Digital PCR: a new DNA quantification tool
Kerry R EmslieNational Measurement Institute
PO Box 264, Lindfield
NSW 2070, Australia
Tel: +61 2 8467 3700
Fax: +61 2 8467 3756
Email: Kerry.emslie@measurement.gov.au
Microbiology Australia 34(4) 178-179 https://doi.org/10.1071/MA13059
Published: 4 September 2013
Digital polymerase chain reaction (PCR) is a quantitative PCR technique with potential to be more accurate and precise than real-time quantitative PCR without the need for a standard curve. The digital PCR sample/reaction mix is randomly distributed into a large number of partitions, such that some partitions contain no copies of the nucleic acid template and others contain at least one copy. Following thermal cycling, partitions containing amplified product can be distinguished by the increased fluorescence generated from the probe. Quantification is achieved by counting the number of positive and negative partitions, followed by Poisson modelling to account for partitions containing more than one template molecule. The partitioning process effectively reduces competition between very similar templates resulting in increased sensitivity for detection of rare mutants in a wild-type background. Digital PCR has been used as a reference method for characterising standards while applications in microbiology include monitoring viral load and detecting bacteria or rare mutant alleles.
The concept of using limiting dilution combined with end-point PCR and Poisson statistics was first introduced in 1992 by Sykes et al.1 from Flinders Medical Centre, South Australia, who measured the amount of rearranged immunoglobulin heavy chain to quantify leukemic cells using a small set of 10 reactions. The term ‘digital PCR’ was later coined by Vogelstein and Kinzler in 19992, reflecting the ability of digital PCR to transform the more familiar exponential real-time PCR signal into a digital readout. However, until the availability of commercial platforms based on microfluidic and/or emulsion technology, advances in digital PCR were limited due to the technical challenges and costs associated with conducting a large number of independent reactions.
Today there are several commercial platforms that automate the partitioning process, splitting the sample into close to 1000 and up to 1,000,000’s of individual partitions. Depending on the platform, the partition volume is in the nanolitre to picolitre range, thus maintaining the total reaction volume in the microlitre range typically used in real-time PCR assays. Following thermal cycling, the fraction of positive partitions is determined using a chip image reader or by streaming emulsion droplets past a droplet reader.
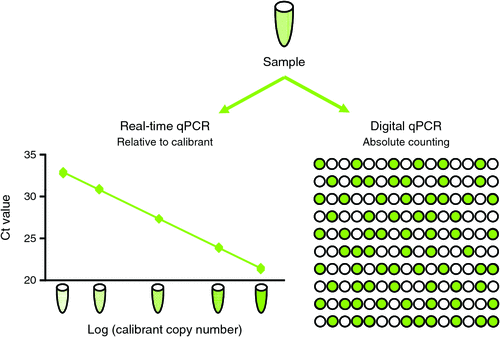
Digital PCR offers certain advantages over real-time quantitative PCR due to three key features (Fig. 1). First, the counting process fundamental to digital PCR is independent of a DNA calibrant and utilises end-point PCR data. This has the potential to improve reproducibility between laboratories and across time, reduce the impact of inhibitors and enable traceability to the International System of Units. Second, since confidence in the Poisson modelling improves with increasing partition number, commercial platforms can generate very precise data enabling discrimination of samples with small differences in template amount. Last, partitioning effectively reduces competition between very similar templates, increasing the sensitivity of detecting rare mutants in a wild-type background. A recent study on human immunodeficiency virus in over 300 clinical samples concluded that digital PCR provided improved sensitivity and precision over real-time PCR and was also less sensitive to mismatches in the primer and probe sequences3. However, accurate digital PCR quantification requires independent partitioning of molecules and successful amplification in partitions containing the target sequence. DNA containing concatemers or linked copies must be digested prior to analysis; otherwise the true copy number will be underestimated since linked copies will partition together. Recently published ‘The digital MIQE guidelines: Guidelines for Minimum Information for Publication of Quantitative Digital PCR Experiments’ contain a checklist for designing and recording digital PCR experiments4.
|
While digital PCR is an absolute measurement technique, this does not obviate the need to conduct a thorough method validation process using well-characterised standards or DNA reference materials with a known number of copies of target DNA. As for real-time PCR, primer design is critical for assay selectivity and thermal cycling conditions must be optimised to maximise the difference in fluorescence intensity between negative and positive partitions. Generally, amplicon cannot be recovered from individual partitions so if any partitions do not fall clearly within the negative or positive population, it is difficult to determine whether this is due to non-specific amplification, inefficient amplification from damaged template or other reasons. This ambiguity is particularly critical for rare mutation detection as misclassification of partitions could lead to a false positive result. Hence, data generated from analysis of DNA standards should be used to determine the assay limit of detection and controls should be used for ongoing assessment of the method performance.
To assess the use of digital PCR as a reference method, the National Measurement Institute of Australia completed extensive validation. Using several different DNA templates, we demonstrated that digital PCR data is in agreement with results obtained using an orthogonal reference measurement technique, isotope dilution mass spectrometry taking into consideration the uncertainty of each measurement technique5–7. A digital PCR reference method is now often used by the National Measurement Institute and other metrology institutes to assign the concentration value of nucleic acid reference materials8. For instance, we have recently prepared a DNA reference material calibrant for real-time PCR assays detecting toxic cyanobacteria. In the short term, increased use of this technology to characterise nucleic acid reference materials will facilitate the availability of appropriate reference materials to support real-time quantitative PCR measurements in clinical and research laboratories.
Currently, implementation of digital PCR in clinical and research laboratories is limited predominantly due to cost and logistics of a high throughput workflow. If second generation digital PCR instruments can overcome these limitations, digital PCR could be integrated into routine testing laboratories alongside or replacing real-time PCR for specific applications.
Acknowledgement
Work at the National Measurement Institute has been conducted with support from the Australian Commonwealth Government’s National Enabling Technology Strategy.
References
[1] Sykes, P.J. et al . (1992) Quantitation of targets for PCR by use of limiting dilution. Biotechniques 13, 444–449.| 1:CAS:528:DyaK3sXhslSksw%3D%3D&md5=6b624f5699339835c819168c94dd8b1bCAS | 1389177PubMed |
[2] Vogelstein, B. and Kinzler, K.W. (1999) Digital PCR. Proc. Natl. Acad. Sci. USA 96, 9236–9241.
| Digital PCR.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXltVequrY%3D&md5=ebd2baf154f7d9bf37884b0748f7cb7bCAS | 10430926PubMed |
[3] Strain, M.C. et al. (2013) Highly precise measurement of HIV DNA by droplet digital PCR. PLoS ONE 8, e55943.
| Highly precise measurement of HIV DNA by droplet digital PCR.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXmtFWrtLY%3D&md5=12802ac8f92cf559829010c8c3403cc2CAS | 23573183PubMed |
[4] Huggett, J.F. et al. (2013) The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin. Chem. 59, 892–902.
| The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXpslKgtbo%3D&md5=2d2ca3075375b6515823200d74c38706CAS | 23570709PubMed |
[5] Bhat, S. et al. (2010) Comparison of methods for accurate quantification of DNA mass concentration with traceability to the International System of Units. Anal. Chem. 82, 7185–7192.
| Comparison of methods for accurate quantification of DNA mass concentration with traceability to the International System of Units.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXpvVGnsLs%3D&md5=16a3b611687416e8e3211ad0e9570d31CAS | 20690645PubMed |
[6] Burke, D.G. et al. (2013) Digital polymerase chain reaction measured pUC19 marker as calibrant for HPLC measurement of DNA quantity. Anal. Chem. 85, 1657–1664.
| Digital polymerase chain reaction measured pUC19 marker as calibrant for HPLC measurement of DNA quantity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhslylur3K&md5=87f6098fc1ce2b23b6ce07445d831c5dCAS | 23215355PubMed |
[7] Pinheiro, L.B. et al. (2012) Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 84, 1003–1011.
| Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhsFaktbzI&md5=53d72f5c50d207961b021e7ff44f20d1CAS | 22122760PubMed |
[8] Haynes, R.J. et al. (2013) Standard reference material 2366 for measurement of human cytomegalovirus DNA. J. Mol. Diagn. 15, 177–185.
| Standard reference material 2366 for measurement of human cytomegalovirus DNA.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXivFyjsr4%3D&md5=a36fcf5869a77eb570d44029c60300ccCAS | 23321018PubMed |
Biography
Kerry Emslie is Manager, Bioanalysis Group at the National Measurement Institute. Kerry’s team address some of the many challenges that face the analytical community in working towards comparable and traceable biomeasurement. Most recently, her laboratory has validated measuring systems and techniques, including digital PCR, and is using these to characterise DNA reference materials for a variety of applications.


