Brucella: not your ‘typical’ intracellular pathogen
Anthony L Keyburn A and Nicky Buller BA CSIRO Australian Animal Health Laboratory (AAHL), Private Bag 24, Geelong, Vic. 3220, Australia. Tel: +61 3 5227 5000, Email: anthony.keyburn@csiro.au
B Department of Primary Industries and Regional Development (DPIRD), Diagnostic and Laboratory Services (DDLS), 3 Baron-Hay Court, South Perth, WA 6151, Australia
Microbiology Australia 41(1) 38-41 https://doi.org/10.1071/MA20010
Published: 28 February 2020
Currently the genus Brucella consists of a group of bacteria that are genetically monospecific yet phenotypically diverse, and a recent genetic and phenotypic divergent group known as ‘atypical’ Brucellae. The host range is extremely varied and includes mammals, including humans, terrestrial animals and marine mammals, but now extends to reptiles and amphibians. Almost all Brucella species are zoonotic. The disease collectively termed Brucellosis leads to abortion and reproductive disease in animals, whereas human infection presents as a non-specific undulating fever accompanied by general malaise, chills, joint pain, muscle aches, genitourinary disease and adverse pregnancy outcomes. These Gram-negative coccobacilli invade and replicate in the host macrophages where they can limit the effects of the host immune system and antibiotic treatment. Due to the phenotypic and genotypic diversity and close relationship with Ochrobactrum species, the genus Brucella presents challenges for accurate identification and recognition of new species.
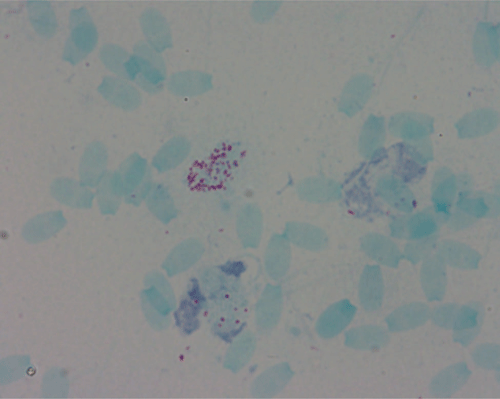
The disease, Brucellosis, affects animals and humans, causing abortions and reproductive disease in animals, and is known as undulating fever, Malta fever or Mediterranean fever in humans1. The causative agent, Brucella, are facultative intracellular, small Gram-negative coccobacilli (Figure 1) 0.5–0.7 µm × 0.5–1.5 µm that survive in the phagocytic cells of the infected host and were first identified in 18872. Transmission occurs through direct contact with infected material via mucus membranes, broken skin, ingestion or inhalation3. Animals tend to recover but can continue to shed the bacterium into the environment, which makes control of the disease difficult1. Brucellosis occurs in most parts of the world, although the disease is rare in Australia with cases usually attributed to international travel in endemic countries or from contact with infected feral pigs particularly in the Eastern states of NSW and QLD. In these areas, hunting of feral pigs has been identified as the principal risk factor for human and dog brucellosis and can result in zoonotic transmission to veterinarians and household contacts4.
|
Brucella species tend to be host specific but can infect other hosts although the disease is usually self-limiting in a non-primary host. Currently, there are 12 species of Brucella and can be divided into classical Brucellae (Brucella melitensis, B. abortus, B. suis, B. canis, B. ovis and B. neotomae), marine mammal (B. ceti and B. pinnipedialis) and recently identified species (B. inopinata, B. microti, B. paponis and B. vulpis). There are also additional strains, awaiting genus affiliation, isolated from human and animal sources. A selection of recent species exhibits different phenotypic traits and greater genetic diversity than those in the classical group and are designated ‘atypical’ Brucellae (B. microti, B. inopinata and B. vulpis).
Most Brucella species pose a significant zoonotic threat to humans most notably B. melitensis, B. suis, B. abortus and B. canis. Other Brucella species including those that affect marine mammals are also zoonotic5. To date only B. ovis, which causes reproductive failure and abortion in sheep, is not zoonotic6,7.
The taxonomy of Brucella presents challenges. Traditionally, new species were named according to the host from which they were isolated, and biovars were assigned to reflect the diverse range of phenotypes within some species. Classical Brucella species are 90% homologous, which means that the genus is monospecific according to the designation of species as having greater than 70% homology using DNA-DNA hybridisation. In this case B. melitensis is the only species, and the rest are biovars8. However, a consensus held by the scientific community has seen the traditional species names retained irrespective of the traditional taxonomic conventions9. Interestingly, application of molecular typing methods such as pulsed field gel electrophoresis, infrequent restriction site polymerase chain reaction, restriction fragment length polymorphisms, insertion sequence site testing, multilocus sequence analysis, variable number tandem repeat analysis and genome sequencing show clustering of genotypes that supports the classical designation of species and biovars10–13.
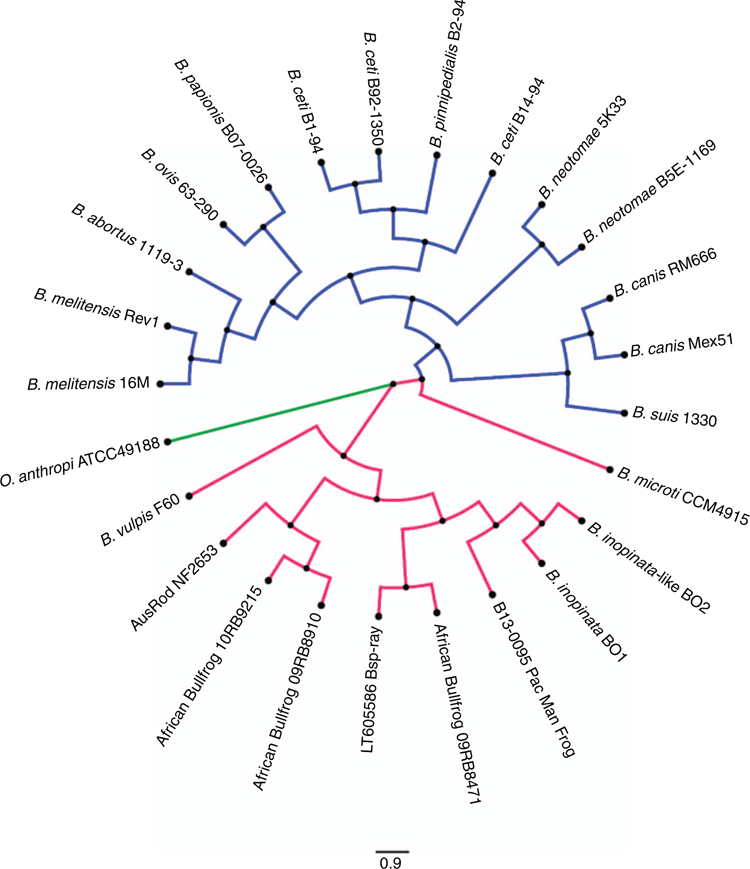
Greater genetic diversity exists among the atypical Brucella clade than in the classical clade (Figure 2). Atypical Brucellae diversity has been attributed to the ability of these basal species to exchange DNA with each other and with other microbes in the environment using horizontal gene transfer15. The atypical group includes designated species (as mentioned earlier) as well as candidate strains like BO2 from a human16, LT605586 from a bluespotted ribbontail ray17 and NF2653 from a rodent18. Recently, atypical Brucella species have been isolated from amphibians15,19–22. Some Brucella isolates obtained from amphibians are most closely related to B. inopinata BO1 and Brucella‐like BO2 strains based on whole genome analysis. Given their similarity to BO isolates, for which the animal reservoir has not yet been identified, amphibians might represent a possible source of these strains. Recently, Brucella from domestic marsh frogs in France were found to be more similar B. microti and not B. inopinata23.
|
Isolation of frog Brucella sp. from Africa, Europe, Australia and America suggests that they may be widespread and highlight a need for a broader assessment of the presence of Brucella in amphibians worldwide. A major aspect of Brucella virulence is their capacity to replicate inside macrophages and escape the host immune system. Recently, in vitro and in vivo infection experiments with amphibian Brucella isolates found that isolates were able to invade and even multiply intracellularly in macrophages and survive in the murine host for up to 12 weeks15. Given the lack of definitive evidence and their proximity with strains associated with human disease, isolates from amphibians should be considered as potential zoonotic pathogens.
Diagnostics
Accurate identification of pathogens is essential for establishing dependable diagnosis, choosing a treatment, and understanding the source of infection. Conventional identification of Brucella is based on modified acid fast staining and phenotypic methods including phage typing and serology that differentiate the species and biotypes24,25. These tests are usually done in a specialist laboratory because of the types of tests conducted and the biohazard of working with the organism. B. melitensis can be identified using matrix-assisted laser desorption ionisation time of flight (MALDI-TOF) mass spectroscopy; however, further differentiation of species is not available using this technology without additional curation of the reference database26. It is important to differentiate Brucella and Ochrobactrum species (close genetic relative within the family Brucellaceae, a genus largely consisting of environmental bacteria occasionally infecting humans) as Brucella has a higher biosafety risk to hospital and laboratory staff and can have different treatment strategies. Due to their similarity, some routine commercial identification systems can mis-identify the two organisms as B. melitensis and O. intermedium have a 98.8% similarity according to the rRNA gene sequence27. Atypical Brucellae further complicate laboratory identification as most members, including amphibian isolates, are motile15,22. Amphibian isolates also exhibit variant lipopolysaccharides (LPS), phage lysis, serum agglutination and dye sensitivities compared to classical Brucella and are often misidentified as Ochrobactrum28,29. Although, when the Brucella reference database is available, MALDI-TOF assays can correctly identify them as Brucella. The differences in LPS of atypical and classical Brucella could result in serological diagnostics being impaired especially for human infections.
Until recently the genus Brucella was considered to represent a genetically homogeneous group of bacteria associated with mammalian hosts. Recently, the situation has become more complex with a rapid increase in the number of novel, genetically divergent, Brucella being isolated from cold-blooded hosts. The zoonotic potential and pathogenicity of these Brucella sp. strains remains unknown. Further studies are required to gain insights on the bacterial carriage and characterisation of these isolates to understand their role in the evolution of the species from being soil bacteria that are characterised by motility and a broad metabolic activity to becoming highly virulent but host-specific clonal pathogens.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This research did not receive any specific funding.
References
[1] Corbel, M.J. et al. (2006) Brucellosis in humans and animals. World Health Organization: Geneva.[2] Bruce, D. (1887) Note on the discovery of a micro-organism in Malta fever. Practitioner 39, 161–170.
[3] Nymo, I.H. et al. (2011) A review of Brucella infection in marine mammals, with special emphasis on Brucella pinnipedialis in the hooded seal (Cystophora cristata). Vet. Res. 42, 93.
| A review of Brucella infection in marine mammals, with special emphasis on Brucella pinnipedialis in the hooded seal (Cystophora cristata).Crossref | GoogleScholarGoogle Scholar | 21819589PubMed |
[4] Mor, S.M. et al. (2016) Emergence of Brucella suis in dogs in New South Wales, Australia: clinical findings and implications for zoonotic transmission. BMC Vet. Res. 12, 199.
| Emergence of Brucella suis in dogs in New South Wales, Australia: clinical findings and implications for zoonotic transmission.Crossref | GoogleScholarGoogle Scholar | 27613248PubMed |
[5] Brew, S.D. et al. (1999) Human exposure to Brucella recovered from a sea mammal. Vet. Rec. 144, 483.
| 10358880PubMed |
[6] Buddle, M.B. (1956) Studies on Brucella ovis (n.sp.), a cause of genital disease of sheep in New Zealand and Australia. J. Hyg. (Lond.) 54, 351–364.
| Studies on Brucella ovis (n.sp.), a cause of genital disease of sheep in New Zealand and Australia.Crossref | GoogleScholarGoogle Scholar | 13367402PubMed |
[7] (2018) Ovine epididymitis (Brucella ovis), in OIE.
[8] Verger, J.M. et al. (1985) Brucella, a monospecific genus as shown by deoxyribonucleic-acid hybridization. Int. J. Syst. Bacteriol. 35, 292–295.
| Brucella, a monospecific genus as shown by deoxyribonucleic-acid hybridization.Crossref | GoogleScholarGoogle Scholar |
[9] Moreno, E. et al. (2002) Brucella evolution and taxonomy. Vet. Microbiol. 90, 209–227.
| Brucella evolution and taxonomy.Crossref | GoogleScholarGoogle Scholar | 12414145PubMed |
[10] Foster, J.T. et al. (2009) Whole-genome-based phylogeny and divergence of the genus Brucella. J. Bacteriol. 191, 2864–2870.
| Whole-genome-based phylogeny and divergence of the genus Brucella.Crossref | GoogleScholarGoogle Scholar | 19201792PubMed |
[11] Le Flèche, P. et al. (2006) Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 6, 9.
| Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay.Crossref | GoogleScholarGoogle Scholar | 16469109PubMed |
[12] Whatmore, A.M. (2009) Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol. 9, 1168–1184.
| Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens.Crossref | GoogleScholarGoogle Scholar | 19628055PubMed |
[13] Whatmore, A.M. et al. (2007) Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 7, 34.
| Characterisation of the genetic diversity of Brucella by multilocus sequencing.Crossref | GoogleScholarGoogle Scholar | 17448232PubMed |
[14] Gardner, S.N. et al. (2015) kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878.
| kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome.Crossref | GoogleScholarGoogle Scholar | 25913206PubMed |
[15] Al Dahouk, S. et al. (2017) Brucella spp. of amphibians comprise genomically diverse motile strains competent for replication in macrophages and survival in mammalian hosts. Sci. Rep. 7, 44420.
| Brucella spp. of amphibians comprise genomically diverse motile strains competent for replication in macrophages and survival in mammalian hosts.Crossref | GoogleScholarGoogle Scholar | 28300153PubMed |
[16] Tiller, R.V. et al. (2010) Identification of an unusual Brucella strain (BO2) from a lung biopsy in a 52 year-old patient with chronic destructive pneumonia. BMC Microbiol. 10, 23.
| Identification of an unusual Brucella strain (BO2) from a lung biopsy in a 52 year-old patient with chronic destructive pneumonia.Crossref | GoogleScholarGoogle Scholar | 20105296PubMed |
[17] Eisenberg, T. et al. (2017) Isolation of a novel ‘atypical’ Brucella strain from a bluespotted ribbontail ray (Taeniura lymma). Antonie van Leeuwenhoek 110, 221–234.
| Isolation of a novel ‘atypical’ Brucella strain from a bluespotted ribbontail ray (Taeniura lymma).Crossref | GoogleScholarGoogle Scholar | 27785661PubMed |
[18] Tiller, R.V. et al. (2010) Characterization of novel Brucella strains originating from wild native rodent species in North Queensland, Australia. Appl. Environ. Microbiol. 76, 5837–5845.
| Characterization of novel Brucella strains originating from wild native rodent species in North Queensland, Australia.Crossref | GoogleScholarGoogle Scholar | 20639360PubMed |
[19] Eisenberg, T. et al. (2012) Isolation of potentially novel Brucella spp. from frogs. Appl. Environ. Microbiol. 78, 3753–3755.
| Isolation of potentially novel Brucella spp. from frogs.Crossref | GoogleScholarGoogle Scholar | 22407680PubMed |
[20] Fischer, D. et al. (2012) Abscesses associated with a Brucellainopinata-like bacterium in a big-eyed tree frog (Leptopelis vermiculatus). J. Zoo Wildl. Med. 43, 625–628.
| Abscesses associated with a Brucellainopinata-like bacterium in a big-eyed tree frog (Leptopelis vermiculatus).Crossref | GoogleScholarGoogle Scholar | 23082529PubMed |
[21] Kimura, M. et al. (2017) Isolation of Brucella inopinata-like bacteria from White’s and Denny’s tree frogs. Vector Borne Zoonotic Dis. 17, 297–302.
| Isolation of Brucella inopinata-like bacteria from White’s and Denny’s tree frogs.Crossref | GoogleScholarGoogle Scholar | 28437181PubMed |
[22] Soler-Lloréns, P.F. et al. (2016) A Brucella spp. isolate from a pac-man frog (Ceratophrys ornata) reveals characteristics departing from classical Brucellae. Front. Cell. Infect. Microbiol. 6, 116.
| A Brucella spp. isolate from a pac-man frog (Ceratophrys ornata) reveals characteristics departing from classical Brucellae.Crossref | GoogleScholarGoogle Scholar | 27734009PubMed |
[23] Jaý, M. et al. (2018) Phenotypic and molecular characterization of Brucellamicroti-like bacteria from a domestic marsh frog (Pelophylax ridibundus). Front. Vet. Sci. 5, 283.
| Phenotypic and molecular characterization of Brucellamicroti-like bacteria from a domestic marsh frog (Pelophylax ridibundus).Crossref | GoogleScholarGoogle Scholar | 30498697PubMed |
[24] (2018) Brucellosis (Brucella abortus, B. melitensis and B. suis) (infection with B. abortus, B. melitensis and B. suis), in OIE.
[25] Alton, G.G. and Jones, L.M. (1967) Laboratory techniques in brucellosis. Monogr. Ser. World Health Organ. 55, 1–92.
| 4963092PubMed |
[26] Mesureur, J. et al. (2018) A MALDI-TOF MS database with broad genus coverage for species-level identification of Brucella. PLoS Negl. Trop. Dis. 12, e0006874.
| A MALDI-TOF MS database with broad genus coverage for species-level identification of Brucella.Crossref | GoogleScholarGoogle Scholar | 30335748PubMed |
[27] Velasco, J. et al. (1998) Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int. J. Syst. Bacteriol. 48, 759–768.
| Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp.Crossref | GoogleScholarGoogle Scholar | 9734029PubMed |
[28] Zygmunt, M.S. et al. (2012) Lipopolysaccharide heterogeneity in the atypical group of novel emerging Brucella species. Clin. Vaccine Immunol. 19, 1370–1373.
| Lipopolysaccharide heterogeneity in the atypical group of novel emerging Brucella species.Crossref | GoogleScholarGoogle Scholar | 22761298PubMed |
[29] Scholz, H.C. et al. (2016) The change of a medically important genus: worldwide occurrence of genetically diverse novel Brucella species in exotic frogs. PLoS One 11, e0168872.
| The change of a medically important genus: worldwide occurrence of genetically diverse novel Brucella species in exotic frogs.Crossref | GoogleScholarGoogle Scholar | 28036367PubMed |
Biographies
Dr Anthony Keyburn is Senior Scientist at the AAHL. His research interests are on brucellosis and veterinary tuberculosis pathogenesis and diagnostic and vaccine development.
Dr Nicky Buller is a Senior Microbiologist at DPIRD. Her interests include identification of aquatic and terrestrial animal bacteria, and is the author of Bacteria and Fungi from Fish and other Aquatic Animals: a Practical Identification Manual. CABI, UK, 2014.


