Chlamydia pecorum: successful pathogen of koalas or Australian livestock?
Martina Jelocnik A B and Adam Polkinghorne AA Centre for Animal Health Innovation (CAHI), Faculty of Science, Health, Education and Engineering, University of the Sunshine Coast, Locked Bag 4, Maroochydore, Qld 4558, Australia
B Tel: +41 7 5456 3585, Email: martina.jelocnik@research.usc.edu.au
Microbiology Australia 38(3) 101-105 https://doi.org/10.1071/MA17042
Published: 9 August 2017
In Australia, the obligate intracellular bacterium Chlamydia pecorum is best known as the notorious koala pathogen that causes debilitating ocular and urogenital tract disease. While globally published data suggests that this species is essentially ubiquitous in livestock, little is known about the epidemiology of livestock C. pecorum infections here in Australia. My research is focused on investigating the genetic diversity and transmission patterns of C. pecorum, and why it causes disease. Using our newly developed C. pecorum-specific molecular epidemiology typing scheme we provided the first epidemiological data on infections in sheep and cattle in Australia, identifying strains associated with a range of diseases in livestock, and uncovering an unexpected level of diversity for this pathogen. Most importantly, we observed that the same strain can infect koala and sheep, indicating on ongoing cross-host transmission and ‘spill-over’ risks to wildlife. Further, by dissecting koala, sheep, cattle and pig C. pecorum strains genomes, we have also identified novel virulence-associated factors that could be explored as vaccine candidates for both livestock and koala infections.
Chlamydiae, known for their obligatory parasitic lifestyle and distinct biphasic life-cycle1, are diverse and enigmatic bacteria that infect an astonishing range of hosts2. This phylum includes Chlamydia pecorum, a species that constantly remains in the spotlight due to its role as a major threat to the survival of the iconic koalas. In koalas, highly prevalent C. pecorum infections cause ocular disease that can eventually lead to blindness, and urogenital disease that can lead to infertility3,4. The pathogenic potential of C. pecorum in economically significant livestock species (cattle, sheep, goats and pigs) has been well documented in Europe and USA, and to a lesser degree elsewhere including Australia. The spectrum of C. pecorum infections in livestock hosts ranges from subclinical to acute disease manifestations such as polyarthritis, sporadic bovine encephalomyelitis, pneumonia and conjunctivitis, with (asymptomatic) gastrointestinal shedding as the common feature5. C. pecorum infections in free-range ruminants such as Alpine ibex and chamois from Europe have also been reported6,7.
The curious case of Chlamydia pecorum infections in Australia
While the koala chlamydial infections are easily the most intensively studied of any wildlife species, almost nothing is known about the prevalence and epidemiology of C. pecorum infections in Australian livestock. Veterinarians from agriculturally productive areas throughout Australia regularly report cases of chlamydiosis in sheep and cattle8,9, but the information about the genetic diversity of strains infecting livestock is lacking. A range of previous molecular studies suggested that C. pecorum strains infecting koala are genetically diverse10,11; however, none of the studies investigated how and whether these strains are related to livestock strains. It has been hypothesised that the origin of koala C. pecorum infections is associated with the import of ‘presumably C. pecorum infected’ livestock in the late 1780s3. The encroachment of koala habitats by sheep and cattle farming along the east coast of Australia is common and raises serious questions over the relationships and potential role that cross-host transmission may have in the epidemiology and origin of chlamydial disease in koalas3. Despite the ongoing field reports, the epidemiology, transmission, and reservoirs of C. pecorum in Australian sheep and cattle remain poorly understood5,12.
C. pecorum molecular epidemiology
Multi-locus sequence typing (MLST) is a widely used epidemiological tool for strain identification and estimation of intra-species relationships for many pathogens. Even today, with whole genome sequencing readily available, MLST is still often used as the first point to barcode a strain13. In an effort to evaluate the overall genetic diversity and relationships between Australian koala, sheep and cattle C. pecorum strains, we developed and applied a C. pecorum-specific MLST to a range of isolates and C. pecorum-positive clinical samples from koalas, Australian sheep and cattle14–16.
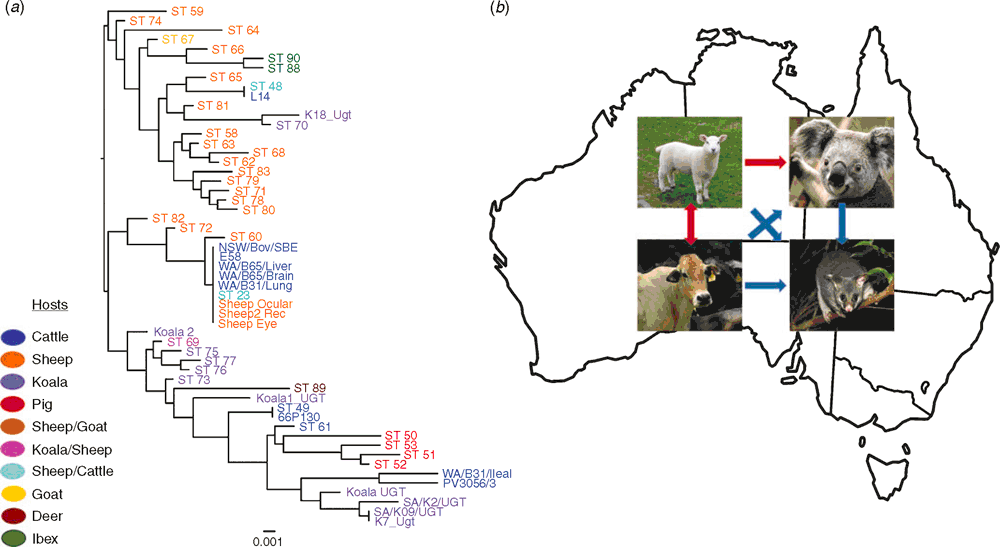
The newly developed MLST proved to be an effective fine-detailed molecular epidemiology tool in characterising novel C. pecorum genotypes infecting livestock and koalas, highlighting an unexpected level of diversity for this pathogen. The genetic diversity of C. pecorum extends from the individual to the population level with repeated molecular evidence that: a) a single host can harbour two distinct C. pecorum strains at different anatomical sites; b) discrete populations can be infected with a mix of genetically diverse strains and c) the same strain can infect two different hosts (e.g. koala and sheep; sheep and cattle)14 (Figure 1a). This observation was also repeated in the recent MLST of C. pecorum infecting koalas from French Island in Victoria. In this study, it was also observed that the certain koala strains were more similar to livestock strains than they were to other koala strains17.
|
C. pecorum-specific MLST and Bayesian phylogenetic analyses of strains infecting Australian sheep with conjunctivitis, polyarthritis, or with no clinical signs of disease and cattle with sporadic encephalomyelitis revealed important observations of the genetic identity and relationships between C. pecorum strains found at different anatomical sites and the range of associated diseases15,16. We observed two distinct C. pecorum genotypes associated with disease, one denoted sequence type (ST) 23 found in association to sheep with polyarthritis and conjunctivitis and cattle with encephalomyelitis, and the other denoted ST 69 found in association with ovine conjunctivitis as well as previously detected in koala urogenital tract and ocular infections (Figure 1a). The majority of C. pecorum strains detected at vaginal and rectal sites of clinically healthy as well as diseased animals clustered together in a distinct and separate clade, away from strains associated with disease. C. pecorum ST 23 strains are also readily found in association with disease in European and USA livestock hosts, indicating a potential global spread of such clonal ‘pathogenic’ C. pecorum14,18. Similarly, genetically identical the ‘non-pathogenic’ vagino-rectal strains characterised in our studies were also detected in the faeces of clinically healthy sheep from Europe (Figure 1a).
Chlamydial infections can spread rapidly in a flock/herd, potentially developing into diseases such as polyarthritis, conjunctivitis and encephalomyelitis, leading to increased morbidity and mortality on farms19. Detection of the genetically different C. pecorum strains at various anatomical sites in a single host and a plausible association of certain genotypes with disease/virulence in our and other studies18,20 raises questions over the existence of ‘pathogenic and non-pathogenic’ C. pecorum and their role in pathogenesis of disease and impact on an animal’s health12.
The role of cross-host transmission in the epidemiology and origin of C. pecorum infections
The repeated evidence that identical or closely related C. pecorum strains infect koalas and livestock raises serious questions over the role of contemporary and historic spill over of this pathogen from domesticated animals to koalas3,21 (Figure 1b). Ongoing work in our laboratory also provided evidence of C. pecorum infections in other Australian native marsupials (unpublished observations), raising the possibility of cross-host transmission to the native wildlife as well (Figure 1b). Although ecologically distinct, livestock and wildlife host species could have an indirect contact allowing an opportunity for infection spill-over, most likely via the faecal-oral route12,22. For livestock C. pecorum infections, sheep and cattle are strong reservoir candidates as evidenced by ubiquitous faecal shedding of presumably ‘non-pathogenic’ strains16,22 (Figure 1b). Evidence for cross-host transmission was also observed in our typing study of C. pecorum strains detected in European free-range ruminants7 where, in limited examples, we observed a phylogenetic relatedness of the Alpine ibex C. pecorum with sheep faecal strains.
The lessons from ongoing C. pecorum genomics
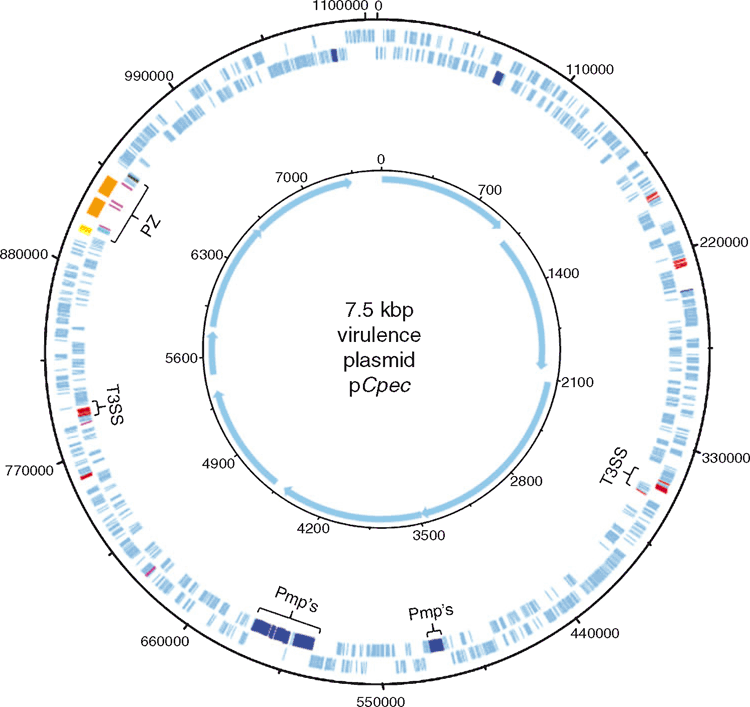
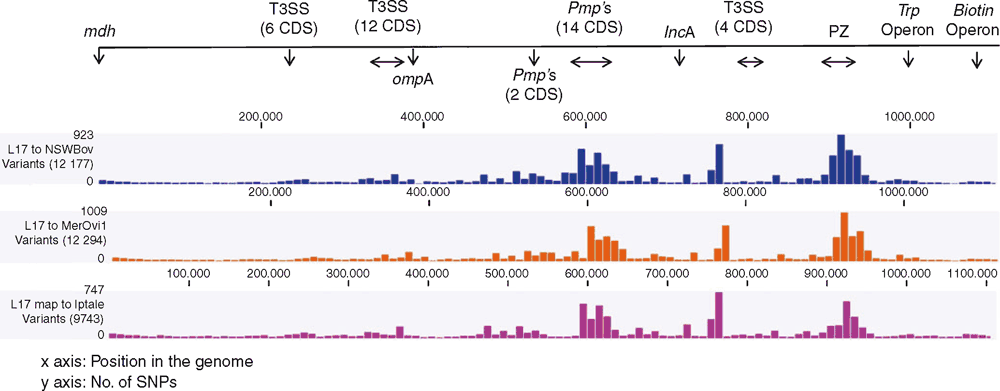
While the MLST provided us with preliminary characterisation of C. pecorum strains from various hosts, comparative genomics provides valuable insights into the lifestyle, within-host adaptation, virulence factors and population structure12,23. The ongoing comparative genomics of koala, sheep, cattle and pig C. pecorum genomes are building the global population framework of this enigmatic species20,23–25. Highly conserved and syntenic, the main genetic differences between C. pecorum strains are limited to single nucleotide polymorphisms (SNPs), mainly accumulating in the highly polymorphic membrane protein (pmp’s) genes and the plasticity zone that harbours virulence factors, such as cytotoxin, phospholipase D and Mac-Perforin genes25 (Figures 2 and 3). The unexpected genomic finding in our studies was the identification of a chlamydial virulence plasmid (pCpec) (Figure 2). pCpec seems to be highly distributed in strains, however plasmidless strains are also common, raising questions over its function in disease pathogenesis26. Culture-independent genome sequencing of sheep, cattle and koala C. pecorum positive clinical swabs demonstrated that not only could multiple strains infect a single host but that a single anatomical site can harbour multiple C. pecorum strains27, somewhat confirming observations from molecular epidemiology but also reiterating complex epidemiology of these infections. C. pecorum genome-derived phylogenies highlighted the polyphyletic history of the pathogen itself, and provided important clues about the evolutionary origins of koala strains, giving more confidence to our hypothesis that the origin of C. pecorum infections in Australia is associated with importation of domesticated animals into Australia with European colonisation3,25,27.
Future directions
Drawing on the knowledge acquired in our studies, we have the opportunity to minimise cross-host transmission and improve on the diagnosis and management of these infections. We have recently developed a prototype anti-C. pecorum sheep28 and improved koala vaccine formulation29 using novel antigens identified in our genomic studies that could be used as a valuable tools to manage and control C. pecorum infections in the future. Improvements to diagnostic testing are now possible and laboratory and Point-of-Care assays for use in the lab and field are now also in development.
References
[1] Hammerschlag, M.R. (2002) The intracellular life of chlamydiae. Semin. Pediatr. Infect. Dis. 13, 239–248.| The intracellular life of chlamydiae.Crossref | GoogleScholarGoogle Scholar |
[2] Taylor-Brown, A. et al. (2015) Twenty years of research into Chlamydia-like organisms: a revolution in our understanding of the biology and pathogenicity of members of the phylum Chlamydiae. Pathog. Dis. 73, 1–15.
| Twenty years of research into Chlamydia-like organisms: a revolution in our understanding of the biology and pathogenicity of members of the phylum Chlamydiae.Crossref | GoogleScholarGoogle Scholar |
[3] Polkinghorne, A. et al. (2013) Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas. Vet. Microbiol. 165, 214–223.
| Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas.Crossref | GoogleScholarGoogle Scholar |
[4] Griffith, J.E. and Higgins, D.P. (2012) Diagnosis, treatment and outcomes for koala chlamydiosis at a rehabilitation facility (1995–2005). Aust. Vet. J. 90, 457–463.
| Diagnosis, treatment and outcomes for koala chlamydiosis at a rehabilitation facility (1995–2005).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhvV2gurjO&md5=9461a12b1231bd2e8a8e79c76a357721CAS |
[5] Walker, E. et al. (2015) Chlamydia pecorum infections in sheep and cattle: a common and under-recognised infectious disease with significant impact on animal health. Vet. J. 206, 252–260.
| Chlamydia pecorum infections in sheep and cattle: a common and under-recognised infectious disease with significant impact on animal health.Crossref | GoogleScholarGoogle Scholar |
[6] Holzwarth, N. et al. (2011) Occurrence of Chlamydiaceae, Mycoplasma Conjunctivae, and pestiviruses in Alpine chamois (Rupicapra r. Rupicapra) of Grisons, Switzerland. J. Vet. Diagn. Invest. 23, 333–337.
| Occurrence of Chlamydiaceae, Mycoplasma Conjunctivae, and pestiviruses in Alpine chamois (Rupicapra r. Rupicapra) of Grisons, Switzerland.Crossref | GoogleScholarGoogle Scholar |
[7] Jelocnik, M. et al. (2015) Novel sequence types of Chlamydia pecorum infect free-ranging Alpine ibex (Capra ibex) and red deer (Cervus elaphus) in Switzerland. J. Wildl. Dis. 51, 479–483.
| Novel sequence types of Chlamydia pecorum infect free-ranging Alpine ibex (Capra ibex) and red deer (Cervus elaphus) in Switzerland.Crossref | GoogleScholarGoogle Scholar |
[8] Watt, B. Chlamydial infection in sheep, Proceedings of the 92nd District Veterinarian’s Conference, District Veterinarian’s Association, Dubbo, New South Wales, Australia.
[9] Pitman, B. (2010) A case of Sporadic Bovine Encephalomyelitis (SBE) in Hume LHPA, Proceedings of the 91st District Veterinarian Conference 2010, Dubbo, New South Wales, Australia.
[10] Marsh, J. et al. (2011) Novel molecular markers of Chlamydia pecorum genetic diversity in the koala (Phascolarctos cinereus). BMC Microbiol. 11, 77.
| Novel molecular markers of Chlamydia pecorum genetic diversity in the koala (Phascolarctos cinereus).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXltFKlt7w%3D&md5=8670ae83ca450c6a81be27b046beded7CAS |
[11] Kollipara, A. et al. (2013) Genetic diversity of Chlamydia pecorum strains in wild koala locations across Australia and the implications for a recombinant C. pecorum major outer membrane protein based vaccine. Vet. Microbiol. 167, 513–522.
| Genetic diversity of Chlamydia pecorum strains in wild koala locations across Australia and the implications for a recombinant C. pecorum major outer membrane protein based vaccine.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhsVSgtLfE&md5=3ba8aee1e125e1782637599d3b835511CAS |
[12] Jelocnik, M. (2016) Molecular epidemiology and comparative genomics of Chlamydia pecorum infecting livestock and koalas. PhD Thesis, University of the Sunshine Coast, Australia.
[13] Maiden, M.C.J. et al. (2013) MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11, 728–736.
| MLST revisited: the gene-by-gene approach to bacterial genomics.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtlalu7nJ&md5=e3c15f2dc4f1fd700274b853350c15e3CAS |
[14] Jelocnik, M. et al. (2013) Multi-locus sequence analysis provides insights into the molecular epidemiology of Chlamydia pecorum infections in Australian sheep, cattle and koalas. J. Clin. Microbiol. 51, 2625–2632.
| Multi-locus sequence analysis provides insights into the molecular epidemiology of Chlamydia pecorum infections in Australian sheep, cattle and koalas.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhsV2lu7fI&md5=006c7ec72ac68d60b1a544968f8a3962CAS |
[15] Jelocnik, M. et al. (2014) Molecular and pathological insights into Chlamydia pecorum-associated sporadic bovine encephalomyelitis (SBE) in Western Australia. BMC Vet. Res. 10, 121.
| Molecular and pathological insights into Chlamydia pecorum-associated sporadic bovine encephalomyelitis (SBE) in Western Australia.Crossref | GoogleScholarGoogle Scholar |
[16] Jelocnik, M. et al. (2014) Evaluation of the relationship between Chlamydia pecorum sequence types and disease using a species-specific multi-locus sequence typing scheme (MLST). Vet. Microbiol. 174, 214–222.
| Evaluation of the relationship between Chlamydia pecorum sequence types and disease using a species-specific multi-locus sequence typing scheme (MLST).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhsV2nsbvJ&md5=13e7a9b384a567bc49c1ae5fc212db88CAS |
[17] Legione, A.R. et al. (2016) Chlamydia pecorum infection in free-ranging koalas (Phascolarctos cinereus) on French Island, Victoria, Australia. J. Wildl. Dis. 52, 426–429.
| Chlamydia pecorum infection in free-ranging koalas (Phascolarctos cinereus) on French Island, Victoria, Australia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXmtFSjsLk%3D&md5=282532fb9094aad8c64a34600b4dcee4CAS |
[18] Mohamad, K.Y. et al. (2014) Host adaptation of Chlamydia pecorum towards low virulence evident in co-evolution of the ompA, incA, and ORF663 Loci. PLoS One 9, e103615.
| Host adaptation of Chlamydia pecorum towards low virulence evident in co-evolution of the ompA, incA, and ORF663 Loci.Crossref | GoogleScholarGoogle Scholar |
[19] Walker, E. et al. (2016) Clinical, diagnostic and pathologic features of presumptive cases of Chlamydia pecorum-associated arthritis in Australian sheep flocks. BMC Vet. Res. 12, 193.
| Clinical, diagnostic and pathologic features of presumptive cases of Chlamydia pecorum-associated arthritis in Australian sheep flocks.Crossref | GoogleScholarGoogle Scholar |
[20] Sait, M. et al. (2014) Genome sequencing and comparative analysis of three Chlamydia pecorum strains associated with different pathogenic outcomes. BMC Genomics 15, 23.
| Genome sequencing and comparative analysis of three Chlamydia pecorum strains associated with different pathogenic outcomes.Crossref | GoogleScholarGoogle Scholar |
[21] Burnard, D. and Polkinghorne, A. (2016) Chlamydial infections in wildlife–conservation threats and/or reservoirs of ‘spill-over’ infections? Vet. Microbiol. 196, 78–84.
| Chlamydial infections in wildlife–conservation threats and/or reservoirs of ‘spill-over’ infections?Crossref | GoogleScholarGoogle Scholar |
[22] Reinhold, P. et al. (2011) Chlamydiaceae in cattle: commensals, trigger organisms, or pathogens? Vet. J. 189, 257–267.
| Chlamydiaceae in cattle: commensals, trigger organisms, or pathogens?Crossref | GoogleScholarGoogle Scholar |
[23] Bachmann, N.L. et al. (2014) Chlamydia genomics: providing novel insights into chlamydial biology. Trends Microbiol. 22, 464–472.
| Chlamydia genomics: providing novel insights into chlamydial biology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXpslCgsLg%3D&md5=9424805400858071e9281b03fb2ff2bcCAS |
[24] Bachmann, N.L. et al. (2014) Comparative genomics of koala, cattle and sheep strains of Chlamydia pecorum. BMC Genomics 15, 667.
| Comparative genomics of koala, cattle and sheep strains of Chlamydia pecorum.Crossref | GoogleScholarGoogle Scholar |
[25] Jelocnik, M. et al. (2015) Genetic diversity in the plasticity zone and the presence of the chlamydial plasmid differentiates Chlamydia pecorum strains from pigs, sheep, cattle, and koalas. BMC Genomics 16, 893.
| Genetic diversity in the plasticity zone and the presence of the chlamydial plasmid differentiates Chlamydia pecorum strains from pigs, sheep, cattle, and koalas.Crossref | GoogleScholarGoogle Scholar |
[26] Jelocnik, M. et al. (2016) Molecular characterisation of the Chlamydia pecorum plasmid from porcine, ovine, bovine, and koala strains indicates plasmid-strain co-evolution. PeerJ 4, e1661.
| Molecular characterisation of the Chlamydia pecorum plasmid from porcine, ovine, bovine, and koala strains indicates plasmid-strain co-evolution.Crossref | GoogleScholarGoogle Scholar |
[27] Bachmann, N.L. et al. (2015) Culture-independent genome sequencing of clinical samples reveals an unexpected heterogeneity of infections by Chlamydia pecorum. J. Clin. Microbiol. 53, 1573–1581.
| Culture-independent genome sequencing of clinical samples reveals an unexpected heterogeneity of infections by Chlamydia pecorum.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXntFOqs70%3D&md5=666080c02e85e6989d4e1631d5062569CAS |
[28] Desclozeaux, M. et al. (2017) Safety and immunogenicity of a prototype anti-Chlamydia pecorum recombinant protein vaccine in lambs and pregnant ewes. Vaccine 35, 3461–3465.
| Safety and immunogenicity of a prototype anti-Chlamydia pecorum recombinant protein vaccine in lambs and pregnant ewes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXotVCgu7k%3D&md5=a59c6cd48082bfff144fd39516d33635CAS |
[29] Desclozeaux, M. et al. (2017) Immunization of a wild koala population with a recombinant Chlamydia pecorum major outer membrane protein (MOMP) or polymorphic membrane protein (PMP) based vaccine: new insights into immune response, protection and clearance. PLoS One 12, e0178786.
| Immunization of a wild koala population with a recombinant Chlamydia pecorum major outer membrane protein (MOMP) or polymorphic membrane protein (PMP) based vaccine: new insights into immune response, protection and clearance.Crossref | GoogleScholarGoogle Scholar |
Biographies
Dr Martina Jelocnik is an Early Career Research Fellow at the University of the Sunshine Coast’s Centre for Animal Health Innovation. In June 2016, she has been awarded PhD in Microbiology from the University of the Sunshine Coast. Her research is focused on investigating the genetic diversity, molecular epidemiology and phylogenomics of chlamydial infections in livestock and wildlife, with particular interest in zoonotic events caused by chlamydial pathogens.
Associate Professor Adam Polkinghorne is a molecular microbiologist and the Director of the University of the Sunshine Coast’s Centre for Animal Health Innovation. His work is primarily focussed on understanding the diagnosis, epidemiology and control of chlamydial infections in a variety of wild and domesticated animal species.