The marine mammal microbiome: current knowledge and future directions
Tiffanie M Nelson A F , Amy Apprill B , Janet Mann C , Tracey L Rogers D and Mark V Brown D EA Department of Animal and Range Sciences, Montana State University, Bozeman, MT 59715, USA
B Woods Hole Oceanographic Institution, 266 Woods Hole Road, Mailstop #4, Woods Hole, MA 02543, USA
C Georgetown University, Regents Hall 516, Washington, DC 20057, USA
D Evolution and Ecology Research Centre, University of New South Wales, Kensington, NSW 2052, Australia
E School of Biotechnology and Biomolecular Sciences, University of New South Wales, Kensington, NSW 2052, Australia
F Corresponding author. Tel: +1 406 539 6898, Email: tiffanie.nelson@gmail.com
Microbiology Australia 36(1) 8-13 https://doi.org/10.1071/MA15004
Published: 6 March 2015
Marine mammals are globally significant because of their sensitivity to environmental change and threatened status, often serving as ‘ecosystem sentinels’1. Disease is a major cause of marine mammal population decline and the role of the microbiome in disease has generated considerable interest. Recent research in humans has greatly enhanced our understanding of how the host-associated microbial community, the microbiome, affects host health. In this review, we provide an overview of the extent of the marine mammal microbiome with a focus on whole community characterisation using genomic methods. This research highlights the overlap in microbial communities between geographically distinct species and populations of marine mammals, suggesting tight links between marine mammals and their microbial symbionts over millions of years of evolution. An understanding of these links in both healthy and compromised hosts is essential to identifying at-risk populations and making ecologically appropriate management decisions. We advocate further development of innovative sampling and analytic techniques that advance the field of microbial ecology of marine mammals.
Recent investigations have highlighted the capacity of the microbiome to act strongly and significantly in maintaining host health with a vital role in disease manifestation and immune system function2,3. Members of the microbial community can directly influence the progression of a disease via infection and also modulate the host’s own immune system regulation and response4. Indeed the host’s microbial partners are essential to immune system function. The microbiome has been observed to be species-specific in a variety of vertebrate hosts5–7 and is influenced by host phylogeny, as a result of millions of years of co-evolution8. Marine mammals represent unique evolutionary lineages and investigations into their associated microbes will provide a deeper understanding of their ecology and evolution.
Marine mammals form a diverse group of 129 species in three orders, and of those, 28 are considered endangered or threatened9. Disease is one of the main causes of death in marine mammals and some populations have suffered mass mortalities caused by bacterial pathogens10. Bacteria exist as part of the normal, or even beneficial, flora associated with a host, fluctuating and changing with a host’s physiology and metabolism11. In mammals, disease can occur under a number of different circumstances, most commonly on occasions when the host’s immune system is compromised. For marine mammals, susceptibility to pathogens may be particularly elevated due to anthropogenic stressors such as depleted food resources, habitat degradation and chemical or sound exposure12–15. Additionally, succession events occurring after an initial bacterial infection may lead to dysbiosis, and alterations in the host’s microbiome may be a better predictor of disease progression than following the presence of individual pathogenic agents16. Hence, we need to establish baseline data on microorganisms commonly associated with marine mammals in order to detect anomalies. In the last decade genomic sequencing technologies have provided a previously unrecognised diversity of microorganisms in numerous diverse habitats. In this brief review we highlight the current knowledge of the microbial composition in associations with marine mammals with a focus on whole community characterisation.
Skin microbiome
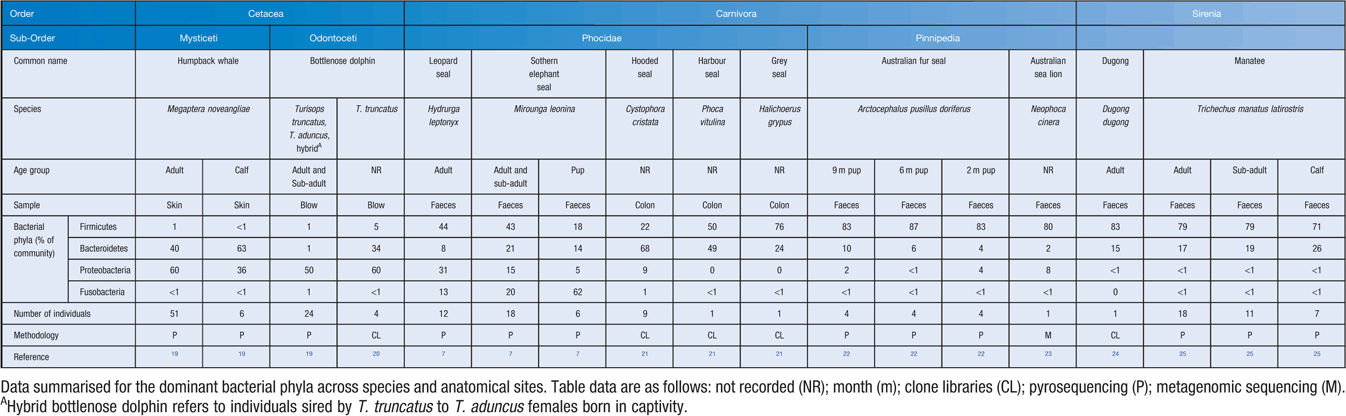
Skin, as the largest organ of mammals, serves as a thick physical barrier that provides defense against the surrounding marine environment. Marine mammal skin is prone to lesions and disorders, however the role of microorganisms in these conditions is still largely unresolved and knowledge is primarily founded on cultivation-based studies17. The recent application of cultivation-independent sequencing-survey approaches to humpback whale (Megaptera novaeangliae) skin has demonstrated that a unique ecosystem of microbes resides on the skin surface (Table 1), which differs from the community present in seawater18.
|
Among populations of humpback whales surveyed in diverse geographic regions, two genera of bacteria (Bacteroidetes genus Tenacibaculum and Gammaproteobacteria genus Psychrobacter) were found to be cosmopolitan and abundant associates on humpback whale skin26. Scanning electron microscopy of humpback whale skin revealed a rich layer of microbial cells on the skin surface26, but as humpback whales regularly undergo skin sloughing through both behavioural27 and physiological activities28 it is possible that the robust Tenacibaculum and Psychrobacter cells may have some means to maintain their residence on the whale skin and could provide benefits to their host. Sequencing survey-based data also demonstrate differences between the skin bacterial associates of healthy and health-compromised humpbacks18,26. Additional data on and study of the skin microbiome might potentially improve our ability to assess health status among free-ranging marine mammals, in particular cetaceans.
Gut microbiome
The gastrointestinal tract is home to an abundant community of microorganisms. The gut microbiome plays a significant role in food breakdown and digestion, the production of essential vitamins and minerals and regulation of the immune system3. In young mammals, the gut microbiome is required for full development of the immune system and maturation of the gut29,30. Studies of the complete gut microbiome of marine mammals include leopard seals (Hydrurga leptonyx), southern elephant seals (Mirounga leonine), grey seals (Halichoerus grypus), hooded seals (Cystophora cristata), harbor seals (Phoca vitulina), Australian fur seals (Arctocephalus pusillus doriferus), Australian sea lions (Neophoca cinerea), Florida manatees (Trichecus manatus latirostris) and dugongs (Dugong dugong). Across all these species the gut microbiome is composed largely of Firmicutes, Bacteroidetes and Proteobacteria (Table 1). Diet and age have been identified as factors that shape the composition of the gut microbiome7,25.
Amongst the seals, the gut microbiome of pinnipeds has a greater abundance of the phylum Firmicutes compared with phocids (Table 1). A ‘core’ group of microorganisms including the genera Ilyobacter, Psychrilyobacter, Fusobacterium, Bacteroides, Subdolingranulum, Sporobacter, Sutterella, Weisella, Anaerococcus and Campylobacter have been observed within phocid seals7,21,22 whilst their herbivorous relatives, within the order Sirenia, shared members from the order Clostridiales, including the genera Clostridium and Ruminococcus24,25,31. The presence of shared bacterial operational taxonomic units (OTUs) in multiple hosts from different studies highlights the strong phylogenetic influence on microbial assembly.
Respiratory microbiome
Respiratory illnesses such as pneumonia are a major cause of mortality in both wild and captive marine mammals32. The cetacean upper respiratory tract terminates in a blowhole, positioned at the top of the head. This feature is a unique adaptation to life in the marine environment, and allows airways to be effectively sealed off from seawater. Upon surfacing, cetaceans forcefully exhale and in the process eject a substance termed blow (also called condensed respiratory vapor or exhaled breath condensate). This material has been shown to harbour potential pathogens in whales33 and has also been used to characterise the normal respiratory-associated microbiome residing in the upper respiratory tract of bottlenose dolphins19,20 (see collection methods in Figure 1). Members of the bacterial genera Plesiomonas, Aeromonas, Escherichia, Clostridium and Pseudomonas, Burkholdaria, Mycobacterium, Haemophylis, Streptococcus and Staphylococcus (including multiple resistant Staphylococcus aureus) have been detected in both sick/dead34 and healthy, free-ranging cetaceans20,33,35.

|
Blow samples from both free-ranging Tursiops truncatus and captive T. aduncus and T. truncatus were dominated by three novel dolphin associated clades (termed DAC 1, 2 and 3) within the Cardiobacteraceae lineage of the Gammaproteobacteria19,20. The Cardiobacteraceae are facultative anaerobic, Gram-negative rod-shaped cells, members of which form part of the commensal microbiome of humans, and whose growth is enhanced by the presence of carbon dioxide36, which occurs in high abundances at the termination of the respiratory tract. Representatives from each of DAC 1, 2, and 3 have been present in every bottlenose dolphin surveyed thus far, although the majority of sequences are associated with DAC 3, indicating this is likely a ubiquitous and critical component of the dolphin respiratory system. Other ‘core’ taxa associated with the dolphin respiratory microbial community appear to include the Arcobacter, Hydrogenimonaceae, Halotalea, Aquimarina, Helococcus, Mycetocola, Methylococcus and Marinimicrobium19. Temporal analysis of captive dolphins suggests community composition in healthy animals is quite stable and that individual dolphins harbour consistently unique microbial communities19.
Sampling techniques
Sampling of material for microbiological analysis from marine mammals is logistically challenging (reviewed by Hunt et al.37), hence the majority of information on microbial disease comes from captive or stranded animals that are not necessarily representative of the greater wild population. However, current sampling methods (see examples in Figures 1 and 2) still provide considerable insight into the microbiome of marine mammals. Capture by sedation or restraint has been employed on smaller species such as seals and dolphins7,38,39 and has recently been used for some larger whales40. However, there are few opportunities to sample using these methods. It is increasingly common to use biopsy darts for collection of skin and blubber samples for genetic and, now, microbiological studies18,41. Permissions for biopsy sampling can be challenging for some species of marine mammals, and repeated samplings are often not possible for the same individuals. In order to increase existing data on the marine mammal microbiome, logistically feasible, non- or minimally-invasive sampling protocols that are easily reproducible and provide biological material suitable for a range of studies are necessary. For example, respiratory blow can be used to examine host DNA42 and hormone levels43,44 as well as respiratory associated microorganisms19,33,37, while non-invasively collected fecal samples can be used to study host DNA45, prey items46 and the gut microbiome22,23.

|
Future research
It appears likely that there are deep branching clades of bacteria that are uniquely associated with marine mammals and have been conserved throughout the evolution of their hosts. Many bacterial sequences obtained from marine mammal studies have close relatives that originate from other marine mammal species. This has significant implications for the transmission of disease amongst these hosts. As they are usually highly social animals, there are numerous opportunities for the transfer of microorganisms between individuals47. Diseases in marine mammals have also been shown to have their roots in other mammals, including dogs48,49 and humans50. In many cases where disease has caused significant mortality in wild marine mammals, it has been linked to viruses, including morbillivirus, phocine distemper and influenza virus51–55. Despite these links being made there is really very little known regarding the ecological role of viruses in marine mammal hosts.
Further investigations into the factors responsible for shaping the marine mammal microbiome need to be made. Designing studies that control for host variation will allow us to make headway in our understanding of disease manifestation. Studies that focus on the functionality of the microbiome will reveal the interactions between host and the microbial community23,56. In human subjects, similar target investigations have allowed for the development of novel metabolites to treat and prevent disease57. Unlike humans, however, to access adequate biological material, strides need to be taken to develop innovative and non-invasive techniques for the collection of relevant samples from wild populations.
Acknowledgements
We thank Dr Ewa Krzyszczyk and Jillian Wisse for allowing us to use their photographs.
References
[1] Moore, S.E. (2008) Marine mammals as ecosystem sentinels. J. Mammal. 89, 534–540.| Marine mammals as ecosystem sentinels.Crossref | GoogleScholarGoogle Scholar |
[2] Hooper, L.V. et al. (2002) How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307.
| How host-microbial interactions shape the nutrient environment of the mammalian intestine.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XmtF2htLs%3D&md5=efb578b1142fe9730af11d2036c27266CAS | 12055347PubMed |
[3] Bäckhed, F. et al. (2005) Host-bacterial mutualism in the human intestine. Science 307, 1915–1920.
| Host-bacterial mutualism in the human intestine.Crossref | GoogleScholarGoogle Scholar | 15790844PubMed |
[4] Maynard, C.L. et al. (2012) Reciprocal interactions of the intestinal microbiota and immune system. Nature 489, 231–241.
| Reciprocal interactions of the intestinal microbiota and immune system.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhtleru7%2FM&md5=9bf7814594021344c182768fbe8fd752CAS | 22972296PubMed |
[5] Yildirim, S. et al. (2010) Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS ONE 5, e13963.
| Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities.Crossref | GoogleScholarGoogle Scholar | 21103066PubMed |
[6] McKenzie, V.J. et al. (2012) Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J. 6, 588–596.
| Co-habiting amphibian species harbor unique skin bacterial communities in wild populations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XisVGgsbc%3D&md5=63ff16bd0ea016e08856514883ede906CAS | 21955991PubMed |
[7] Nelson, T.M. et al. (2013) Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ. Microbiol. 15, 1132–1145.
| Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXlsFSisbc%3D&md5=88fcb07dbad3902580f87d34259b735aCAS | 23145888PubMed |
[8] Ley, R.E. et al. (2008) Evolution of mammals and their gut microbes. Science 320, 1647–1651.
| Evolution of mammals and their gut microbes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXnt1Oju7o%3D&md5=8c5b2610e301dfa736c5617e679e8279CAS | 18497261PubMed |
[9] Pompa, S. et al. (2011) Global distribution and conservation of marine mammals. Proc. Natl. Acad. Sci. USA 108, 13600–13605.
| Global distribution and conservation of marine mammals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtV2ms7vF&md5=205185faa0c791ff51cec95057cdd079CAS | 21808012PubMed |
[10] Waltzek, T.B. et al. (2012) Marine mammal zoonoses: a review of disease manifestations. Zoonoses Public Health 59, 521–535.
| Marine mammal zoonoses: a review of disease manifestations.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC38jhslWhtQ%3D%3D&md5=50077cf7973e6a47f633e470bc6457deCAS | 22697432PubMed |
[11] Pamer, E.G. (2007) Immune responses to commensal and environmental microbes. Nat. Immunol. 8, 1173–1178.
| Immune responses to commensal and environmental microbes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtF2itbvI&md5=2ccaef33da2f6b994a2b87c427b7b904CAS | 17952042PubMed |
[12] Mos, L. et al. (2006) Chemical and biological pollution contribute to the immunological profiles of free-ranging harbor seals. Environ. Toxicol. Chem. 25, 3110–3117.
| Chemical and biological pollution contribute to the immunological profiles of free-ranging harbor seals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtlSmsLvL&md5=5be9be5c9555f9d676d7af43772c9d10CAS | 17220078PubMed |
[13] Fair, P.A. et al. (2013) Associations between perfluoroalkyl compounds and immune and clinical chemistry parameters in highly exposed bottlenose dolphins (Tursiops truncatus). Environ. Toxicol. Chem. 32, 736–746.
| Associations between perfluoroalkyl compounds and immune and clinical chemistry parameters in highly exposed bottlenose dolphins (Tursiops truncatus).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXkslSltrc%3D&md5=47f311f04cbcf68cc0376f72c0891975CAS | 23322558PubMed |
[14] Kight, C.R. and Swaddle, J.P. (2011) How and why environmental noise impacts animals: an integrative, mechanistic review. Ecol. Lett. 14, 1052–1061.
| How and why environmental noise impacts animals: an integrative, mechanistic review.Crossref | GoogleScholarGoogle Scholar | 21806743PubMed |
[15] Kannan, K. et al. (2007) A comparative analysis of polybrominated diphenyl ethers and polychlorinated biphenyls in southern sea otters that died of infectious diseases and noninfectious causes. Arch. Environ. Contam. Toxicol. 53, 293–302.
| A comparative analysis of polybrominated diphenyl ethers and polychlorinated biphenyls in southern sea otters that died of infectious diseases and noninfectious causes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmvVygsLY%3D&md5=e5aa23f15adcc57f0f9dca7ca94d5919CAS | 17587145PubMed |
[16] Klepac-Ceraj, V. et al. (2010) Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ. Microbiol. 12, 1293–1303.
| Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXntFyitL0%3D&md5=f3467d10cc80aa19fd7ac8159f6f7c8aCAS | 20192960PubMed |
[17] Mouton, M. and Botha, A. (2012) Cutaneous lesions in cetaceans: an indicator of ecosystem status? in New Approaches to the Study of Marine Mammals, A. Romero and E.O. Keith, Editors. InTech.
[18] Apprill, A. et al. (2011) Humpback whales harbour a combination of specific and variable skin bacteria. Environ. Microbiol. Rep. 3, 223–232.
| Humpback whales harbour a combination of specific and variable skin bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXlvFyhtrY%3D&md5=7e74d531e887d510695be0ced9e460c2CAS | 23761254PubMed |
[19] Lima, N. et al. (2012) Temporal stability and species specificity in bacteria associated with the bottlenose dolphins respiratory system. Environ. Microbiol. Rep. 4, 89–96.
| Temporal stability and species specificity in bacteria associated with the bottlenose dolphins respiratory system.Crossref | GoogleScholarGoogle Scholar | 23757234PubMed |
[20] Johnson, W.R. et al. (2009) Novel diversity of bacterial communities associated with bottlenose dolphin upper respiratory tracts. Environ. Microbiol. Rep. 1, 555–562.
| Novel diversity of bacterial communities associated with bottlenose dolphin upper respiratory tracts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXjtFWns7w%3D&md5=3f6c57b1614e6d422b22ef7e18dd61f4CAS | 23765934PubMed |
[21] Glad, T. et al. (2010) Ecological characterisation of the colonic microbiota in arctic and sub-arctic seals. Microb. Ecol. 60, 320–330.
| Ecological characterisation of the colonic microbiota in arctic and sub-arctic seals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtFOqsr%2FN&md5=0835b32ba428cbcf5ae2908a9d7c81d0CAS | 20523986PubMed |
[22] Smith, S.C. et al. (2013) Age-related differences revealed in Australian fur seal Arctocephalus pusillus doriferus gut microbiota. FEMS Microbiol. Ecol. 86, 246–255.
| Age-related differences revealed in Australian fur seal Arctocephalus pusillus doriferus gut microbiota.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhs1eksb7N&md5=a05e448126dbb5e6acfa0f08b0c4c58bCAS | 23746080PubMed |
[23] Lavery, T.J. et al. (2012) High nutrient transport and cycling potential revealed in the microbial metagenome of Australian sea lion (Neophoca cinerea) faeces. PLoS ONE 7, e36478.
| High nutrient transport and cycling potential revealed in the microbial metagenome of Australian sea lion (Neophoca cinerea) faeces.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XnslOqs7s%3D&md5=9aeb567ca83adabccd3cc43e776ab321CAS | 22606263PubMed |
[24] Tsukinowa, E. et al. (2008) Fecal microbiota of a dugong (Dugong dugong) in captivity at Toba Aquarium. J. Gen. Appl. Microbiol. 54, 25–38.
| Fecal microbiota of a dugong (Dugong dugong) in captivity at Toba Aquarium.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXjsVKhu7c%3D&md5=1740a8b3f3bbadf4a3c08b9f5df8da13CAS | 18323679PubMed |
[25] Merson, S.D. et al. (2014) Variation in the hindgut microbial communities of the Florida manatee, Trichechus manatus latirostris over winter in Crystal River, Florida. FEMS Microbiol. Ecol. 87, 601–615.
| Variation in the hindgut microbial communities of the Florida manatee, Trichechus manatus latirostris over winter in Crystal River, Florida.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXjsFGgtr8%3D&md5=85929a59134422c0b3013f6504cdd1efCAS | 24215517PubMed |
[26] Apprill, A. et al. (2014) Humpback whale populations share a core skin bacterial community: towards a health index for marine mammals? PLoS ONE 9, e90785.
| Humpback whale populations share a core skin bacterial community: towards a health index for marine mammals?Crossref | GoogleScholarGoogle Scholar | 24671052PubMed |
[27] Clapham, P.J. et al. (1993) High-energy behaviors in humpback whales as a source of sloughed skin for molecular analysis. Mar. Mamm. Sci. 9, 213–220.
| High-energy behaviors in humpback whales as a source of sloughed skin for molecular analysis.Crossref | GoogleScholarGoogle Scholar |
[28] Durban, J.W. and Pitman, R.L. (2012) Antarctic killer whales make rapid, round-trip movements to subtropical waters: evidence for physiological maintenance migrations? Biol. Lett. 8, 274–277.
| Antarctic killer whales make rapid, round-trip movements to subtropical waters: evidence for physiological maintenance migrations?Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC383psFagtw%3D%3D&md5=9307aa5e673b16468736bded5413507bCAS | 22031725PubMed |
[29] Palmer, C. et al. (2007) Development of the human infant intestinal microbiota. PLoS Biol. 5, e177.
| Development of the human infant intestinal microbiota.Crossref | GoogleScholarGoogle Scholar | 17594176PubMed |
[30] Vael, C. and Desager, K. (2009) The importance of the development of the intestinal microbiota in infancy. Curr. Opin. Pediatr. 21, 794–800.
| The importance of the development of the intestinal microbiota in infancy.Crossref | GoogleScholarGoogle Scholar | 19770768PubMed |
[31] Eigeland, K. (2012) Bacterial community structure in the hindgut of wild and captive dugongs (Dugong dugon). Aquat. Mamm. 38, 402–411.
| Bacterial community structure in the hindgut of wild and captive dugongs (Dugong dugon).Crossref | GoogleScholarGoogle Scholar |
[32] Venn-Watson, S. et al. (2012) Thirty year retrospective evaluation of pneumonia in a bottlenose dolphin Tursiops truncatus population. Dis. Aquat. Organ. 99, 237–242.
| Thirty year retrospective evaluation of pneumonia in a bottlenose dolphin Tursiops truncatus population.Crossref | GoogleScholarGoogle Scholar | 22832722PubMed |
[33] Acevedo-Whitehouse, K. et al. (2010) A novel non-invasive tool for disease surveillance of free-ranging whales and its relevance to conservation programs. Anim. Conserv. 13, 217–225.
| A novel non-invasive tool for disease surveillance of free-ranging whales and its relevance to conservation programs.Crossref | GoogleScholarGoogle Scholar |
[34] Buck, C.D. and Schroeder, J.P. (1990) Public health significance of marine mammal disease, in Handbook of Marine Mammal Medicine, L.A. Dierauf, Editor. CRC Press, Boca Raton, FL. pp. 163–173.
[35] Morris, P.J. et al. (2011) Isolation of culturable microorganisms from free-ranging bottlenose dolphins (Tursiops truncatus) from the southeastern United States. Vet. Microbiol. 148, 440–447.
| Isolation of culturable microorganisms from free-ranging bottlenose dolphins (Tursiops truncatus) from the southeastern United States.Crossref | GoogleScholarGoogle Scholar | 20888150PubMed |
[36] Savage, D.D. et al. (1977) Cardiobacterium hominis endocarditis: description of two patients and characterization of the organism. J. Clin. Microbiol. 5, 75–80.
| 1:STN:280:DyaE2s%2FpsVWmuw%3D%3D&md5=ccc6b33a63536ccc63dee335ea4efd76CAS | 833269PubMed |
[37] Hunt, K.E. et al. (2013) Overcoming the challenges of studying conservation physiology in large whales: a review of available methods. Conservation Physiology 1, cot006.
| Overcoming the challenges of studying conservation physiology in large whales: a review of available methods.Crossref | GoogleScholarGoogle Scholar |
[38] Ortiz, R.M. and Worthy, G.A.J. (2000) Effects of capture on adrenal steroid and vasopressin concentrations in free-ranging bottlenose dolphins (Tursiops truncatus). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 125, 317–324.
| Effects of capture on adrenal steroid and vasopressin concentrations in free-ranging bottlenose dolphins (Tursiops truncatus).Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3czosFGrsw%3D%3D&md5=81ee0fe9bd68b3a7614be471b81cdb33CAS | 10794960PubMed |
[39] Fair, P.A. et al. (2014) Stress response of wild bottlenose dolphins (Tursiops truncatus) during capture-release health assessment studies. Gen. Comp. Endocrinol. 206, 203–212.
| Stress response of wild bottlenose dolphins (Tursiops truncatus) during capture-release health assessment studies.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhsV2itr7O&md5=325338b2089ea37a7cf166c9c41a75deCAS | 25019655PubMed |
[40] St Aubin, D.J. et al. (2001) Hematology and plasma chemistry as indicators of health and ecological status in beluga whales, Delphinapterus leucas. Arctic 54, 317–331.
| Hematology and plasma chemistry as indicators of health and ecological status in beluga whales, Delphinapterus leucas.Crossref | GoogleScholarGoogle Scholar |
[41] Palsbøll, P.J. et al. (1997) Genetic tagging of humpback whales. Nature 388, 767–769.
| Genetic tagging of humpback whales.Crossref | GoogleScholarGoogle Scholar | 9285587PubMed |
[42] Frère, C.H. et al. (2010) Thar she blows! A novel method for DNA collection from cetacean blow. PLoS ONE 5, e12299.
| Thar she blows! A novel method for DNA collection from cetacean blow.Crossref | GoogleScholarGoogle Scholar | 20811619PubMed |
[43] Hogg, C.J. et al. (2005) Determination of testosterone in saliva and blow of bottlenose dolphins (Tursiops truncatus) using liquid chromatography-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 814, 339–346.
| Determination of testosterone in saliva and blow of bottlenose dolphins (Tursiops truncatus) using liquid chromatography-mass spectrometry.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhvFyisw%3D%3D&md5=334644c1a012bc6ba3e7578869b14210CAS | 15639457PubMed |
[44] Hunt, K.E. et al. (2014) Detection of steroid and thyroid hormones via immunoassay of North Atlantic right whale (Eubalaena glacialis) respiratory vapor. Mar. Mamm. Sci. 30, 796–809.
| Detection of steroid and thyroid hormones via immunoassay of North Atlantic right whale (Eubalaena glacialis) respiratory vapor.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXlvFWhtbc%3D&md5=67a44cdeaf1a1cdd59c8e7275148f83aCAS |
[45] Green, M.L. et al. (2007) Noninvasive methodology for the sampling and extraction of DNA from free-ranging Atlantic spotted dolphins (Stenella frontalis). Mol. Ecol. Notes 7, 1287–1292.
| Noninvasive methodology for the sampling and extraction of DNA from free-ranging Atlantic spotted dolphins (Stenella frontalis).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXlslSlsg%3D%3D&md5=ea1b17de2d57700be19a04ed71eb6c4eCAS |
[46] Deagle, B.E. et al. (2009) Analysis of Australian fur seal diet by pyrosequencing prey DNA in faeces. Mol. Ecol. 18, 2022–2038.
| Analysis of Australian fur seal diet by pyrosequencing prey DNA in faeces.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXmsVektLY%3D&md5=9aa0c0fef4b3199e887fae5eeb87b34aCAS | 19317847PubMed |
[47] Lombardo, M. (2008) Access to mutualistic endosymbiotic microbes: an underappreciated benefit of group living. Behav. Ecol. Sociobiol. 62, 479–497.
| Access to mutualistic endosymbiotic microbes: an underappreciated benefit of group living.Crossref | GoogleScholarGoogle Scholar |
[48] Butina, T.V. et al. (2010) Canine distemper virus diversity in Lake Baikal seal (Phoca sibirica) population. Vet. Microbiol. 144, 192–197.
| Canine distemper virus diversity in Lake Baikal seal (Phoca sibirica) population.Crossref | GoogleScholarGoogle Scholar | 20083361PubMed |
[49] Greig, D.J. et al. (2014) Surveillance for zoonotic and selected pathogens in harbor seals Phoca vitulina from central California. Dis. Aquat. Organ. 111, 93–106.
| Surveillance for zoonotic and selected pathogens in harbor seals Phoca vitulina from central California.Crossref | GoogleScholarGoogle Scholar | 25266897PubMed |
[50] Osterhaus, A.D. (2000) Influenza B virus in seals. Science 288, 1051–1053.
| Influenza B virus in seals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXjsVSnsbo%3D&md5=dd20fcfd46a74023944c805a8d7c3cd5CAS | 10807575PubMed |
[51] Thompson, P.M. and Miller, D. (1992) Phocine distemper virus outbreak in the Moray Firth common seal population: an estimate of mortality. Sci. Total Environ. 115, 57–65.
| Phocine distemper virus outbreak in the Moray Firth common seal population: an estimate of mortality.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK383nvFOlsQ%3D%3D&md5=675d35b37ea1da225a7a2817977f9d6bCAS | 1594935PubMed |
[52] Van Bressem, M.F. et al. (2014) Cetacean morbillivirus: current knowledge and future directions. Viruses 6, 5145–5181.
| 1:CAS:528:DC%2BC2MXhvVGmtL4%3D&md5=f7af76ee527ced39a79aac9b0ee8c650CAS | 25533660PubMed |
[53] Pollack, J.D. (2001) Caspian seal die-off is caused by canine distemper virus. Trends Microbiol. 9, 108.
| Caspian seal die-off is caused by canine distemper virus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXlvFentLY%3D&md5=dd1298cf67e41da635716289ec9ad795CAS | 11239773PubMed |
[54] Anthony, S.J. et al. (2012) Emergence of fatal avian influenza in New England harbor seals. MBio 3, e00166–12.
| Emergence of fatal avian influenza in New England harbor seals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhtlaht7nF&md5=0ea14bbf58973014ba369aa49a9852c2CAS | 22851656PubMed |
[55] Ramis, A.J. et al. (2012) Influenza A and B virus attachment to respiratory tract in marine mammals. Emerg. Infect. Dis. 18, 817–820.
| Influenza A and B virus attachment to respiratory tract in marine mammals.Crossref | GoogleScholarGoogle Scholar | 22516350PubMed |
[56] Stewart, J.R. et al. (2014) Survey of antibiotic-resistant bacteria isolated from bottlenose dolphins Tursiops truncatus in the southeastern USA. Dis. Aquat. Organ. 108, 91–102.
| Survey of antibiotic-resistant bacteria isolated from bottlenose dolphins Tursiops truncatus in the southeastern USA.Crossref | GoogleScholarGoogle Scholar | 24553415PubMed |
[57] Donia, M.S. et al. (2014) A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414.
| A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhsFCgtLvP&md5=12eafae0d6ac6f77c152d8395756ede3CAS | 25215495PubMed |
Biographies
Tiffanie Nelson is a researcher from Australia currently undertaking a postdoctoral fellowship at Montana State University, Bozeman, USA. Tiff is a microbial ecologist, who focuses on the microbiome of marine mammals as well as humans and environmental samples. Her interests are in health and disease associated with the microbiome. Tiff’s current project is investigating the vaginal tract microbiome of women in relation to bacterial vaginosis using both culture-dependant and -independent methods.
Amy Apprill is a researcher at the Woods Hole Oceanographic Institution in Massachusetts, USA. Amy is a marine microbiologist researching questions that focus on the contribution of microorganisms to the health and ecology of marine animals. Amy is also interested in how animal-associated microbes reflect the alterations occurring in their surrounding marine environment. Her current research uses a combination of field measurements and observations and laboratory experiments and relies on diverse methodology (cultivation, genomic, metagenomic and bioinformatic) to examine the microbiomes of reef-building corals and marine mammals.
Janet Mann is professor of biology and psychology and vice provost for research at Georgetown University, Washington DC, USA. Janet has expertise in the field of animal behavior with extensive research focusing on marine mammals. Her work has focused on social networks, female reproduction, calf development, life history, conservation, tool-use, social learning and culture among bottlenose dolphins in Shark Bay, Australia. Her long-term study ‘The Shark Bay Dolphin Research Project’, tracks over 1600 dolphins throughout their lives and includes an international team on three continents where each group studies different aspects of delphinid biology.
Tracey Rogers is associate professor at the University of New South Wales. Tracey works across a diverse range of research fields with many years of experience working in Antarctica with marine mammals. The common theme in Tracey’s research is in attempting to understand how mammals respond to change. Tracey uses multidisciplinary approaches to understand the ecology of mammals. Most of her work uses models and techniques with captive populations for applications in field settings. Other techniques include stable isotope analysis, satellite telemetry and acoustics.
Mark Brown is a senior research fellow at the University of New South Wales, Sydney, Australia. He has extensive expertise in research that focuses on microbes (Bacteria, Archaea and microbial Eukaryotes), primarily from marine environments. Mark’s main interest is in investigating how microbes interact with each other and their environment to form communities that sustain critical ecosystem processes. His current research couples innovative in situ sampling methods, genetic tools, bioinformatics and ecological theory to elucidate and predict the form, function and impact of microbes in rapidly changing ecosystems.


