Staphylococcus aureus and Streptococcus pyogenes in the north: distinctively different
Deborah Holt A B * and Philip Giffard A BA College of Health and Human Sciences, Charles Darwin University, Darwin, NT 0811, Australia.
B Tropical and Emerging Infectious Diseases Division, Menzies School of Health Research, Charles Darwin University, Darwin, NT 0811, Australia.

Deborah Holt is a senior lecturer at Charles Darwin University and Honorary Research Fellow at the Menzies School of Health Research. She is a molecular biologist whose research focuses on the molecular epidemiology and pathogenesis of skin pathogens with importance in Indigenous communities in northern Australia. |

Phil Giffard is Head of Laboratory Science at the Menzies School of Health Research and Head of Biomedical Science in the College of Health and Human Science at Charles Darwin University. He has a long-standing research interested in microbial genotyping technology and associated bioinformatic methods. |
Microbiology Australia 43(3) 104-107 https://doi.org/10.1071/MA22034
Submitted: 28 June 2022 Accepted: 28 August 2022 Published: 20 September 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Staphylococcus aureus and Streptococcus pyogenes are important contributors to disease in northern Australia. Both are opportunistic pathogens, frequently carried on the skin or in the respiratory tract in the absence of disease. A large proportion of the S. aureus strains causing infection in northern Australia possess the Panton Valentine (PVL) toxin, with ST93, ST5, and ST121 being significant. PVL+ strains are associated with both community- and healthcare-associated infections, and a large proportion are methicillin-resistant S. aureus (MRSA). MRSA strains known to be healthcare associated (ST239 and ST22) are not prevalent. CC1 PVL− MRSA continue to cause infections. The diversity of S. pyogenes emm types in northern Australia is high with skin tropic and non-tropic emm types predominating. This contrasts with other parts of Australia where emm diversity is lower and rates of pharyngitis higher. The high diversity raises concerns for the likely efficacy of vaccines based on the variable region of the M protein, the nucleotide sequence of which underpins emm typing. It is likely that complex interactions occur between these two important bacterial pathogens, and other important skin pathogens in the region such as the scabies mite.
Keywords: emm typing, M protein, Panton Valentine leucocidin, PVL, pyoderma, rheumatic heart disease, skin infections, Staphylococcus aureus, Streptococcus pyogenes.
Staphylococcus aureus and Streptococcus pyogenes are firmicutes, within the Bacillales and Lactobacillales respectively. Both are associated with asymptomatic colonisation but are also human pathogens of global significance, causing a range of infections from superficial to life-threatening invasive disease. They are important causes of skin infections in northern Australia1 with distinct differences in epidemiology to that seen in other parts of the country.
S. aureus has long been one of the most notorious agents of nosocomial infections, but also causes disease in the general community. A dominant concept is the dichotomy between β-lactam resistant (known as methicillin resistant S. aureus (MRSA)) and sensitive (methicillin susceptible S. aureus (MSSA)) strains. The MRSA phenotype is homoplastic, due to mobility of the SCCmec genetic determinant of β-lactam resistance.2 Also important is the dichotomy between community-associated MRSA (CA-MRSA), and healthcare-associated MRSA (HA-MRSA).3 HA-MRSA strains are well adapted to the health care environment, are typically multiresistant, and in general do not cause community onset infections. CA-MRSA cause community onset infections, but also now predominate in healthcare facilities. CA-MRSA frequently carry a bacteriophage specifying the pore-forming toxin Panton-Valentine leukocidin (PVL). Phage mobility results in PVL+ CA-MRSA being within several evolutionary lineages, and PVL+ MRSA and MSSA are often closely related. The broad framework for understanding the core genome population structure S. aureus is based on ‘clonal complexes’ (CCs) consisting of closely related sequence types (STs) defined by the multi-locus sequence typing (MLST) scheme.2
Australia is similar to rest of the world in that the major HA-MRSA strain for many years was ST239. Recently, ST239 has become less prominent, with the most prevalent lineage now being ST22/CC22 ‘EMRSA-15’.4–6 Northern Australia appears to have experienced the steep decline of ST239, but with only limited replacement with ST22.7,8
CA-MRSA strains in northern Australia are similar to Australia as a whole, but a contributor to this is expansion of strains originating in remote areas into urban Australia.4,9,10 The most prominent is ST93. Essentially all ST93 are PVL+ and the strain is regarded as highly virulent. ST93 MRSA and MSSA co-exist. Genomic studies indicate that ST93 arose from a point source/genetic bottleneck in north-western Australia in the 1990s. ST93 is divergent from all other S. aureus evolutionary lineages, and the nature of the point source remains enigmatic.10 Consequently, ST93 does not define an observed CC of a substantial number of related STs. ST93 has disseminated throughout Australia and much of New Zealand10 and is often isolated from health care facilities as well as the general community.6 More recent is the emergence of a CC5 PVL+ CA-MRSA strain, also in north-western or northern Australia, and this appears to be in the expansion phase11 perhaps repeating what was seen with ST93. Other PVL+ strains of current or recent significance in northern Australia include a ST30 MRSA that has long been associated with the Pacific region, and has reduced in prevalence in recent years,8,12 and ST121 (CC121), which is a PVL+ strain mainly associated with south-east and east Asia, and almost always MSSA.8,13,14 CA-MRSA strains are not always PVL+. In particular, PVL− CC1 MRSA, often designated as WA-1, has been causing community-acquired infections in north-western Australia for many years.6,15
We recently reported a multi-year longitudinal genomics-based study of S. aureus in Top End dialysis clinic clients, staff, and researcher.8 The objective was identification of reservoirs underpinning infection. The genetic nature of the recovered isolates are largely consistent with other studies and provide ‘in miniature’ a picture of S. aureus epidemiology in the Top End (Fig. 1). The infections were nearly all ‘skin and soft tissue’, with a large proportion caused by the PVL+ MRSA (ST93 and ST5), PVL+ MSSA (ST121), and PVL− MRSA (ST762 (CC1)). Strikingly, asymptomatic carriage of the PVL+ strains was vanishingly rare. In contrast, the PVL− MRSA strains were associated with both carriage and infection, and putative transmission from carriage to infection was identified. ST762 is single locus variant of ST1. ST762 isolates are very rare in the MLST database but were prevalent in the STARRS isolates, and putatively transmitted from carriage to infection. These are potentially WA-1 or similar. The STARRs PVL− MSSA isolates largely reflect CCs found globally. They were stably carried, largely in the nasal cavity, are under-represented in the infection isolates, and the only putative transmission events identified were ‘carriage to carriage’. HA-MRSA were rare, with ST239 represented by one carriage and one clinical isolate. No ST22 isolates were recovered.

|
It is of interest that the species Staphylococcus argenteus was first identified during a survey of S. aureus carriage and infections in a remote community in the Northern Territory. The species is now known to be globally distributed, with similar pathogenic properties and potential to acquire antimicrobial resistance as S. aureus.16 It is one of five known species in the ‘S. aureus complex’ with the others being S. aureus, Staphylococcus schweitzeri, Staphylococcus singaporensis, and Staphylococcus roterodami. In general, these newly defined species appear rare in humans, and in the case of S. schweitzeri are overwhelmingly associated with non-human animals. In the STARRs study a small minority of the carriage isolates and none of the clinical isolates were S. argenteus.8 The current clinical impact of S. argenteus in northern Australia is unclear.
Streptococcus pyogenes (Group A streptococci (GAS)) are β-haemolytic, pyogenic organisms which cause a range of infections from pyoderma and pharyngitis, to invasive diseases including necrotising fasciitis and toxic shock syndrome. Importantly, autoimmune responses post-infection can lead to significant sequelae including acute rheumatic fever (ARF), rheumatic heart disease (RHD), and acute post-streptococcal glomerulonephritis (ASPGN). It has long been recognised that the burden of ARF and RHD in northern Australia is among the highest reported in the world.17
The N-terminal region of the surface streptococcal M protein is highly variable and has been the basis of both serotyping and genetic typing methods. emm typing has classically been the main approach to genetic typing and involves sequence analysis of the region of the emm gene encoding the variable N-terminal region. Further analysis combining emm sequence type with other information including the arrangement of emm and emm-like genes, defines emm pattern types. Population based studies have indicated that emm patterns A–C show throat tropism, emm pattern D shows skin tropism, while emm pattern E does not appear to have a predilection for either site. Over 200 emm sequence types have been defined that correlate well with the emm patterns.18
In the temperate regions of Australia the molecular epidemiology of S. pyogenes is similar to that of other developed countries, where throat tropic emm types dominate and more restricted emm diversity is observed.19 However, in northern Australia, the prevalence of S. pyogenes pyoderma is high, while pharyngitis is low compared with other areas of the country.20 The emm types belonging to the skin tropic emm pattern D and non-tropic emm pattern E predominate.21 Outbreaks of invasive disease related to a subset of emm types are seen on a background of high diversity and rapid turnover in the circulating emm types.22–25 While novel emm sequence types have been found in remote communities in northern Australia, the overall diversity does not seem to be the result of local diversification but rather the circulating population represents a large subset of known global S. pyogenes diversity.26,27 The pattern seen in northern Australia is consistent with what is seen in developing countries,19 as well as in Indigenous populations in other developed countries such as New Zealand where the burden of ARF and RHD is similarly high.17,28,29 In these regions, skin and throat tropic emm types co-exist, increasing the total diversity of circulating emm types.
The variable N-terminal region of the M protein is considered a useful vaccine target due to its immunogenic epitopes and low propensity to induce antibodies which cross react with human tissues. However, the high diversity of emm types in northern Australia and developing regions creates concerns for the likely efficacy of such vaccines in these areas.21 An emm cluster system has been developed which is an elaboration of the emm typing scheme and is based on the surface exposed portion of the M protein and its binding capacity with six human serum proteins, and emm cluster can be inferred from the emm type.30 This can serve as a resource for vaccine development as cross-opsonisation experiments have demonstrated cross-protection between some emm types.30
A 30mer M protein vaccine which has reached human trials is largely based on emm types prevalent in North America and Europe. It contains only a single emm pattern D sequence, which did not demonstrate a high proportion of cross protection for other pattern D emm types.30 A study examining over 1700 isolates from northern Australia collected over more than 20 years demonstrated that over 50% of the isolates were cluster D or outlier emm types. Based on the cross-opsonisation data,30 the 30mer vaccine may not provide good protection against these isolates. In addition, poor coverage of the 30mer vaccine against APSGN associated strains such as emm55 is predicted21 (Fig. 2). Broader protection may be elicited by the 26mer precursor of the 30mer vaccine, which contained two additional cluster D4 emm types that are prevalent in northern Australia, or by utilising a combination vaccine approach.21 Alternatively, other vaccine strategies targeting more conserved regions of the M protein and non-M protein targets29 may prove more effective in regions with high emm diversity.
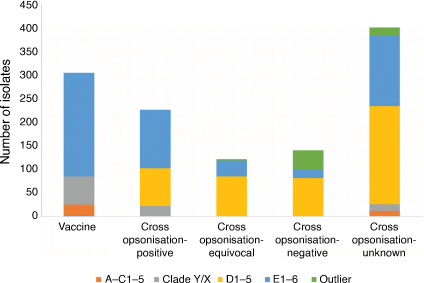
|
S. aureus and S. pyogenes are significant pathogens causing substantial health impacts from acute infections and their serious sequelae. In northern Australia, rates of skin infections are high and there are likely complex interactions with other skin pathogens such as the scabies mite Sarcoptes scabiei and the dermatophyte Trichophyton rubrum.1 S. aureus and S. pyogenes are commonly both recovered from impetigo lesions,31,32 and S. pyogenes is more likely to be recovered from impetigo lesions if scabies infection is also present.31,32 This dynamic creates particular challenges for public health programs targeting skin health in this region.1
Data availability
Data sharing is not applicable as no new data were generated or analysed during this study.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This work did not receive any specific funding.
References
[1] Currie, BJ and Carapetis, JR (2000) Skin infections and infestations in Aboriginal communities in northern Australia. Australas J Dermatol 41, 139–143.| Skin infections and infestations in Aboriginal communities in northern Australia.Crossref | GoogleScholarGoogle Scholar |
[2] Turner, KME and Feil, EJ (2007) The secret life of the multilocus sequence type. Int J Antimicrob Agents 29, 129–135.
| The secret life of the multilocus sequence type.Crossref | GoogleScholarGoogle Scholar |
[3] Henderson, A and Nimmo, GR (2018) Control of healthcare- and community-associated MRSA: recent progress and persisting challenges. Br Med Bull 125, 25–41.
| Control of healthcare- and community-associated MRSA: recent progress and persisting challenges.Crossref | GoogleScholarGoogle Scholar |
[4] Coombs, GW et al.. (2014) Community-onset Staphylococcus aureus Surveillance Programme annual report, 2012. Commun Dis Intell Q Rep 38, E59–E69.
[5] Dotel, R et al.. (2019) Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates in New South Wales, Australia, 2012–2017. Infect Dis Health 24, 134–140.
| Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates in New South Wales, Australia, 2012–2017.Crossref | GoogleScholarGoogle Scholar |
[6] Coombs, GW et al.. (2020) Australian Group on Antimicrobial Resistance (AGAR) Australian Enterococcal Sepsis Outcome Programme (AESOP) Annual Report 2019. Commun Dis Intell 44, 1–12.
| Australian Group on Antimicrobial Resistance (AGAR) Australian Enterococcal Sepsis Outcome Programme (AESOP) Annual Report 2019.Crossref | GoogleScholarGoogle Scholar |
[7] Brennan, L et al.. (2013) Community-associated meticillin-resistant Staphylococcus aureus carriage in hospitalized patients in tropical northern Australia. J Hosp Infect 83, 205–211.
| Community-associated meticillin-resistant Staphylococcus aureus carriage in hospitalized patients in tropical northern Australia.Crossref | GoogleScholarGoogle Scholar |
[8] Holt, DC et al.. (2021) Longitudinal whole-genome based comparison of carriage and infection associated Staphylococcus aureus in northern Australian dialysis clinics. PLoS One 16, e0245790.
| Longitudinal whole-genome based comparison of carriage and infection associated Staphylococcus aureus in northern Australian dialysis clinics.Crossref | GoogleScholarGoogle Scholar |
[9] Tong, SYC et al.. (2015) Progressive increase in community-associated methicillin-resistant Staphylococcus aureus in Indigenous populations in northern Australia from 1993 to 2012. Epidemiol Infect 143, 1519–1523.
| Progressive increase in community-associated methicillin-resistant Staphylococcus aureus in Indigenous populations in northern Australia from 1993 to 2012.Crossref | GoogleScholarGoogle Scholar |
[10] van Hal, SJ et al.. (2018) Global scale dissemination of ST93: a divergent Staphylococcus aureus epidemic lineage that has recently emerged from remote northern Australia. Front Microbiol 9, 1453.
| Global scale dissemination of ST93: a divergent Staphylococcus aureus epidemic lineage that has recently emerged from remote northern Australia.Crossref | GoogleScholarGoogle Scholar |
[11] McGuinness, SL et al.. (2021) Clinical and molecular epidemiology of an emerging Panton-Valentine leukocidin-positive ST5 methicillin-resistant Staphylococcus aureus clone in Northern Australia. mSphere 6, e00651-20.
| Clinical and molecular epidemiology of an emerging Panton-Valentine leukocidin-positive ST5 methicillin-resistant Staphylococcus aureus clone in Northern Australia.Crossref | GoogleScholarGoogle Scholar |
[12] Tong, SYC et al.. (2009) Community-associated strains of methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus in indigenous Northern Australia: epidemiology and outcomes. J Infect Dis 199, 1461–1470.
| Community-associated strains of methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus in indigenous Northern Australia: epidemiology and outcomes.Crossref | GoogleScholarGoogle Scholar |
[13] Rao, Q et al.. (2015) Staphylococcus aureus ST121: a globally disseminated hypervirulent clone. J Med Microbiol 64, 1462–1473.
| Staphylococcus aureus ST121: a globally disseminated hypervirulent clone.Crossref | GoogleScholarGoogle Scholar |
[14] Harch, SAJ et al.. (2017) High burden of complicated skin and soft tissue infections in the Indigenous population of Central Australia due to dominant Panton Valentine leucocidin clones ST93-MRSA and CC121-MSSA. BMC Infect Dis 17, 405.
| High burden of complicated skin and soft tissue infections in the Indigenous population of Central Australia due to dominant Panton Valentine leucocidin clones ST93-MRSA and CC121-MSSA.Crossref | GoogleScholarGoogle Scholar |
[15] Coombs, GW et al.. (2004) Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J Clin Microbiol 42, 4735–4743.
| Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia.Crossref | GoogleScholarGoogle Scholar |
[16] Tong, SYC et al.. (2015) Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int J Syst Evol Microbiol 65, 15–22.
| Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov.Crossref | GoogleScholarGoogle Scholar |
[17] Carapetis, JR et al.. (1996) Acute rheumatic fever and rheumatic heart disease in the top end of Australia’s Northern Territory. Med J Aust 164, 146–149.
| Acute rheumatic fever and rheumatic heart disease in the top end of Australia’s Northern Territory.Crossref | GoogleScholarGoogle Scholar |
[18] McMillan, DJ et al.. (2013) Updated model of group A Streptococcus M proteins based on a comprehensive worldwide study. Clin Microbiol Infect 19, E222–E229.
| Updated model of group A Streptococcus M proteins based on a comprehensive worldwide study.Crossref | GoogleScholarGoogle Scholar |
[19] Steer, AC et al.. (2009) Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis 9, 611–616.
| Global emm type distribution of group A streptococci: systematic review and implications for vaccine development.Crossref | GoogleScholarGoogle Scholar |
[20] McDonald, MI et al.. (2006) Low rates of streptococcal pharyngitis and high rates of pyoderma in Australian aboriginal communities where acute rheumatic fever is hyperendemic. Clin Infect Dis 43, 683–689.
| Low rates of streptococcal pharyngitis and high rates of pyoderma in Australian aboriginal communities where acute rheumatic fever is hyperendemic.Crossref | GoogleScholarGoogle Scholar |
[21] Giffard, PM et al.. (2019) Concerns for efficacy of a 30-valent M-protein-based Streptococcus pyogenes vaccine in regions with high rates of rheumatic heart disease. PLoS Negl Trop Dis 13, e0007511.
| Concerns for efficacy of a 30-valent M-protein-based Streptococcus pyogenes vaccine in regions with high rates of rheumatic heart disease.Crossref | GoogleScholarGoogle Scholar |
[22] Bessen, DE et al.. (2000) Contrasting molecular epidemiology of group A streptococci causing tropical and nontropical infections of the skin and throat. J Infect Dis 182, 1109–1116.
| Contrasting molecular epidemiology of group A streptococci causing tropical and nontropical infections of the skin and throat.Crossref | GoogleScholarGoogle Scholar |
[23] McDonald, MI et al.. (2007) Molecular typing of Streptococcus pyogenes from remote Aboriginal communities where rheumatic fever is common and pyoderma is the predominant streptococcal infection. Epidemiol Infect 135, 1398–1405.
| Molecular typing of Streptococcus pyogenes from remote Aboriginal communities where rheumatic fever is common and pyoderma is the predominant streptococcal infection.Crossref | GoogleScholarGoogle Scholar |
[24] McDonald, MI et al.. (2008) The dynamic nature of group A streptococcal epidemiology in tropical communities with high rates of rheumatic heart disease. Epidemiol Infect 136, 529–539.
| The dynamic nature of group A streptococcal epidemiology in tropical communities with high rates of rheumatic heart disease.Crossref | GoogleScholarGoogle Scholar |
[25] Richardson, LJ et al.. (2011) Preliminary validation of a novel high-resolution melt-based typing method based on the multilocus sequence typing scheme of Streptococcus pyogenes. Clin Microbiol Infect 17, 1426–1434.
| Preliminary validation of a novel high-resolution melt-based typing method based on the multilocus sequence typing scheme of Streptococcus pyogenes.Crossref | GoogleScholarGoogle Scholar |
[26] McGregor, KF et al.. (2004) Group A streptococci from a remote community have novel multilocus genotypes but share emm types and housekeeping alleles with isolates from worldwide sources. J Infect Dis 189, 717–723.
| Group A streptococci from a remote community have novel multilocus genotypes but share emm types and housekeeping alleles with isolates from worldwide sources.Crossref | GoogleScholarGoogle Scholar |
[27] Towers, RJ et al.. (2013) Extensive diversity of Streptococcus pyogenes in a remote human population reflects global-scale transmission rather than localised diversification. PLoS One 8, e73851.
| Extensive diversity of Streptococcus pyogenes in a remote human population reflects global-scale transmission rather than localised diversification.Crossref | GoogleScholarGoogle Scholar |
[28] Williamson, DA et al.. (2015) M-Protein analysis of Streptococcus pyogenes isolates associated with acute rheumatic fever in New Zealand. J Clin Microbiol 53, 3618–3620.
| M-Protein analysis of Streptococcus pyogenes isolates associated with acute rheumatic fever in New Zealand.Crossref | GoogleScholarGoogle Scholar |
[29] Good, MF et al.. (2015) Strategic development of the conserved region of the M protein and other candidates as vaccines to prevent infection with group A streptococci. Expert Rev Vaccines 14, 1459–1470.
| Strategic development of the conserved region of the M protein and other candidates as vaccines to prevent infection with group A streptococci.Crossref | GoogleScholarGoogle Scholar |
[30] Sanderson-Smith, M et al.. (2014) A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis 210, 1325–1338.
| A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development.Crossref | GoogleScholarGoogle Scholar |
[31] Valery, PC et al.. (2008) Skin infections among Indigenous Australians in an urban setting in far North Queensland. Epidemiol Infect 136, 1103–1108.
| Skin infections among Indigenous Australians in an urban setting in far North Queensland.Crossref | GoogleScholarGoogle Scholar |
[32] Bowen, AC et al.. (2014) The microbiology of impetigo in indigenous children: associations between Streptococcus pyogenes, Staphylococcus aureus, scabies, and nasal carriage. BMC Infect Dis 14, 727.
| The microbiology of impetigo in indigenous children: associations between Streptococcus pyogenes, Staphylococcus aureus, scabies, and nasal carriage.Crossref | GoogleScholarGoogle Scholar |


