From isolate to answer: how whole genome sequencing is helping us rapidly characterise nosocomial bacterial outbreaks
Leah RobertsSchool of Chemistry and Molecular Biosciences (SCMB), University of Queensland, St Lucia, Qld, Australia
Tel: +61 7 3365 8549
Email: l.roberts3@uq.edu.au
Microbiology Australia 38(3) 127-130 https://doi.org/10.1071/MA17047
Published: 18 August 2017
The occurrence of highly resistant bacterial pathogens has risen in recent years, causing immense strain on the healthcare industry. Hospital-acquired infections are arguably of most concern, as bacterial outbreaks in clinical settings provide an ideal environment for proliferation among vulnerable populations. Understanding these outbreaks beyond what can be determined with traditional clinical diagnostics and implementing these new techniques routinely in the hospital environment has now become a major focus. This brief review will discuss the three main whole genome sequence techniques available today, and how they are being used to further discriminate bacterial outbreaks in nosocomial settings.
In recent decades, society has witnessed a rapid and alarming increase in highly resistant pathogens causing human disease, to the extent that the World Health Organization (WHO) has labelled antimicrobial resistance (AMR) as, ‘a serious threat to global public health’1. The challenge of AMR is arguably greatest in health-care facilities, which present a unique environment for pathogens to proliferate and infect those most vulnerable. Once within the hospital, these pathogens can be readily spread due to the close proximity of patients and the mass of shared vectors in the environment (including bathrooms, wards, healthcare workers, trolleys, etc.). With limited treatment options, patient outcomes during these nosocomial outbreaks decline, leading to increased mortality and a substantial economic burden2–4. Traditional techniques employed in routine hospital diagnostics focus mainly on phenotypic and gene-centric analyses, which are useful for inexpensive and rapid surveillance in the preliminary stages of an outbreak. However, these techniques are unable to render fine-scale analyses capable of detecting transmission pathways and otherwise unexpected epidemiological connections. As such, new high-resolution technologies, such as whole genome sequencing (WGS), are now being used in response to outbreak investigations. This article will discuss the three main WGS technologies available today and briefly review their existing and future impact in clinical settings.
Short-read sequencing in nosocomial settings
Many hospitals worldwide already appreciate the power of WGS analysis, having integrated it into several published investigations5–7. The advantages of WGS are most accessible through the use of short-read sequencing as exemplified by the Illumina platform. In addition to being both high-throughput and cost-effective, analysis tools specific for short-read sequencing are well established, making it a useful and reliable research tool. One of the main advantages of WGS is the ability to detect single nucleotide variants (SNV). Detection of SNV can allow prediction of phenotypic changes, such as enhanced resistant to antibiotics (for example, via the loss of a functional outer membrane porin cause by indels or point mutations8–10). When coupled with metadata, SNV data can also be used to predict transmission pathways by inferring transmission directionality via the accumulation of SNV over time. Short-read data are also routinely used to determine presence or absence of genes using read mapping or short-read assembly techniques.
Carbapenem-resistant Enterobacteriaceae (CRE) are among the most prevalent clinically relevant organisms, designated as an urgent threat by the Centers for Disease Control and Prevention11. Snitkin et al.12 were amongst the first to apply WGS to identify transmission pathways in a CRE outbreak that could not be resolved using traditional epidemiological investigations alone. Recently, we have used WGS to investigate a suspected CRE outbreak in a Brisbane hospital13. Using Illumina short-read sequencing, we were able to determine transmission of a carbapenemase-producing Enterobacter cloacae sequence type (ST) 90 strain within the intensive care unit (ICU) between three patients over a three-month period. Of the 10 isolates analysed, we detected only 4 SNV overall, indicative of direct transmission. Patients 1 and 3 were found to have identical isolates at the core genome level, despite not being in the ICU at the same time. This suggested a probable environmental source of the infection, rather than direct patient-to-patient transmission. This was supported by comparison of the isolates to publicly available genomes, which identified a near-identical E. cloacae from 2013 (isolated in the same ward and hospital). This E. cloacae differed by only one SNV from the 2015 isolates, suggesting a reservoir in the hospital environment since at least 2013. Despite efforts to identify the environmental source in the hospital through extensive screening of all possible environmental vectors (excluding healthcare workers), a source was not found. Overall, WGS was able to accurately determine the relationship of E. cloacae over the course of the outbreak, providing unambiguous evidence of environmental cross-transmission over at least two years, prompting further surveillance and re-imposing infection control standards.
Beyond clonal transmission: tracking mobile genetic elements with long-read sequencing
The most significant drawback of using short-read sequencing is the inability to accurately characterise mobile genetic elements (MGE) such as insertion sequences (IS), genomic islands and plasmids. MGE are often associated with important elements such as virulence factors and antibiotic resistance genes14–16, but are also known to comprise numerous repetitive regions, the main culprit being IS17. These repetitive regions are unable to be traversed by short-read sequencing, causing ‘collapsed repeats’ in final assemblies that ultimately impedes the contextualisation of important genomic regions18–20. While tools exist to patch together these assemblies, such as Bandage21, increasing the sequence read length to span these repetitive regions is the only unambiguous solution for resolving MGEs22.
Currently, the most established long-read sequencing technology is Pacific Biosciences (PacBio) Single Molecule Real-Time (SMRT) sequencing. This technology can now routinely provide complete bacterial chromosomes and plasmids23, allowing contextualisation of important genomic regions, tracking of plasmids and completion of high-quality reference genomes24. One of the most progressive examples of PacBio integration into clinical settings comes from Mount Sinai, New York, where Sullivan et al.25 successively sequenced 137 methicillin-resistant Staphylococcus aureus (MRSA) isolates using PacBio to allow high-precision surveillance and outbreak control. In our investigation of the 2015 carbapenemase-producing E. cloacae, we used PacBio SMRT sequencing to completely characterise a large ~330 kb IncHI2 plasmid carrying a complex ~55 kb multidrug resistant (MDR) region that contained the carbapenemase gene blaIMP4. Once characterised, we were able to undertake broader epidemiological surveillance for this plasmid, resulting in its identification in E. cloacae patient isolates (and an Escherichia coli) from other South-East Queensland hospitals. Comparison of our plasmid to publicly available data also found a remarkably similar plasmid carried by a Salmonella enterica isolate from a cat26. Similarly, Sheppard et al.27 were able to use a combination of long- and short-read sequencing to track blaKPC positive Enterobacteriaceae in a single hospital over 5 years, ultimately showing the promiscuity of blaKPC within several different hosts and in several different plasmids. These studies highlight the importance of tracking not only outbreaks focused on clonal transmission, but also MGE transmission between bacterial strains or species in hospital settings (Figure 1).
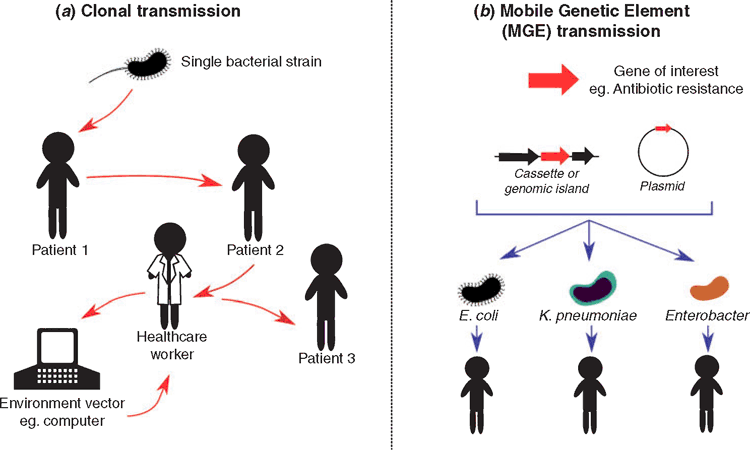
|
While this technology is effective at producing complete genomes, it does come at a price. A single bacterial genome with PacBio SMRT sequencing can cost 20 times that of an Illumina sequence28. Additionally, the amount of DNA required, the time needed for the library preparation (Figure 2), as well as the relatively low throughput system, makes routine sequencing with PacBio less attractive than short-read sequencing due to the time required per sample. Ultimately, to implement long-read sequencing into the clinical setting, the turn-around time from sample isolation to analysis results needs to be within a workable timeframe.
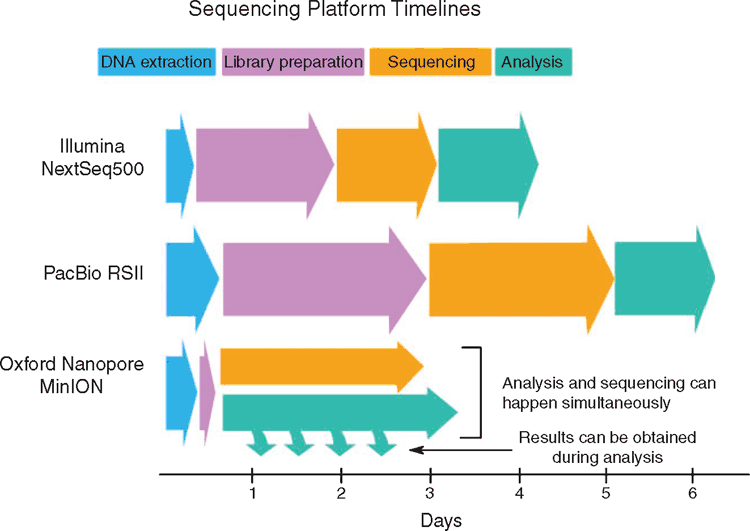
|
Nanopore: a new and emerging technology for real-time analysis
Oxford Nanopore MinION sequencing is a relatively new sequencing platform designed to couple long-read sequencing with a rapid turn-around time. Its portability and capacity to perform real-time analysis during sequencing has made it an attractive companion for outbreak investigations in remote regions, as was the case with the ZIBRA project29. Clinically, MinION sequencing has been used to retrospectively analyse S. enterica isolates in relation to a European-wide outbreak30, which showed that the outbreak strain could be identified from less than 2 hours of sequencing, despite not having the resolution to elucidate transmission pathways. Viral pathogens have also been identified from clinical metagenomic samples, with a projected turn-around time of less than 6 hours31. While this new technology is promising, some hurdles remain before MinION sequencing can be used routinely. These include the lower base level accuracy, as well as the current difficulty in sequencing more than 12 isolates in parallel (as compared to Illumina)28,32.
Overall, WGS technology has advanced considerably in the last decade, with an equivalently rapid decline in price33–35. While still being more expensive than traditional diagnostics, if successful in preventing further transmission during an outbreak this cost becomes negligible compared to the cost of continued patient treatment and repeated infection control. WGS also retrospectively provides a large wealth of information, allowing the hospital to progressively catalogue past and ongoing infection occurrences, ultimately providing a highly detailed epidemiological map of pathogen movement in the hospital and from the community that can quickly be referred to in the case of new infections. Implementation of WGS in response to an outbreak guarantees an in-depth high-resolution analysis that cannot be determined using traditional phenotypic and genotypic methods alone. Currently, all three sequencing technologies presented in this article can be used complimentarily to produce a comprehensive outbreak understanding and provide ongoing genomic surveillance of the strain or element. Routine implementation of WGS in healthcare settings will undoubtedly become widespread in the near future, aiding clinicians, patients, infection control and researchers alike.
References
[1] WHO (2016) World Health Organization Antimicrobial resistance facts sheet. http://www.who.int/mediacentre/factsheets/fs194/en/ (accessed 15 June 2017).[2] Kaye, K.S. et al. (2014) Effect of nosocomial bloodstream infections on mortality, length of stay, and hospital costs in older adults. J. Am. Geriatr. Soc. 62, 306–311.
| Effect of nosocomial bloodstream infections on mortality, length of stay, and hospital costs in older adults.Crossref | GoogleScholarGoogle Scholar |
[3] Kollef, M.H. (2000) Inadequate antimicrobial treatment: an important determinant of outcome for hospitalized patients. Clin. Infect. Dis. 31, S131–S138.
| Inadequate antimicrobial treatment: an important determinant of outcome for hospitalized patients.Crossref | GoogleScholarGoogle Scholar |
[4] Sydnor, E.R. and Perl, T.M. (2011) Hospital epidemiology and infection control in acute-care settings. Clin. Microbiol. Rev. 24, 141–173.
| Hospital epidemiology and infection control in acute-care settings.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjtFSgu7Y%3D&md5=7b7e48a9b0c88ee37e7e68f136acfeccCAS |
[5] Ben Zakour, N.L. et al. (2012) Analysis of a Streptococcus pyogenes puerperal sepsis cluster by use of whole-genome sequencing. J. Clin. Microbiol. 50, 2224–2228.
| Analysis of a Streptococcus pyogenes puerperal sepsis cluster by use of whole-genome sequencing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFCmsL7O&md5=b50ba83f7d5e3fe5dabee03799888b0eCAS |
[6] Köser, C.U. et al. (2012) Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 366, 2267–2275.
| Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak.Crossref | GoogleScholarGoogle Scholar |
[7] Lewis, T. et al. (2010) High-throughput whole-genome sequencing to dissect the epidemiology of Acinetobacter baumannii isolates from a hospital outbreak. J. Hosp. Infect. 75, 37–41.
| High-throughput whole-genome sequencing to dissect the epidemiology of Acinetobacter baumannii isolates from a hospital outbreak.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3c3lsVGgtg%3D%3D&md5=a4b03d0512dba57ad8943e145ee8f9a2CAS |
[8] Babouee Flury, B. et al. (2016) Association of novel nonsynonymous single nucleotide polymorphisms in ampD with cephalosporin resistance and phylogenetic variations in ampC, ampR, ompF, and ompC in Enterobacter cloacae isolates that are highly resistant to carbapenems. Antimicrob. Agents Chemother. 60, 2383–2390.
| Association of novel nonsynonymous single nucleotide polymorphisms in ampD with cephalosporin resistance and phylogenetic variations in ampC, ampR, ompF, and ompC in Enterobacter cloacae isolates that are highly resistant to carbapenems.Crossref | GoogleScholarGoogle Scholar |
[9] Ocampo-Sosa, A.A. et al. (2012) Alterations of OprD in carbapenem-intermediate and -susceptible strains of Pseudomonas aeruginosa isolated from patients with bacteremia in a Spanish multicenter study. Antimicrob. Agents Chemother. 56, 1703–1713.
| Alterations of OprD in carbapenem-intermediate and -susceptible strains of Pseudomonas aeruginosa isolated from patients with bacteremia in a Spanish multicenter study.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XltV2rurg%3D&md5=e930eb91d5bb0c7bace31893e0cc600cCAS |
[10] uz Zaman, T. et al. (2014) Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 28, 186–192.
| Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhs1KgtrvE&md5=78f93f116213591b5accdfab6c6f6367CAS |
[11] CDC (2017) Antibiotic/antimicrobial resistance. https://http://www.cdc.gov/drugresistance/biggest_threats.html (accessed 23 June 2017).
[12] Snitkin, E.S. et al. (2012) Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 4, 148ra116.
| Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing.Crossref | GoogleScholarGoogle Scholar |
[13] Roberts, L.W. et al. (2017) Genomic investigation of an outbreak of carbapenemase-producing Enterobacter cloacae: long-read sequencing reveals the context of blaIMP4 on a widely distributed IncHI2 plasmid. BioRxiv. , .
| Genomic investigation of an outbreak of carbapenemase-producing Enterobacter cloacae: long-read sequencing reveals the context of blaIMP4 on a widely distributed IncHI2 plasmid.Crossref | GoogleScholarGoogle Scholar |
[14] Brown-Jaque, M. et al. (2015) Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 79, 1–7.
| Transfer of antibiotic-resistance genes via phage-related mobile elements.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtVGgurk%3D&md5=9c2d28db8455df111e7161c008beb458CAS |
[15] Liu, Y.Y. et al. (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168.
| Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study.Crossref | GoogleScholarGoogle Scholar |
[16] Wellington, E.M. et al. (2013) The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 13, 155–165.
| The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXht1Ghsb0%3D&md5=92a212e456cd1706ab0c065fb7a0ee8bCAS |
[17] Siguier, P. et al. (2014) Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol. Rev. 38, 865–891.
| Bacterial insertion sequences: their genomic impact and diversity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhsFOgu7vP&md5=de76a93e07958f822ef6df52599426d9CAS |
[18] Kingsford, C. et al. (2010) Assembly complexity of prokaryotic genomes using short reads. BMC Bioinformatics 11, 21.
| Assembly complexity of prokaryotic genomes using short reads.Crossref | GoogleScholarGoogle Scholar |
[19] Ricker, N. et al. (2012) The limitations of draft assemblies for understanding prokaryotic adaptation and evolution. Genomics 100, 167–175.
| The limitations of draft assemblies for understanding prokaryotic adaptation and evolution.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtVKntbnE&md5=12b6c3e6a68904eae6d45137b7e292bbCAS |
[20] Toussaint, A. and Chandler, M. (2012) Prokaryote genome fluidity: toward a system approach of the mobilome. Methods Mol. Biol. 804, 57–80.
| Prokaryote genome fluidity: toward a system approach of the mobilome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhslaitL3P&md5=353e77b99febf8b606f392e00cb9cd76CAS |
[21] Wick, R.R. et al. (2015) Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352.
| Bandage: interactive visualization of de novo genome assemblies.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xhs1Git7%2FJ&md5=361f32e83a52d5aeefc661a595e0ceb5CAS |
[22] Koren, S. and Phillippy, A.M. (2015) One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr. Opin. Microbiol. 23, 110–120.
| One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhvF2hu7jF&md5=909e8064d6e4826b6f4ce45bddfb5c54CAS |
[23] Stoesser, N. et al. (2016) Complete sequencing of plasmids containing bla OXA-163 and bla OXA-48 in Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 60, 6948–6951.
| Complete sequencing of plasmids containing bla OXA-163 and bla OXA-48 in Escherichia coli sequence type 131.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXitlSqsrc%3D&md5=98cd199d3f2865525631d9eec60148e8CAS |
[24] Rasko, D.A. et al. (2011) Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365, 709–717.
| Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtV2lu7jJ&md5=1e7c2af1a19520b4354e32a1f4a7f685CAS |
[25] Sullivan, M. et al. (2016) Continuous surveillance by whole-genome sequencing to identify and manage methicillin-resistant Staphylococcus aureus outbreaks. Open Forum Infectious Diseases 3, 942.
[26] Abraham, S. et al. (2016) Isolation and plasmid characterization of carbapenemase (IMP-4) producing Salmonella enterica Typhimurium from cats. Sci. Rep. 6, 35527.
| Isolation and plasmid characterization of carbapenemase (IMP-4) producing Salmonella enterica Typhimurium from cats.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhslGrtr%2FF&md5=ed77178a0603a34c1de3c37a6c3449caCAS |
[27] Sheppard, A.E. et al. (2016) Nested Russian Doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob. Agents Chemother. 60, 3767–3778.
| Nested Russian Doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhslKrsLvF&md5=3d8145847e5d5ddbdc736928eb662439CAS |
[28] Goodwin, S. et al. (2016) Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351.
| Coming of age: ten years of next-generation sequencing technologies.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XnvFOksL8%3D&md5=bad76d9c276dbc8f2ea01cc4042626f4CAS |
[29] Faria, N.R. et al. (2017) Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546, 406–410.
| Establishment and cryptic transmission of Zika virus in Brazil and the Americas.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXosVagt7s%3D&md5=4b8f107f4fee23f95bebbb72d5d3bc50CAS |
[30] Quick, J. et al. (2015) Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol. 16, 114.
| Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella.Crossref | GoogleScholarGoogle Scholar |
[31] Greninger, A.L. et al. (2015) Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 7, 99.
| Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis.Crossref | GoogleScholarGoogle Scholar |
[32] Lu, H. et al. (2016) Oxford Nanopore MinION Sequencing and Genome Assembly. Genomics Proteomics Bioinformatics 14, 265–279.
| Oxford Nanopore MinION Sequencing and Genome Assembly.Crossref | GoogleScholarGoogle Scholar |
[33] Mardis, E.R. (2011) A decade’s perspective on DNA sequencing technology. Nature 470, 198–203.
| A decade’s perspective on DNA sequencing technology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhslWktbk%3D&md5=9361ed7363868580e4c5721e03a1863dCAS |
[34] Muir, P. et al. (2016) The real cost of sequencing: scaling computation to keep pace with data generation. Genome Biol. 17, 53.
| The real cost of sequencing: scaling computation to keep pace with data generation.Crossref | GoogleScholarGoogle Scholar |
[35] Wetterstrand, K.A. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) 2016. http://www.genome.gov/sequencingcostsdata/ (accessed 15 June 2017).
Biography
Leah Roberts is currently a third-year PhD candidate completing her studies at the University of Queensland, Brisbane, Australia. Under the supervision of Associate Professor Scott A Beatson and Professor Mark A Schembri, her research interests have mainly focused on the use of whole genome sequencing and its application in clinical settings. Using a range of sequencing technologies such as Illumina, PacBio and Nanopore, Leah has analysed a variety of bacterial outbreaks, including Acinetobacter baumannii, Klebsiella pneumoniae and Enterobacter cloacae. She has presented her work at both domestic and international conferences, and has won several awards including the 2016 ASM BD award for Queensland, as well as the 2017 Applied Bioinformatics and Public Health Microbiology (ABPHM) student poster prize.


