Public health impacts of culture independent diagnostic testing in Australia
Fiona J MayMetro North Public Health Unit
Queensland Health
Windsor, Qld, Australia
Tel: +61 7 3624 1212
Email: Fiona.May@health.qld.gov.au
Microbiology Australia 38(4) 162-164 https://doi.org/10.1071/MA17058
Published: 9 November 2017
Culture independent diagnostic tests (CIDT) for detection of pathogens in clinical specimens have become widely adopted in Australian pathology laboratories. Pathology laboratories are the primary source of notification of pathogens to state and territory surveillance systems. Monitoring and analysis of surveillance data is integral to guiding public health actions to reduce the incidence of disease and respond to outbreaks. As with any change in testing protocol, the advantages and disadvantages of the change from culture based testing to culture independent testing need to be weighed up and the impact on surveillance and outbreak detection assessed. This article discusses the effect of this change in testing on surveillance and public health management of pathogens in Australia, with specific focus on gastrointestinal pathogens.
What is CIDT and how is it being used in Australia?
A CIDT is any diagnostic test that is performed directly on the clinical specimen without first requiring laboratory culture of a pathogen isolate1,2. Common CIDT methods diagnose an infectious agent by detecting the nucleic acids of the pathogen (e.g. polymerase chain reaction [PCR]) or the pathogen antigen (e.g. enzyme immunoassays). Commercial and in-house CIDTs have been developed for a range of pathogens.
Many pathology laboratories across Australia have introduced PCR panels for identification of gastrointestinal pathogens (bacteria, parasites and viruses) in faecal specimens (John Bates for the Public Health Laboratory Network, personal communication). Most of these laboratories are continuing to perform culture of specimens, either concurrent with PCR or reflexively only on specimens that test positive by PCR. As with any change in testing protocol, the addition of PCR for pathogen diagnosis bears advantages and disadvantages, both for the diagnostic laboratory and for end users of laboratory testing data, such as public health professionals and clinicians. As PCRs for gastrointestinal pathogens are the primary CIDT in use in Australian pathology laboratories, and gastrointestinal disease causes a significant burden of illness3, these will provide the focus for examples used in this article. However, PCR is becoming common for the diagnosis of a range of pathogens and most elements of this discussion are applicable across all pathogens.
CIDT has revolutionised pathogen detection
Many pathology laboratories have embraced PCR due to the low ongoing cost, speed, and ease of use1,4–6. PCR requires less training and expertise, and is less subjective than culture, for which experience is required to identify appropriate colonies to select for further characterisation1,6. PCR is generally more sensitive than culture, and can detect pathogens that do not grow easily in culture, have been treated with antibiotics or are in low numbers in the specimen5,6. PCR is more likely to detect a pathogen in specimens with decreased viability due to a delay before testing7. PCR can also identify polymicrobial infections whereas culture would likely only detect the fastest growing pathogen5.
PCR also benefits public health. Faster identification of the causative agent in patients who would otherwise have gone undiagnosed due to a negative culture can result in earlier prevention of transmission. Outbreaks may be identified sooner1. The use of tests with a higher sensitivity, such as PCR, provides a more accurate estimate of the burden of disease, which can inform targeted response and control measures4.
CIDT must be interpreted with caution
One of the most important uses of surveillance data is for the detection of an increase in disease through comparison with historical data5,6. Therefore, any change in testing method will need to be reviewed in the context of previous results, to enable valid comparison of data from before and after the change in method. Since PCR is more sensitive than traditional culture methods, the detected incidence of pathogens will likely increase after introduction of PCR5,7. However, other factors can also cause an increase in the detected incidence. Due to the rapid turnaround of results for PCR, clinicians may submit specimens from patients where previously they would not (e.g. a case where mild illness is likely to resolve before results would available by culture). Pathology laboratories may advertise the introduction of a new technology, such as PCR, resulting in an increase in specimen submission. Finally, there may be a real increase in the incidence of disease due to another cause.
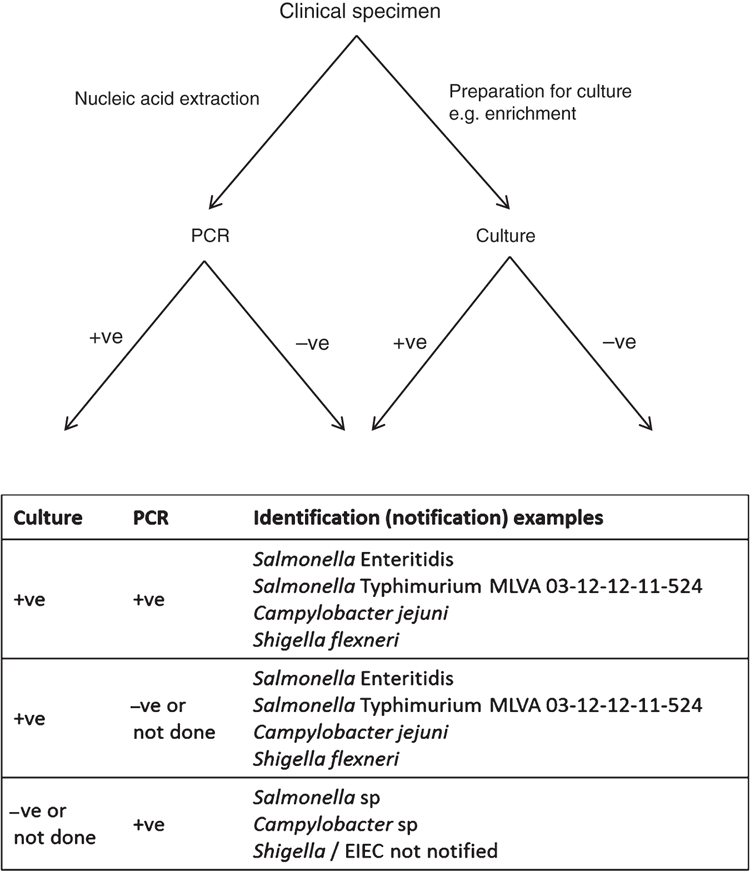
Current commercially available diagnostic PCR kits are generally unable to distinguish between dead and living cells5 or identify a pathogen beyond the genus level. Current methods for typing of pathogens, such as serotyping and genotyping, and assessment of antimicrobial susceptibility require a cultured isolate (Figure 1)2,5,6. For common pathogens, such as Salmonella, this makes it difficult to detect outbreaks of a particular genotype against background surveillance data6. For outbreaks of foodborne gastroenteritis, this limits detection of outbreaks of common pathogens to those identified via complaints from the public or notifications from an event, food business or facility. Community outbreaks where cases appear to be unrelated (such as those involving commercial foods not eaten on premises) will not be detected if relying on PCR diagnostics alone. Without information on the specific genotype of the pathogen, it is difficult to prove the linkage of cases to pathogen isolates obtained after traceback to the potential food source. This can affect the success of interventions or litigation1,6.
The impact of the introduction of PCR is different for each pathogen. For example, Salmonella is easy to grow in culture, so concurrent or reflex culture will result in an isolate for further characterisation most of the time. In contrast, as Campylobacter is fastidious8, culture has a much lower sensitivity than PCR, resulting in a higher proportion of specimens positive only by PCR even when culture is attempted.
If the specificity of the primers used in the PCR is not limited to the pathogen in question, the PCR may overestimate incidence of disease. For example, Shigella is genetically closely related to enteroinvasive Escherichia coli (EIEC)9 and the PCR primers used in commercial multiplex PCRs amplify the ipaH gene, which is common between Shigella and EIEC10,11. Thus, culture-based phenotypic tests are required to differentiate between the two genera. However, Shigella can be difficult to culture12, making it difficult to determine how many of the culture negative/PCR positive specimens are from true cases of shigellosis. While shigellosis is a notifiable disease in Australia, gastroenteritis due to EIEC is not, and the true incidence of EIEC and Shigella in Australia is unknown. However, studies from other developed countries show a higher incidence of Shigella than EIEC13,14 and a study from Victoria suggested that Shigella may be more common than EIEC in Australia15. In Queensland, the number of stools positive for Shigella/EIEC increased dramatically following the introduction of PCR16 and a similar increase is likely where PCR has been introduced into public health laboratories in other jurisdictions. Most jurisdictional public health guidelines require notification of only those Shigella positive stools with a confirmatory culture15. This minimises unnecessary follow up of cases of gastroenteritis caused by EIEC, but may result in loss of information about the true incidence and risk of Shigella transmission. Addition of primers that can distinguish between Shigella and EIEC to the gastroenteritis multiplex PCR is essential for accurate delivery of public health response.
Whole genome sequencing (WGS) of pathogens is currently under development for use in public health (see article by A. Jennison in this issue). WGS allows further characterisation of isolates and prediction of antimicrobial sensitivity which can be useful for public health and clinical treatment. However, the WGS techniques currently in use for high throughput testing of many clinical samples still require a pure culture so the introduction of PCR and subsequent reduced availability of cultures from clinical specimens in laboratories that have introduced CIDT will hinder the development of this technology.
Conclusion and future
Although it varies for each pathogen, the impact of the introduction of CIDT has both benefits and drawbacks for the laboratory, clinical diagnosis of disease and public health surveillance. It remains important that laboratories continue to perform concurrent or reflex culture in order to inform public health action, especially as WGS becomes more common. This period of extra testing is likely to be transitional, as techniques for sequencing directly from clinical specimens (metagenomics) are developed and neither culture nor PCR will be required for diagnosis of pathogens. In addition to bypassing the requirement for culture, metagenomics will allow detection of previously unknown pathogens via sequencing of all nucleic acid present in a specimen, expanding our knowledge of the agents causing infectious disease.
Conflicts of interest
The author declares no conflicts of interest.
Acknowledgements
Thank you to Daniel Francis and Cassie Jansen for discussion and comments on the manuscript.
References
[1] Janda, J.M. and Abbott, S.A. (2014) Culture-independent diagnostic testing: have we opened Pandora’s box for good? Diagn. Microbiol. Infect. Dis. 80, 171–176.| Culture-independent diagnostic testing: have we opened Pandora’s box for good?Crossref | GoogleScholarGoogle Scholar |
[2] Jones, T.F. and Gerner-Smidt, P. (2012) Nonculture diagnostic tests for enteric diseases. Emerg. Infect. Dis. 18, 513–514.
| Nonculture diagnostic tests for enteric diseases.Crossref | GoogleScholarGoogle Scholar |
[3] OzFoodNet Working Group (2015) Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet network, 2011. Commun. Dis. Intell. Q. Rep. 39, E236–E264.
[4] Atkinson, R. et al. (2013) A challenge and an opportunity to improve patient management and public health surveillance for food-borne infections through culture-independent diagnostics. J. Clin. Microbiol. 51, 2479–2482.
| A challenge and an opportunity to improve patient management and public health surveillance for food-borne infections through culture-independent diagnostics.Crossref | GoogleScholarGoogle Scholar |
[5] Langley, G. et al. (2015) Effect of culture-independent diagnostic tests on future emerging infections program surveillance. Emerg. Infect. Dis. J. 21, 1582.
| 1:CAS:528:DC%2BC2sXivVCksQ%3D%3D&md5=4ed046f434dcaefa0a34544743c91d14CAS |
[6] Cronquist, A.B. et al. (2012) Impacts of culture-independent diagnostic practices on public health surveillance for bacterial enteric pathogens. Clin. Infect. Dis. 54, S432–S439.
| Impacts of culture-independent diagnostic practices on public health surveillance for bacterial enteric pathogens.Crossref | GoogleScholarGoogle Scholar |
[7] Van Lint, P. et al. (2015) Evaluation of a real-time multiplex PCR for the simultaneous detection of Campylobacter jejuni, Salmonella spp., Shigella spp./EIEC, and Yersinia enterocolitica in fecal samples. Eur. J. Clin. Microbiol. Infect. Dis. 34, 535–542.
| Evaluation of a real-time multiplex PCR for the simultaneous detection of Campylobacter jejuni, Salmonella spp., Shigella spp./EIEC, and Yersinia enterocolitica in fecal samples.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhvVent7nO&md5=5095aa9c7e33ec83b1c4067754c27161CAS |
[8] Bessède, E. et al. (2011) New methods for detection of Campylobacters in stool samples in comparison to culture. J. Clin. Microbiol. 49, 941–944.
| New methods for detection of Campylobacters in stool samples in comparison to culture.Crossref | GoogleScholarGoogle Scholar |
[9] Lan, R. and Reeves, P.R. (2002) Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 4, 1125–1132.
| Escherichia coli in disguise: molecular origins of Shigella.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XnsVansb0%3D&md5=6b82d78d771c0a67322d351a8a36a4e0CAS |
[10] Sethabutr, O. et al. (1993) Detection of Shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J. Infect. Dis. 167, 458–461.
| Detection of Shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK3s7jt1Cjsw%3D%3D&md5=e9c4fe009bfa0c82cad128d97e7e9e08CAS |
[11] Thiem, V.D. et al. (2004) Detection of Shigella by a PCR assay targeting the ipaH gene suggests increased prevalence of shigellosis in Nha Trang, Vietnam. J. Clin. Microbiol. 42, 2031–2035.
| Detection of Shigella by a PCR assay targeting the ipaH gene suggests increased prevalence of shigellosis in Nha Trang, Vietnam.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXkvFOjtrw%3D&md5=0edd3fbac551174de1b7a606c9280c23CAS |
[12] Dutta, S. et al. (2001) Sensitivity and performance characteristics of a direct PCR with stool samples in comparison to conventional techniques for diagnosis of Shigella and enteroinvasive Escherichia coli infection in children with acute diarrhoea in Calcutta, India. J. Med. Microbiol. 50, 667–674.
| Sensitivity and performance characteristics of a direct PCR with stool samples in comparison to conventional techniques for diagnosis of Shigella and enteroinvasive Escherichia coli infection in children with acute diarrhoea in Calcutta, India.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXlvVGqu7Y%3D&md5=416e3677ae602c45132ce435808dd791CAS |
[13] Escher, M. et al. (2014) A severe foodborne outbreak of diarrhoea linked to a canteen in Italy caused by enteroinvasive Escherichia coli, an uncommon agent. Epidemiol. Infect. 142, 2559–2566.
| A severe foodborne outbreak of diarrhoea linked to a canteen in Italy caused by enteroinvasive Escherichia coli, an uncommon agent.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC2cvlslWrsA%3D%3D&md5=b3d876f42a9252b4d83fc85f38cf6e15CAS |
[14] Svenungsson, B. et al. (2000) Enteropathogens in adult patients with diarrhea and healthy control subjects: a 1-year prospective study in a Swedish clinic for infectious diseases. Clin. Infect. Dis. 30, 770–778.
| Enteropathogens in adult patients with diarrhea and healthy control subjects: a 1-year prospective study in a Swedish clinic for infectious diseases.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3cvgtFSqsQ%3D%3D&md5=201d2e90a109438da2e46e44322b8ee9CAS |
[15] Tai, A.Y. et al. (2016) A review of the public health management of shigellosis in Australia in the era of culture-independent diagnostic testing. Aust. N. Z. J. Public Health 40, 588–591.
| A review of the public health management of shigellosis in Australia in the era of culture-independent diagnostic testing.Crossref | GoogleScholarGoogle Scholar |
[16] May, F.J. et al. (2017) The effects of culture independent diagnostic testing on the diagnosis and reporting of enteric bacterial pathogens in Queensland, 2010 to 2014. Commun. Dis. Intell. , .
Biography
Dr Fiona May BSc(Hons), PhD, GCPH, MPhil(App Epid) is an epidemiologist at Metro North Public Health Unit in Metro North Hospital and Health Service, Queensland Health. She began her career with a PhD and post-doctoral research in molecular virology before working as a sequencing manager. She entered the world of public health epidemiology via the Master of Philosophy in Applied Epidemiology field epidemiology training program. Her interests include foodborne, sexually transmitted and mosquito-borne diseases and applying her molecular and sequencing background to her current role in public health epidemiology.