The role of the gut microbiome in host systems
Clarissa Febinia A , Connie Ha A , Chau Le A and Andrew Holmes A BA School of Molecular Bioscience and Charles Perkins Centre, University of Sydney, Sydney, NSW, Australia
B Corresponding author. Email: andrew.holmes@sydney.edu.au
Microbiology Australia 36(1) 14-17 https://doi.org/10.1071/MA15005
Published: 6 March 2015
The presence of microbes exerts such a profound influence on animals that they are best considered holobionts – an organism comprised of multiple biological partners. The concept of dysbiosis is disease states that result from undesirable interactions between the partners in a holobiont. Many molecular mechanisms that link the gut microbiome with host health and disease have now been established and these are giving rise to new insights in healthcare. In essence these studies show that our microbiome is so closely intertwined with our physiology that microbiome composition is reflective of many aspects of our health. Of special importance is recognition of the intersection between chronic diet habits and the microbiome in driving changes in our physiological state. In the foreseeable future it is likely microbiome profiling will be a standard diagnostic test in diverse areas of medicine and that interventions targeting the microbiome will be developed.
All animals are associated with microorganisms for the majority of their life, only embryonic stages are microbe-free. However the complexity of animal-microbe interactions and the nature of their outcomes vary. For some animals their associations with microbes include obligate partnerships with a specific microbe that has obvious benefits for the animals life history (e.g. Coral:Zooxanthellae, Squid:Vibrio, Aphid:Buchnera). For others, the animal may have a specialised structure in which it receives obvious benefits from microbes (e.g. the rumen), but these arise via a community of many microbial species. In contrast microbes can also interact with animals to cause disease. For the majority of animals such specific pathogens have historically been the focus of scientific attention. The remaining microbes were traditionally viewed as commensals. The past decade has seen a dramatic, and ongoing, revision of this view with recognition that those microbes that form communities of stable composition at various body sites (our microbiomes) influence many aspects of our postnatal development and physiology. This is especially true of the gut microbiome.
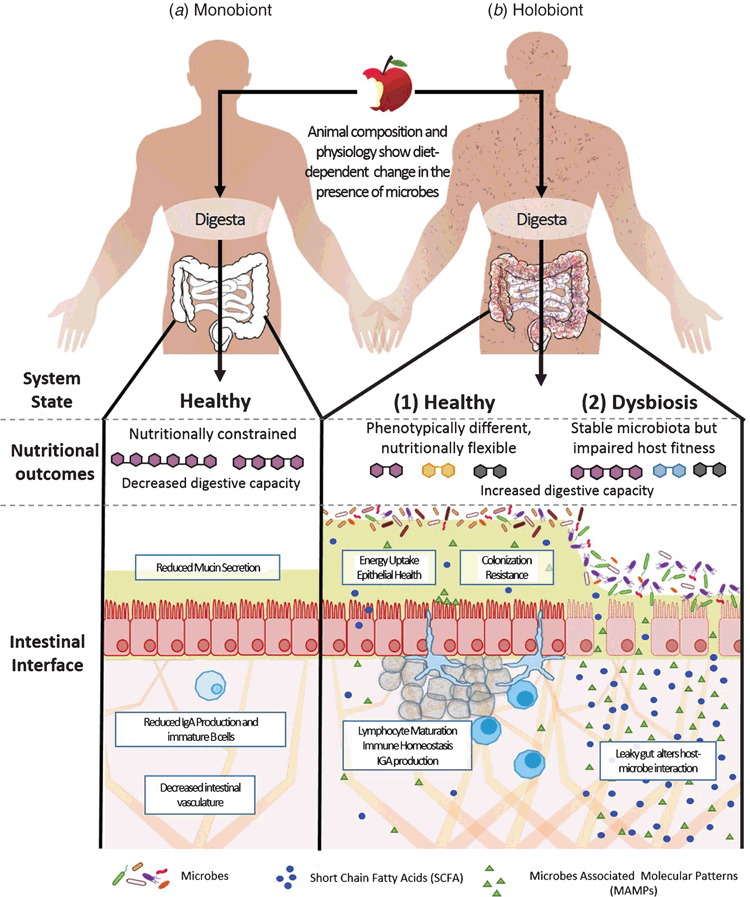
The links between the gut microbiome and host physiological properties are now known to be important to the pathophysiology of metabolic and immunological diseases. Studies of germ-free (GF) animals have demonstrated robust connections between the gut microbiome and host development and physiology. These include roles in vascular development1 and immune cell maturation2,3. A consequence of such developmental effects is that emergent aspects of animal health and physiology such as inflammatory tone4, energy balance5,6, feeding behaviour and even mood and gross anatomy can differ in germ-free animals. Three key points that have emerged from these studies are schematically represented as they might apply to human biology in Figure 1. First, the existence of GF animals indicates the presence of microbes is not essential for the viable development and physiology of an animal. However, GF animals are different, with significant constraints on their environmental fitness, including susceptibility to systemic infection should they be exposed to pathogens and having additional nutritional demands (Figure 1a). Second, if microbes are non-essential to normal physiological processes of animals it is arguable that their most fundamental contribution to the animals state is alteration of how the animal system perceives and responds to its environment, both internal and external. Finally, both microbiome association studies and transplant studies show that different compositions of the microbiome are associated with different host states. Where the microbiome composition gives benefits to desirable host functions such as improved nutrition on available foods or reduced immunopathology we may view the stable host-microbiome system as a healthy holobiont (Figure 1b1). Where the microbiome composition is stable, but results in undesirable host functions such as impaired energy balance or inflammation we may view the holobiont as being in a state of dysbiosis (Figure 1b2).
Although various studies have proposed some key beneficial microbes for gut health (e.g. Faecalibacterium prausnitzii, Coprococcus sp., Ruminococcus bromii, Bacteroidetes spp.), it is rare that presence or absence of any one microbe is specifically associated with health. This reflects a high degree of functional redundancy in the gut community, whereby multiple microbes with similar functions comprise ‘guilds’ that have broadly similar ecological roles within the community. Thus either benefit or detriment to the host system is typically an emergent property of the whole microbial community. Details of the mechanisms by which variations in the gut community impact host nutrition and physiology are now emerging. Broadly speaking microbes contribute to nutrition through the production of metabolites and impact physiology through both metabolites and structural components. Microbial conversion of digestion resistant carbohydrate to short chain fatty acids (SCFAs) or production of vitamins both result in increased capacity for the host system to extract nutrition from food6,7. The SCFAs also exert other effects in the host system, particularly butyrate which is a primary energy source for colonocytes, and therefore important for maintaining epithelial health. SCFAs also impact the function of other tissues and organs in the host by acting as signalling molecules for G-coupled protein receptors (e.g. GPR41, GPR43). Known regulatory roles of SCFAs include: appetite regulation, epigenetic state, gut motility, energy metabolism, endocrine functions, and immune regulation8–10. Since these SCFAs are primarily microbial metabolites it could be argued that the host is monitoring the activity of its microbiome via metabolite sensors and integrating this information into homeostatic regulation. Similarly the host also monitors microbial presence via pattern recognition receptors and signalling pathways contribute to regulation of diverse aspects of immune and metabolic functions. Significantly, disruption of the key metabolite receptors (e.g. GPR4111) or PRR receptors (e.g. TLR512) in mouse knockout models is capable of eliciting disease states, highlighting the importance of microbial signalling for dysbiosis. Collectively these observations show that change in the nature or strength of various microbiome signals, rather than presence/absence of specific microbes, is the primary determinant of health or dysbiosis. Given this, in order to understand dysbioses we must ask what drives disturbance to this?
In wild type animals this signalling-based disturbance to host-microbiome interactions is thought to primarily arise through changes in microbial composition or activity. Since different microbes (e.g. Gram-positive vs Gram-negative) contain different microbe-associated molecular patterns (MAMPs) they drive different PRR-signalling pathways. Similarly since microbes differ in their capacities to degrade macromolecules and which metabolites they produce, changes in community composition will also drive changes in metabolite-signalling pathways. Although diverse factors including anatomical, genotypic, cultural and environmental factors can influence the gut microbial community13–15, it is chronic diet patterns that are thought to be the dominant factor, since what we eat, and the pattern of food consumption, impact the availability of nutrients to gut bacteria for their growth and metabolism. Arguably, the key insight is not the role of the microbes, but rather the role of diet as a key modulator of the interaction between microbes and the host6,14,16. This reflects that major mechanisms of microbial influence are via small molecules that are uniquely microbial cellular components or metabolites. In summary, the concepts of dysbiosis, and animals as holobionts, are changing the way we view human biology, especially modern diseases with a lifestyle component.
In general terms there are two routes to improve health via understanding of the microbiome; diagnostics and interventions. In diagnostics, microbiome signals are included in our evaluation of the host state to inform disease prognosis or intervention plans. In intervention the microbiome is itself the target of manipulation (e.g. prebiotics or probiotics). Greater understanding of host-microbiome interactions can inform both routes through: (1) identification of biomarkers of health or disease in microbiome association studies (e.g. cancer diagnostics17); (2) identification of specific microbes or consortia of microbes that are capable of effecting change if introduced18–20; or (3) intervention in signalling pathways that derive from microbes21. Progress toward these objectives could be achieved across a very wide range of diseases and conditions if microbial community profiling were broadly adopted as a standard test. However, standardised experimental protocols and metadata collection (e.g. sample collection, DNA extraction method22, diet formula) need to be implemented in order to discern patterns that are robust across geographically and culturally diverse populations23.
References
[1] Stappenbeck, T.S. et al. (2002) Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 99, 15451–15455.| Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXjvVKg&md5=1a1e9ec8c0f8c1c1433c4b91a184e016CAS | 12432102PubMed |
[2] Cebra, J.J. (1999) Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69, 1046s–1051s.
| 1:CAS:528:DyaK1MXislGntbk%3D&md5=6120d77dca8db13044a4b40d57c6b71dCAS | 10232647PubMed |
[3] Rhee, K.-J. et al. (2004) Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J. Immunol. 172, 1118–1124.
| Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXnvVSh&md5=124adc02478164a0369a92bd1bea6903CAS | 14707086PubMed |
[4] Lam, Y.Y. et al. (2011) Role of the gut in visceral fat inflammation and metabolic disorders. Obesity (Silver Spring) 19, 2113–2120.
| Role of the gut in visceral fat inflammation and metabolic disorders.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtlyjs7bP&md5=90241429edeed1b2d37753d63e99d342CAS | 21881620PubMed |
[5] Bäckhed, F. et al. (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 101, 15718–15723.
| The gut microbiota as an environmental factor that regulates fat storage.Crossref | GoogleScholarGoogle Scholar | 15505215PubMed |
[6] Turnbaugh, P.J. et al. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031.
| An obesity-associated gut microbiome with increased capacity for energy harvest.Crossref | GoogleScholarGoogle Scholar | 17183312PubMed |
[7] LeBlanc, J.G. et al. (2013) Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 24, 160–168.
| Bacteria as vitamin suppliers to their host: a gut microbiota perspective.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht12js77E&md5=25489da3c446eade1ec317a36659fa23CAS | 22940212PubMed |
[8] Brown, A.J. et al. (2003) The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 278, 11312–11319.
| The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXit1Kgsbc%3D&md5=2787d42ba823200796ad0edaed4f4216CAS | 12496283PubMed |
[9] Licciardi, P.V. et al. (2011) Histone deacetylase inhibition and dietary short-chain Fatty acids. ISRN Allergy 2011, 869647.
| Histone deacetylase inhibition and dietary short-chain Fatty acids.Crossref | GoogleScholarGoogle Scholar | 23724235PubMed |
[10] Kumar, H. et al. (2014) Gut microbiota as an epigenetic regulator: pilot study based on whole-genome methylation analysis. mBio 5.
[11] Samuel, B.S. et al. (2008) Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA 105, 16767–16772.
| Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtlersrvP&md5=2968dad45f2460b5f16d015e922e4829CAS | 18931303PubMed |
[12] Vijay-Kumar, M. et al. (2010) Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231.
| Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXkt1Cgt7o%3D&md5=7694a3a1f3bbe9b396b89e3a51fcdfb3CAS | 20203013PubMed |
[13] Ley, R.E. et al. (2008) Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788.
| Worlds within worlds: evolution of the vertebrate gut microbiota.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFWitLrN&md5=6dab818add424b9f0324ce386f661bf7CAS | 18794915PubMed |
[14] Swartz, T.D. et al. (2013) Preserved adiposity in the Fischer 344 rat devoid of gut microbiota. FASEB J. 27, 1701–1710.
| Preserved adiposity in the Fischer 344 rat devoid of gut microbiota.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXmtFegtbw%3D&md5=820c7c2fd6e281edbdbf78f014159c0aCAS | 23349551PubMed |
[15] Yatsunenko, T. et al. (2012) Human gut microbiome viewed across age and geography. Nature 486, 222–227.
| 1:CAS:528:DC%2BC38Xos1emtLc%3D&md5=b67ba3c98c472c92bddb9439d84e0c4aCAS | 22699611PubMed |
[16] Fleissner, C.K. et al. (2010) Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br. J. Nutr. 104, 919–929.
| Absence of intestinal microbiota does not protect mice from diet-induced obesity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtFCktL3O&md5=90a4e43e91a9ac57c1717911cee58fd7CAS | 20441670PubMed |
[17] Zackular, J.P. et al. (2014) The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. (Phila. Pa.) 7, 1112–1121.
| 1:CAS:528:DC%2BC2cXhvVyjur3J&md5=5ccca942a048d248d02644339746eee8CAS |
[18] Lawley, T.D. et al. (2012) Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8, e1002995.
| Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhs1OisLbO&md5=1eaeb2b5d7218c3c87a0b88344fb31f5CAS | 23133377PubMed |
[19] Adamu, B.O. and Lawley, T.D. (2013) Bacteriotherapy for the treatment of intestinal dysbiosis caused by Clostridium difficile infection. Curr. Opin. Microbiol. 16, 596–601.
| Bacteriotherapy for the treatment of intestinal dysbiosis caused by Clostridium difficile infection.Crossref | GoogleScholarGoogle Scholar | 23866975PubMed |
[20] Atarashi, K. et al. (2013) Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236.
| Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtFShsbfJ&md5=0dcce03f9c770d0383b9d1e8c6341dabCAS | 23842501PubMed |
[21] Ryan, K.K. et al. (2014) FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 509, 183–188.
| FXR is a molecular target for the effects of vertical sleeve gastrectomy.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXns1Wqsr8%3D&md5=95be88fd77bc21fbc5eaa37aaa781d7cCAS | 24670636PubMed |
[22] Kennedy, N.A. et al. (2014) The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS ONE 9, e88982.
| The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing.Crossref | GoogleScholarGoogle Scholar | 24586470PubMed |
[23] Finucane, M.M. et al. (2014) A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS ONE 9, e84689.
| A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter.Crossref | GoogleScholarGoogle Scholar | 24416266PubMed |
Biographies
The authors are all members of the School of Molecular Bioscience and the Microbiome node of the Charles Perkins Centre at the University of Sydney. Their research is focussed on understanding the dynamics of gut microbial community composition, the mechanisms of host-microbe interaction in the gut and development of tools to enable management of the gut microbial ecosystem for health. A particular focus is the relationship between our nutrient environment and its effect on host-microbiome interactions in health. We gratefully acknowledge funding support from the ARC and NHMRC.
Clarissa Febinia is a postgraduate student and the recipient of an Australia Awards Scholarship with affiliations to the Eijkman Institute for Molecular Biology, Jakarta. Her project is on the intersection between cultural, genetic and diet factors in lifestyle disease.
Connie Ha is in the final year of PhD studies on the mechanisms of diet-induced obesity and the recipient of an APRA.
Chau Le is a PhD student working on the influence of gut microbes on regulation of feeding behaviour. She is the recipient of an APRA.
Andrew Holmes is currently an Associate Professor in the School of Molecular Bioscience at the University of Sydney and leads research programs in the Charles Perkins Centre and Marie Bashir Institute. He was the recipient of the 2006 Fenner Prize from the Australian Society for Microbiology. He has general interests in microbial diversity, its evolutionary origins and ecological applications. He is a Senior Editor for Microbiology and The ISME Journal, and a member of the Editorial Boards of Applied and Environmental Microbiology and Environmental Microbiology.