) Promoter analysis of cytolethal distending toxin genes (CdtA, B, and C) and effect of a luxS mutation on CDT production in Campylobacter jejuni. Microbiol Immunol 49, 599–603.
| Promoter analysis of cytolethal distending toxin genes (CdtA, B, and C) and effect of a luxS mutation on CDT production in Campylobacter jejuni.Crossref | GoogleScholarGoogle Scholar |
[21] Quiñones, B et al. (2009) Autoinducer-2 production in Campylobacter jejuni contributes to chicken colonization. Appl Environ Microbiol 75, 281–5.
| Autoinducer-2 production in Campylobacter jejuni contributes to chicken colonization.Crossref | GoogleScholarGoogle Scholar |
[22] Plummer, P et al. (2011) Identification of a key amino acid of LuxS involved in AI-2 production in Campylobacter jejuni. PLoS One 6, e15876.
| Identification of a key amino acid of LuxS involved in AI-2 production in Campylobacter jejuni.Crossref | GoogleScholarGoogle Scholar |
[23] Macfarlane, S et al. (2007) Microbial colonization of the upper gastrointestinal tract in patients with Barrett’s esophagus. Clin Infect Dis 45, 29–38.
| Microbial colonization of the upper gastrointestinal tract in patients with Barrett’s esophagus.Crossref | GoogleScholarGoogle Scholar |
[24] Blackett, KL et al. (2013) Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther 37, 1084–92.
| Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: association or causality?Crossref | GoogleScholarGoogle Scholar |
[25] Lavrencic, P et al. (2012) Investigation of motility and biofilm formation by intestinal Campylobacter concisus strains. Gut Pathog 4, 22.
| Investigation of motility and biofilm formation by intestinal Campylobacter concisus strains.Crossref | GoogleScholarGoogle Scholar |
[26] Pereira, VT et al. (2014) The association between detectable plasmatic human immunodeficiency virus (HIV) viral load and different subgingival microorganisms in Brazilian adults with HIV: a multilevel analysis. J Periodontol 85, 697–705.
| The association between detectable plasmatic human immunodeficiency virus (HIV) viral load and different subgingival microorganisms in Brazilian adults with HIV: a multilevel analysis.Crossref | GoogleScholarGoogle Scholar |
[27] Marchesan, J et al. (2015) TLR4, NOD1 and NOD2 mediate immune recognition of putative newly identified periodontal pathogens. Mol Oral Microbiol 31, 243–58.
| TLR4, NOD1 and NOD2 mediate immune recognition of putative newly identified periodontal pathogens.Crossref | GoogleScholarGoogle Scholar |
[28] Le Roux E, Lastovica AJ (1998) The Cape Town Protocol: how to isolate the most campylobacters for your dollar, pound, franc, yen, etc. In International Workshop on Campylobacter, Helicobacter and Related Organisms (Lastovica AJ, et al., eds). pp. 30–3. Institute of Child Heath, Cape Town, South Africa.
[29] Russell J (1995) Campylobacter like organisms: investigation of clinical and phenotypical aspects. MSc thesis, RMIT University, Melbourne, Vic., Australia.
[30] Reuter, M et al. (2010) Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl Environ Microbiol 76, 2122–8.
| Biofilm formation by Campylobacter jejuni is increased under aerobic conditions.Crossref | GoogleScholarGoogle Scholar |
[31] Huq M (2016) Molecular characterization of biofilm production and whole genome sequencing of selected Campylobacter concisus oral and clinical strains. PhD thesis, RMIT University, Melbourne, Vic., Australia. https://researchrepository.rmit.edu.au/esploro/outputs/doctoral/Molecular-%20through%20characterization-of-biofilm-production-and/9921863647601341


