Arrays in clinical microbiology
Joanne MercerDepartment of Microbiology and Infectious Diseases
Sydney South West Pathology, Liverpool
Locked Bag 7090
Liverpool BC, NSW 1871, Australia
Tel: +61 2 9828 5124
Fax: +61 2 9828 5129
Email: joanne.mercer@sswahs.nsw.gov.au
Microbiology Australia 34(4) 184-185 https://doi.org/10.1071/MA13063
Published: 31 October 2013
Application of molecular techniques in the diagnostic microbiology laboratory for the detection of pathogens continues to develop and expand. Typically, nucleic acid amplification is utilised to generate large amounts of target nucleic acid to facilitate detection. Amplification of either universal targets by broad range PCR or specific targets may be combined with detection in real time or post-amplification hybridisation. Simultaneous detection of large numbers of microbial targets or resistance genes may be achieved using microarray technology, or the newly developed FilmArray system. This automated nested PCR with melt curve analysis is a promising tool for the diagnostic microbiology laboratory.
Detection of amplified products in real-time PCR with the use of fluorescent dyes or labelled probes has increased substantially in recent years and provides rapid, sensitive and specific detection of either single or multiple targets. A major advantage of real-time PCR is that handling of amplified material is not required, substantially reducing the risk of amplicon contamination in the laboratory.
A number of commercially available platforms (e.g. Genexpert, BDmax) that enable automated extraction, amplification and analyte detection in the one system are now available, and reduce hands-on time for laboratory scientists. Generally these systems are simple to use and provide rapid (<2 hours) results making them suitable to integrate into non-specialised molecular laboratories. Multiplexing large numbers of targets may however be technically difficult and be largely dictated by the capabilities of detection instrumentation. Detection of an extended number of targets may require multiple amplification reactions increasing setup time and reducing throughput.
An alternate detection methodology that is gaining increased interest is that of microarrays. Generally, a DNA array contains hundreds to thousands of DNA fragments attached at fixed locations on a solid support such as a glass slide, silicon chip or bead. Hybridisation with DNA, mRNA or amplified products from the test sample allows the analysis of many genes or targets. This technique has proved extremely valuable in gene expression analysis (both within a single sample and as a comparison between cell types) where the expression of thousands of genes may be examined at once1. Research in the microbiology field has also been enhanced with microarrays used for studies of microbial expression, host gene expression, mutation analysis, identification of emerging pathogens, detection of known pathogens, typing of organisms and drug resistance detection2.
Application of array technology in the diagnostic microbiology laboratory is also promising with the availability of commercially manufactured low and middle density arrays for the detection of large numbers of microbial genetic targets. Detection follows PCR amplification of specific gene targets, often covering both bacterial and viral pathogens in the one assay. The potential to detect 100 or more targets in the one assay has broadened the utility of molecular testing considerably. Assays are commonly ‘specimen or syndrome based’ in design and enable multiple results to be available to the clinician from the one assay on a single sample.
The NanoCHIP XL system (Savyondiagnostics), for example, offers several diagnostic panels including gastrointestinal pathogens (bacteria and parasites), sexually transmitted infections, infection control and respiratory pathogens. This system is an electronic microarray using electric fields to control nucleic acid transport. A positive current is applied to designated test sites on the array attracting negatively charged nucleic acids (probes). Bonds are formed between a streptavidin coating on the array surface and the biotinylated probes. Probes for additional targets may be added in the same manner. Fluorescently labelled target DNA is then added and, if present, hybridises with the appropriate immobilised probes on the array. Fluorescent reporters on bound DNA identify positive tests. This flexible system allows one specimen to be hybridised to multiple test sites and multiple samples to be analysed on the same array cartridge.
The use of the ‘liquid-bead suspension’ array of the Luminex system has also proved of great benefit in identifying microbial pathogens. This three-dimensional, low-density array utilises microscopic spheres or beads as the solid support and may be probed for up to 100 targets at a time using xTAG technology. Beads containing dyes of a unique spectral profile are bound to oligonucleotides of defined sequences. A mixture of such bead complexes may then be used to test for targets of interest generated by multiple PCR (and containing sequences complimentary to the bound oligonucleotides). Solutions containing hybridised bead–oligo–target complexes are subjected to flow cytometry and identified by the emission spectrum of the dye within the bead.
Array technologies are most valuable when screening for a large number of pathogens (e.g. detection of multiple respiratory viruses), or when multiple methodologies can be replaced by using one array assay (e.g. gastrointestinal pathogens, including bacteria, viruses and parasites). Processing relies less on individual test requests and reduces the need for complex laboratory testing algorithms. Comparable performance with that of real-time PCR is reported3 (though real-time PCR appears superior for adenovirus detection), and the detection of multiple pathogens in the one sample is not uncommon. Availability of a comprehensive range of results in 5 hours after minimal hands-on time provides substantial improvement to both laboratory workflow and turnaround time, and enables timely patient management.
However, one potential disadvantage of the microarray when compared with real-time PCR, is the contamination risk introduced through manipulation of amplified material during hybridisation and washing steps. Stringent decontamination and quality control procedures are required to ensure the validity of results.
A recently described system that obviates this problem and offers the detection of an exciting range of pathogens in a diagnostic setting is the FilmArray (Biofire Diagnostics)4. Nucleic acid purification, reverse transcription, nested multiplex PCR and amplicon melting curve analysis of more than 100 different nucleic acid targets at one time may be performed in an automated, self-enclosed system (Figure 1). Sample processing and detection is conducted in the FilmArray pouch, which contains multiple reagent reservoirs, channel-linked processing stations and a final stage 102 welled array for the second PCR stage. Freeze-dried reagents remain under vacuum, maintaining the integrity of the reagents and allowing simple delivery of sample and hydration solution. Movement of liquid through the pouch is controlled by pneumatic elements within the FilmArray instrument. Minimal sample setup time is required (<5 minutes), and results of a comprehensive range of pathogen targets are available in about an hour. System performance has been validated with the detection of a panel of respiratory pathogens, with performance/sensitivity generally comparable to real-time PCR5 and Luminex microarray detection6. Panels for the detection of gastrointestinal pathogens (viral, bacterial, protozoan) and blood pathogens (bacteria, fungi, resistance genes) are under development.
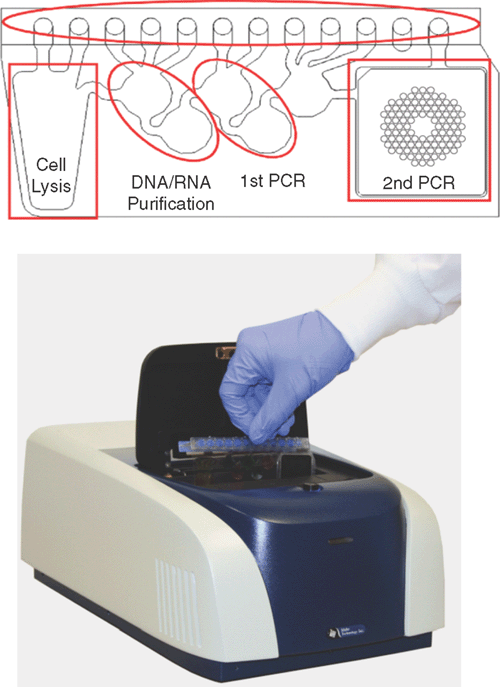
|
Although the limited throughput of this system in its current form may preclude replacing existing methods in high throughput laboratories, its ease of use and rapid turnaround time make it an attractive option for rapid testing in select patient groups, out of hours testing or processing in the smaller laboratory. The full potential of this system may be assessed as further applications are developed. Certainly, the increased availability of array style detection products enables the diagnostic laboratory to consider comprehensive multi-target detection approaches.
References
[1] Miller, M. and Tang, Y. (2009) Basic concepts of microarrays and potential applications in clinical microbiology. Clin. Microbiol. Rev. 22, 611–633.| Basic concepts of microarrays and potential applications in clinical microbiology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsVyhsrfP&md5=405debb2d022fcb77ebabde0ac0d1325CAS | 19822891PubMed |
[2] Stover, A. et al. (2004) Hybridization array technology. In Molecular Microbiology, Diagnostic Principles and Practice (Persing, D., ed), 619–639, ASM Press.
[3] Pabbaraju, K. et al. (2008) Comparison of the Luminex xTAG respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J. Clin. Microbiol. 46, 3056–3062.
| Comparison of the Luminex xTAG respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFOku7vK&md5=e79054dc843b70801d1a862f075e90ddCAS | 18632902PubMed |
[4] Poritz, M. et al. (2011) FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS ONE 6, e26047.
| FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhsVaksr3F&md5=7df20960d63e5632626717b2e6c4ff1bCAS | 22039434PubMed |
[5] Loeffelholz, M. et al. (2011) Comparison of the FilmArray respiratory panel and Prodesse real-time PCR assays for the detection of respiratory pathogens. J. Clin. Microbiol. 49, 4083–4088.
| Comparison of the FilmArray respiratory panel and Prodesse real-time PCR assays for the detection of respiratory pathogens.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC38%2FjtVGqug%3D%3D&md5=901169fe54ea48b11c9454652dd1ac01CAS | 21998418PubMed |
[6] Rand, K. et al. (2011) Comparison of two multiplex methods for detection of respiratory viruses: Filmarray RP and xTAG RVP. J. Clin. Microbiol. 49, 2449–2453.
| Comparison of two multiplex methods for detection of respiratory viruses: Filmarray RP and xTAG RVP.Crossref | GoogleScholarGoogle Scholar | 21508156PubMed |
Biography
Joanne Mercer is a senior scientist in the Department of Microbiology and Infectious Diseases, Sydney South West Pathology Service, Liverpool. She is responsible for the implementation and performance of diagnostic molecular microbiology assays and has a particular interest in enteric parasitology and bacteriology.


