New insights into chlamydial zoonoses
Adam Polkinghorne A B C and James Branley A B DA Department of Microbiology and Infectious Diseases, New South Wales Health Pathology, Nepean Blue Mountains Pathology Service, Penrith, NSW 2751, Australia
B The University of Sydney Medical School, Nepean Clinical School, Faculty of Medicine and Health, The University of Sydney, Penrith, NSW 2751, Australia
C Email: adam.polkinghorne@health.nsw.gov.au
D Email: james.branley@health.nsw.gov.au
Microbiology Australia 41(1) 14-18 https://doi.org/10.1071/MA20005
Published: 28 February 2020
Chlamydiae are obligate intracellular bacterial pathogens of humans. Infections in animals are also widespread with some species, such as Chlamydia psittaci, long recognised as a serious threat to human health. Critical to the public health response of any zoonotic disease outbreaks is reliable and up-to-date information on the epidemiology of the target pathogen. Aided by advances in the use of quantitative PCR, molecular typing and culture-independent genomic studies, significant recent work has highlighted an expanded diversity and host range of chlamydial pathogens in animals. New and unexpected cases of chlamydial zoonoses have now been recently documented in Australia and elsewhere, emphasising the importance of multi-disciplinary ‘One Health’ collaboration and the use of standardised methods to detect and characterise chlamydial pathogens in humans and animals.
A brief history of chlamydial zoonosis
The first recognition of the zoonotic potential of chlamydial infections predates the actual description of the bacteria1. In 1879, Jacob Ritter described an epidemic of fatal respiratory disease in humans associated with contact with caged parrots and finches. At the time, the aetiological agent of this disease, later coined psittacosis, was unclear although it was suspected that it was of viral origin. Interest in the disease re-emerged in 1929–1930, with epidemics of human disease reported in Europe and the Americas, again linked to infected and imported parrots. The global attention generated from these outbreaks prompted several years of fruitful research, including important studies by Australia’s Sir Frank Macfarlane-Burnet2, 3, ultimately leading to the description and characterisation of a bacterium, Chlamydia psittaci, with a complex biphasic developmental cycle1. Since this time, C. psittaci has been considered the classical chlamydial zoonotic pathogen, with zoonotic transmission and acute, serious disease (in the form of atypical pneumonia) resulting from direct contact with infected birds or their contaminated excreta4. Historically, most of the attention is rightly placed on the pathogenic potential of C. psittaci; however, isolated cases of the zoonotic transmission of closely related Chlamydia abortus from sheep have also been documented, the latter linked to subsequent abortion in pregnant women that are exposed to the secretions of C. abortus infected ewes5.
Growing recognition of the diversity of chlamydial infections in animals
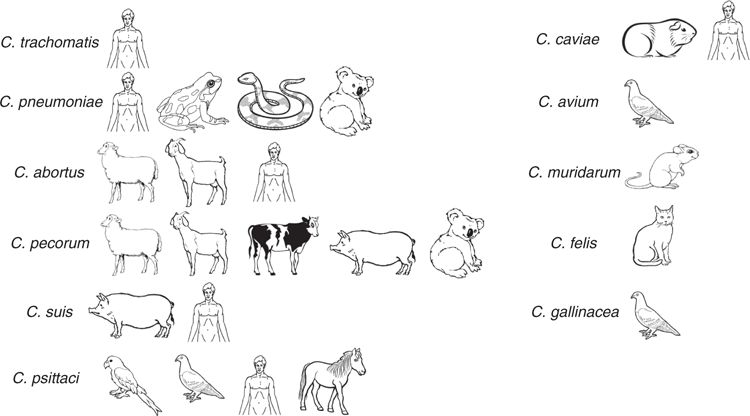
Bacterial adaptation to an obligate intracellular niche would typically imply genetic conservation and a restricted host range. As we have learned more about the diversity of taxa within the phylum Chlamydiae, new surprises continue to emerge. Recent years have seen the proposal and description of several new order and family level linages of chlamydiae6. Nevertheless, most attention remains on the genus Chlamydia since it comprises a number of important human and animal pathogens. Since the relatively recent (re-) classification of chlamydial species into a single genus7, a plethora of new species (14 total species in the genus) have now been proposed or formally classified6 beyond the 11 that were initially included. The documented host range of some of the most well described of these chlamydial species in the genus Chlamydia is shown in Figure 1. Supported by the use of molecular tools such as family- and genus-specific qPCR assays targeting conserved chlamydial sequences8 of widely conserved eubacterial ribosomal rRNA genes, these discoveries have primarily come on the back of veterinary investigations into previously recognised warm-blooded hosts of chlamydial infections as well as new hosts such as snakes9–12. While the pathogenic potential of many of these newly described chlamydial pathogens remains unclear, their discovery has highlighted how little is still yet known about the diversity and epidemiology of chlamydiae.
Advances in molecular tools to study chlamydial epidemiology and zoonotic events
A number of technical innovations over the past 20 years in the chlamydial research community have paved the way for a greater understanding of chlamydial epidemiology and the documentation of infection spill-over events from recognised and emerging chlamydial zoonotic agents. The first of these is the shift from laborious culture-based methods for detecting chlamydial infections to the use of conventional and quantitative PCR-based methods that detect the presence of chlamydial DNA13. Coupled with the use of the aforementioned broad-range order and family-specific primers8 to ‘cast a wide net’ in the screening of clinical specimens, these approaches have revolutionised the sensitive detection for chlamydial pathogens in human and animal samples.
An interesting example of the use of these approaches to uncover an unexpected potential for chlamydial zoonoses comes from studies in Belgian pig farms and slaughterhouses14–16, including one study where researchers have documented the presence of Chlamydia suis, a ubiquitous pig pathogen, in the mucosal swab samples (conjunctival and rectal) collected from farmers and slaughterhouse workers15. This discovery is of particular concern given that C. suis harbours the only known naturally occurring antibiotic resistance cassette in the Chlamydiaceae, raising fears over the potential to transfer this genetic element to the closely related human chlamydial pathogen, Chlamydia trachomatis17,18. A potentially legitimate criticism of the use of DNA-based methods for detection of chlamydial pathogens in new hosts is that the detection simply reflects contamination or exposure and not genuine infection that might lead to disease and/or subsequent chlamydial shedding. To rebut this criticism, the Belgian team also detected the presence of species-specific antibodies in the human subjects, providing stronger evidence for the zoonotic potential for C. suis16. The challenge in doing so for other chlamydial species suspected of zoonotic transfer is an almost complete lack of serological tools for measuring specific human host responses to the growing and diverse range of chlamydial pathogens infecting animals. In an exciting advance to the field, this may become easier in the future with the recent development of highly specific peptide microarray assays for detecting Chlamydia-species specific antibodies in human and animal sera19. Even though it is only pilot data, it is interesting to note that studies of small selections of samples from livestock with these assays uncovered specific antibody responses to a diverse range of chlamydial pathogens, potentially suggesting that the previously postulated host barriers for most species in the genus Chlamydia do not actually exist19.
Another important technical advance has been the development and application of multi-locus sequence typing schemes (MLST) to study the fine-detailed molecular epidemiology of chlamydial pathogens. This standardised approach utilises DNA sequences of conserved ‘house-keeping’ genes that, when combined, create a unique profile for each genetically distinct strain sequenced20. Schemes have now been developed for most species in the genus Chlamydia, creating opportunities to interrogate strains of the same chlamydial species from different hosts to gain insight into their relationship and the potential for cross-host transmission, as will be discussed below. These approaches have become even more powerful when coupled with the use of culture-independent genome sequencing technologies to obtain the full genome sequence from strains in clinical samples, opening the possibilities for interrogating ‘field-relevant’ strains21.
‘One Health’ investigations document new cases of chlamydial zoonoses in Australia and the rest of the world
While technological advances have opened up new opportunities to perform surveillance of potential chlamydial zoonotic events, they are only useful if employed as a part of multidisciplinary teams of experts including doctors, veterinarians, public health staff and microbiologists.
The best example of a recent ‘One Health’ partnership to conduct surveillance of zoonotic chlamydial infections comes from The Netherlands. In this report, Chlamydia caviae, a guinea pig pathogen was found to be the causative agent of severe pneumonia in three unrelated human cases22. In all cases, the humans had developed symptoms after being exposed to ill guinea pigs. Courtesy of an agreement between veterinary and human diagnostic laboratories to use the same molecular detection and typing methods23, the authorities were able to confirm the suspected transmission by showing that the strains from the human subject and diseased guinea pig were identical in at least one case.
Closer to home, a team of human and veterinary clinicians and researchers have provided important new insight into the zoonotic potential of the avian and zoonotic pathogen, C. psittaci. A cluster of cases of probable psittacosis were detected amongst veterinary students and staff at regional university veterinary school in New South Wales24. The ensuing public health investigation revealed shared contact with the infected placental membranes of a mare that had delivered a foal that died prematurely. Fine-detailed molecular analysis by our team, including application of a species-specific C. psittaci MLST scheme, revealed that the sample contained a C. psittaci strain belonging to the avian 6BC clade25, responsible for the global epidemics in the 1930s and found in Australian native parrots26. The genetic relationships of the newly detected equine C. psittaci strains to strains from other hosts is illustrated in Figure 2. This discovery provided the first evidence of a potential mammal to mammal transmission of C. psittaci and revealed a new zoonotic risk by this chlamydial agent. Subsequent studies by our team have further revealed that: (1) avian C. psittaci infections are a more common cause of reproductive loss in mares than previously thought27; (2) the subsequent occupational risk to veterinarians may be significant; and (3) other C. psittaci strains beyond those found in parrots may also infect horses and, hence, potentially pose a risk to human health28.

|
Future directions in understanding chlamydial zoonoses
This review has highlighted a growing awareness of the zoonotic potential of a broad range of chlamydial pathogens both in Australia and abroad. This new information comes largely on the back of improvements in the surveillance of chlamydial pathogens with new technologies increasing our ability to detect and monitor these intracellular bacteria in different hosts. An awareness of the host range and pathogenic potential of animal chlamydiae in humans is important information that can be used to guide surveillance efforts by public health authorities.
As the next step in efforts to predict and minimise the risk of chlamydial zoonoses, one area of research that needs significant more work is in understanding what factors influence chlamydial spill-over events between animals and humans. For example, in the case of equine C. psittaci infections, what specific factors influence transmission of the pathogen in birds and, hence, present a risk of infection to humans? Prior to the detection of C. psittaci infections in horses, have infection spill-over always occurred historically or are there specific changes in the local bird ecology that increase the risk of C. psittaci shedding in the environment? One Health partnerships between human and veterinary stakeholders will continue to be at the forefront of efforts to answer these questions for chlamydiae and other zoonotic agents.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This research did not receive any specific funding.
References
[1] Pospischil, A. (2009) From disease to etiology: historical aspects of Chlamydia-related diseases in animals and humans. Drugs Today (Barc) 45, 141–146.| 20011706PubMed |
[2] Burnet, F.M. (1935) Enzootic psittacosis amongst wild Australian parrots. J. Hyg. (Lond.) 35, 412–420.
| Enzootic psittacosis amongst wild Australian parrots.Crossref | GoogleScholarGoogle Scholar | 20475292PubMed |
[3] Polkinghorne, A. et al. (2020) The recent history of psittacosis in Australia: expanding our understanding of the epidemiology of this important globally-distributed zoonotic disease. Intern. Med. J. , .
| 32037712PubMed |
[4] Stewardson, A.J. and Grayson, M.L. (2010) Psittacosis. Infect. Dis. Clin. North Am. 24, 7–25.
| Psittacosis.Crossref | GoogleScholarGoogle Scholar | 20171542PubMed |
[5] Pospischil, A. (2002) Abortion in humans caused by Chlamydophila abortus (Chlamydia psittaci serovar 1). Schweiz. Arch. Tierheilkd. 144, 463–466.
| Abortion in humans caused by Chlamydophila abortus (Chlamydia psittaci serovar 1).Crossref | GoogleScholarGoogle Scholar | 12677684PubMed |
[6] Taylor-Brown, A. and Polkinghorne, A. (2017) New and emerging chlamydial infections of creatures great and small. New Microbes New Infect. 18, 28–33.
| New and emerging chlamydial infections of creatures great and small.Crossref | GoogleScholarGoogle Scholar | 28560043PubMed |
[7] Sachse, K. et al. (2015) Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognised species. Syst. Appl. Microbiol. 38, 99–103.
| Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognised species.Crossref | GoogleScholarGoogle Scholar | 25618261PubMed |
[8] Everett, K.D. et al. (1999) Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49, 415–440.
| Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms.Crossref | GoogleScholarGoogle Scholar | 10319462PubMed |
[9] Taylor-Brown, A. et al. (2015) Characterisation of Chlamydia pneumoniae and other novel chlamydial infections in captive snakes. Vet. Microbiol. 178, 88–93.
| Characterisation of Chlamydia pneumoniae and other novel chlamydial infections in captive snakes.Crossref | GoogleScholarGoogle Scholar | 25944652PubMed |
[10] Taylor-Brown, A. et al. (2017) Culture-independent metagenomics supports discovery of uncultivable bacteria within the genus Chlamydia. Sci. Rep. 7, 10661.
| Culture-independent metagenomics supports discovery of uncultivable bacteria within the genus Chlamydia.Crossref | GoogleScholarGoogle Scholar | 28878306PubMed |
[11] Taylor-Brown, A. et al. (2016) Culture-independent genomic characterisation of Candidatus Chlamydia sanzinia, a novel uncultivated bacterium infecting snakes. BMC Genomics 17, 710.
| Culture-independent genomic characterisation of Candidatus Chlamydia sanzinia, a novel uncultivated bacterium infecting snakes.Crossref | GoogleScholarGoogle Scholar | 27595750PubMed |
[12] Staub, E. et al. (2018) Novel Chlamydia species isolated from snakes are temperature-sensitive and exhibit decreased susceptibility to azithromycin. Sci. Rep. 8, 5660.
| Novel Chlamydia species isolated from snakes are temperature-sensitive and exhibit decreased susceptibility to azithromycin.Crossref | GoogleScholarGoogle Scholar | 29618824PubMed |
[13] Sachse, K. et al. (2009) Recent developments in the laboratory diagnosis of chlamydial infections. Vet. Microbiol. 135, 2–21.
| Recent developments in the laboratory diagnosis of chlamydial infections.Crossref | GoogleScholarGoogle Scholar | 18986778PubMed |
[14] De Puysseleyr, K. et al. (2014) Evaluation of the presence and zoonotic transmission of Chlamydia suis in a pig slaughterhouse. BMC Infect. Dis. 14, 560.
| Evaluation of the presence and zoonotic transmission of Chlamydia suis in a pig slaughterhouse.Crossref | GoogleScholarGoogle Scholar | 25358497PubMed |
[15] De Puysseleyr, L. et al. (2017) Assessment of Chlamydia suis infection in pig farmers. Transbound. Emerg. Dis. 64, 826–833.
| Assessment of Chlamydia suis infection in pig farmers.Crossref | GoogleScholarGoogle Scholar | 26576707PubMed |
[16] Kieckens, E. et al. (2018) Co-occurrence of Chlamydia suis DNA and Chlamydia suis-specific antibodies in the human eye. Vector Borne Zoonotic Dis. , .
| Co-occurrence of Chlamydia suis DNA and Chlamydia suis-specific antibodies in the human eye.Crossref | GoogleScholarGoogle Scholar | 30251925PubMed |
[17] Dugan, J. et al. (2007) Functional characterisation of IScs605, an insertion element carried by tetracycline-resistant Chlamydia suis. Microbiology 153, 71–79.
| Functional characterisation of IScs605, an insertion element carried by tetracycline-resistant Chlamydia suis.Crossref | GoogleScholarGoogle Scholar | 17185536PubMed |
[18] Seth-Smith, H.M. et al. (2017) The Chlamydia suis genome exhibits high levels of diversity, plasticity, and mobile antibiotic resistance: comparative genomics of a recent livestock cohort shows influence of treatment regions. Genome Biol. Evol. 9, 750–760.
| The Chlamydia suis genome exhibits high levels of diversity, plasticity, and mobile antibiotic resistance: comparative genomics of a recent livestock cohort shows influence of treatment regions.Crossref | GoogleScholarGoogle Scholar | 28338777PubMed |
[19] Sachse, K. et al. (2018) A novel synthetic peptide microarray assay detects Chlamydia species-specific antibodies in animal and human sera. Sci. Rep. 8, 4701.
| A novel synthetic peptide microarray assay detects Chlamydia species-specific antibodies in animal and human sera.Crossref | GoogleScholarGoogle Scholar | 29549361PubMed |
[20] Jelocnik, M. et al. (2019) Multilocus sequence typing (MLST) of Chlamydiales. Methods Mol. Biol. 2042, 69–86.
| Multilocus sequence typing (MLST) of Chlamydiales.Crossref | GoogleScholarGoogle Scholar | 31385271PubMed |
[21] Taylor-Brown, A. et al. (2018) Culture-independent approaches to chlamydial genomics. Microb. Genom. 4, 2.
| Culture-independent approaches to chlamydial genomics.Crossref | GoogleScholarGoogle Scholar |
[22] Ramakers, B.P. et al. (2017) Zoonotic Chlamydia caviae presenting as community-acquired pneumonia. N. Engl. J. Med. 377, 992–994.
| Zoonotic Chlamydia caviae presenting as community-acquired pneumonia.Crossref | GoogleScholarGoogle Scholar | 28877022PubMed |
[23] Roest, H.J. et al. (2018) An integrated human-animal health approach to reduce the disease burden of psittacosis. In Chlamydial infections: Proceedings of the Fourteenth International Symposium on Human Chlamydial Infections, The Netherlands (Chernesky, M. et al. eds), pp. 709–712.
[24] Chan, J. et al. (2017) An outbreak of psittacosis at a veterinary school demonstrating a novel source of transmission. One Health 3, 29–33.
| An outbreak of psittacosis at a veterinary school demonstrating a novel source of transmission.Crossref | GoogleScholarGoogle Scholar | 28616500PubMed |
[25] Jelocnik, M. et al. (2017) Multi-locus sequence typing identifies an avian-like Chlamydia psittaci strain involved in equine placentitis and associated with subsequent human psittacosis. Emerg. Microbes Infect. 6, e7.
| Multi-locus sequence typing identifies an avian-like Chlamydia psittaci strain involved in equine placentitis and associated with subsequent human psittacosis.Crossref | GoogleScholarGoogle Scholar | 28196971PubMed |
[26] Branley, J. et al. (2016) Australian human and parrot Chlamydia psittaci strains cluster within the highly virulent 6BC clade of this important zoonotic pathogen. Sci. Rep. 6, 30019.
| Australian human and parrot Chlamydia psittaci strains cluster within the highly virulent 6BC clade of this important zoonotic pathogen.Crossref | GoogleScholarGoogle Scholar | 27488134PubMed |
[27] Jenkins, C. et al. (2018) An epizootic of Chlamydia psittaci equine reproductive loss associated with suspected spillover from native Australian parrots. Emerg. Microbes Infect. 7, 88.
| An epizootic of Chlamydia psittaci equine reproductive loss associated with suspected spillover from native Australian parrots.Crossref | GoogleScholarGoogle Scholar | 29765033PubMed |
[28] Jelocnik, M. et al. (2018) Molecular evidence to suggest pigeon-type Chlamydia psittaci in association with an equine foal loss. Transbound. Emerg. Dis. 65, 911–915.
| Molecular evidence to suggest pigeon-type Chlamydia psittaci in association with an equine foal loss.Crossref | GoogleScholarGoogle Scholar | 29352509PubMed |
Biographies
Dr Adam Polkinghorne is a Senior Hospital Scientist at NSW Health Pathology and a Honorary Senior Principle Research Fellow in the University of Sydney Nepean Clinical School. His research interests are primarily focussed on the (a) diagnosis, management and control of chlamydial infections in humans and animals and (b) the detection and control of hospital-acquired infections in infants and at-risk patients.
Dr James Branley is an infectious disease physician and Head of Department, Infectious Diseases and Microbiology at Nepean Hospital, Penrith. He is also the Local Pathology Director, NSW Health Pathology Nepean and an adjunct Associate Professor in the University of Sydney Nepean Clinical School. He has a long-standing interest in psittacosiss, recently completing a PhD on this topic at the University of Sydney.