Active screening for multiresistant Enterobacteriaceae
Paul R Ingram A B E and Jon Iredell C D FA Department of Microbiology and Infectious Diseases, Royal Perth Hospital
B School of Pathology and Laboratory Medicine, University of Western Australia
C Centre for Infectious Diseases and Microbiology, Westmead Hospital, NSW 2145, Australia
D Marie Bashir and Westmead Millennium Institutes, University of Sydney
E Email: paul.ingram@health.wa.gov.au
F Email: jonathan.iredell@sydney.edu.au
Microbiology Australia 35(1) 13-16 https://doi.org/10.1071/MA14005
Published: 5 February 2014
The control of multiresistant organisms (MROs) is made difficult by a large reservoir of unrecognised, asymptomatic colonised patients. Hence, active screening is generally used as part of a multifaceted infection control intervention. Active screening for multiresistant Gram-negative bacteria (MRGNB) involves collection of screening specimens from patients with relevant risk factors. Positive results may result in institution of contact precautions, cohorting of patients and enhanced cleaning and surveillance. Active decolonisation is used for some Gram-positive bacteria (e.g. Staphylococcus aureus) but is not thought effective for Gram-negative bacteria, especially those in which resistance is highly transmissible (e.g. plasmid-borne). Accurate and rapid identification of positive specimens allows prompt intervention to interrupt the transmission cycle, and exclusion of MRO colonisation reduces the adverse impact on patient care and hospital workflow. Knowledge of MRO status may also improve the appropriateness of empiric antibiotic prescribing1.
For MRGNB the ratio of colonised to infected patients has been estimated at ~8:1 for extended spectrum beta-lactamases2 and 9:1 for carbapenem-resistant Enterobacteriaceae (CRE)3. In addition to this, the very mechanism by which antibiotic resistance is transferred between common species like Escherichia coli and Klebsiella pneumoniae is such that resistance acquisition can be highly variable in phenotype and often relatively ‘silent’4. Antibiotic susceptible organisms can acquire resistance mechanisms very quickly5, so that the potential for acquisition by gene transfer is itself an important risk to understand.
The most appropriate specimen type for screening of MRGNBs depends on the organism’s likely habitat on the human body or clinical environment. Thus, the most sensitive specimens for detecting Enterobacteriaceae may be rectal swab, peri-rectal swab or faeces, but wounds and respiratory samples and even environmental sampling (sinks, drains, surfaces) may be more important for the control of outbreaks due to organisms such as Pseudomonas or Acinetobacter6.
Increased specimen volume and a higher MRO faecal density both increase yield of screening7,8. For this reason faecal samples may perform better than rectal swabs9, although the latter is more practical. Using more than one screening specimen or specimen type may increase sensitivity6, but again for practical purposes one specimen is usual. Concurrent antibiotic use has a variable impact on MRO faecal density10 but logic dictates that detection of a reservoir of resistance is more likely in the presence of relevant selection pressure.
Active screening specimens may be processed in the laboratory using phenotypic (culture based) or direct genotypic (amplification and detection of bacterial DNA) methods. There is no consensus as to the optimal laboratory processing method but factors that should be considered are local molecular epidemiology (including the likely minimum inhibitory concentration (MIC) of relevant antibiotics against target isolates), availability of expertise and equipment, cost, capacity for integration into laboratory workflow and test performance including sensitivity, specificity, limit of detection and turnaround time. Currently, phenotypic methods are more widely used, although we anticipate that with advances in automation, data processing and reductions in cost these will be replaced by genotypic methods in the future. The main advantages of genotypic methods are shorter achievable turnaround times (and potential for point-of-care testing), improved sensitivity and reduced need for human resources. Potential disadvantages of genotypic methods include higher costs, a need for expertise/complex equipment and the lack of an isolate for susceptibility testing or typing if using direct screening without culture.
Regardless of which screening method (phenotypic or genotypic) is used for detecting transmissible antibiotic resistance, one must accept a trade-off between (a) the risk of overlap between the highest naturally occurring MIC of wild-type organisms (that is, those without the targeted resistance) and those with a low MIC in the presence of a relevant mechanism, especially one that is readily transmissible to an organism in which it facilitates high levels of resistance and (b) the failure to identify a new or variant mechanism. This is illustrated by the overlapping ceftazidime MICs of local E. coli populations with common transmissible ESBL genes (Figure 1).
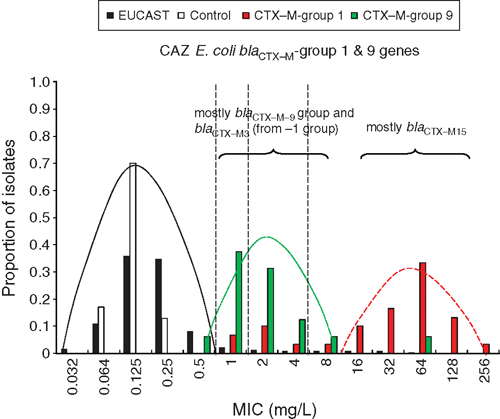
|
We will now use the example of CRE to examine the spectrum of active screening options. CRE are currently infrequent in Australia; in the 2011 AGAR study of hospital-acquired Enterobacteriaceae, eight of 2633 isolates possessed a carbapenemase11. However, recent importation of blaNDM12 and blaOXA-4813 and the development of endemic CRE in Australia (primarily blaIMP-4)14 prompted the Australian Commission on Safety and Quality in Health Care to release national guidelines recommending active screening for CRE15. This document suggests screening high-risk patients using a rectal swab, peri-rectal swab or faeces with or without a swab from open wounds or urinary catheter specimens.
These specimens may be processed using a variety of phenotypic methods that have a turnaround time of 24–48 h. Inhouse methods include a MacConkey plate impregnated with meropenem (1 ug/mL) or the ‘CDC Method’16, which involves inoculation of TSB broth containing a 10 µg meropenem disc, then subculture the next day onto MacConkey agar. Although cheap and readily available, these methods do not perform as well as commercial chromagars17. At the time of writing, only three commercial chromagars designed specifically for CRE detection were available in Australia, all of which contain proprietary ingredients and cost approximately $3–4 per plate. Comparative studies are hampered by a lack of an agreed gold standard but commercial chromagars appear to be highly specific and have reasonable sensitivity17. A new agar medium not yet widely available, known as SuperCarba, may be a promising alternative18. Sensitivity of screening agars is less for isolates with a lower carbapenem MIC19. Phenotypic screens may be made more sensitive by deploying low-level screening cut-offs, although the latter needs understanding of other contributive factors that are more important in some organisms than others (Figure 2). For example, it would be most appropriate to screen with a low antibiotic concentration for a highly transmissible resistance plasmid with commonly low MICs in an organism like E. coli (e.g. typical plasmids carrying blaIMP, blaNDM) but a resistance gene with limited capacity to move beyond the host bacterial species (e.g. blaOXA-23 in Acinetobacter baumannii) might be more efficiently screened using a much higher antibiotic concentration. Screening with meropenem 1 μg/mL is almost certainly too high in E. coli; a cut-off of meropenem or ertapenem ≥0.5 μg/mL has been recommended20.
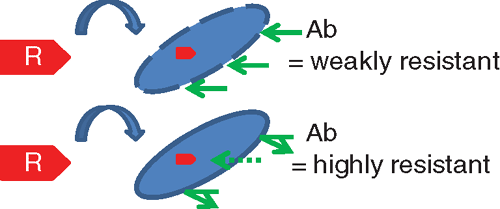
|
The cefotaxime epidemiological cut-off (ECOFF) of 0.25 μg/mL appears to be reliable for metallo-betalactamases and blaKPC and can also be expected to additionally identify plasmid AmpC and ESBL gene carriage. For emerging blaOXA-48/-121-containing strains reported cefotaxime MICs are lower21. Although ESBLs are not uncommon accompaniments, there is no strong genetic link to blaOXA-4822 so that active screening with cefotaxime only may not suffice. An alternative approach with piperacillin/tazobactam and temocillin discs for screening of suspected isolates appeared effective in a large recent European study23. Regardless of the culture-based method being used, laboratories should evaluate test performance in their local setting using clinical specimens and/or stored genotypically confirmed CRE isolates.
Genotypic screening may be conducted by manual or semiautomated nucleic acid test methods such as PCR, gene probe, or some combination or variation of these, either direct or following an antibiotic-containing growth step, as above. Several direct methods are practical for laboratories with basic molecular biological expertise (Table 1), but rely on either a known target or at least an understanding of the likely mechanism. These can be better focused by the judicious use of phenotypic screens, including specific detection of relevant (e.g. carbapenem hydrolysis) activity such as the NP Carba test24.
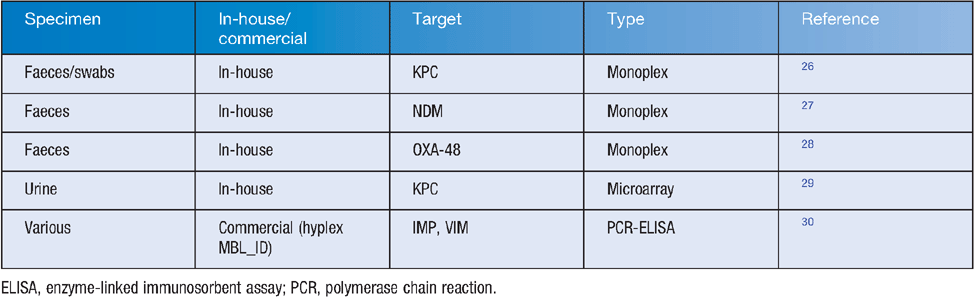
|
Genotypic screens (even direct PCR) can have high predictive value if informed by local epidemiology. The negative predictive value of genetic testing for resistance to aminoglycosides and/or major beta-lactams can be greater than 99.5% in a country like Australia with relatively low pre-test probability25, but the optimal frequency with which this must be surveyed and the applicability of this approach in other countries, including high-prevalence settings, remains to be tested.
In summary, when used alongside other infection control measures, active screening is a useful tool that may assist with limiting the spread of MRGNBs. There is no consensus as to the optimal laboratory processing method, although genotypic methods are likely to become the most frequently used in the future. An understanding of local molecular epidemiology and MICs will assist laboratories in selecting the most appropriate screening assay for their setting.
References
[1] Robinson, J.O. et al. (2013) Knowing prior methicillin-resistant Staphylococcus aureus (MRSA) infection or colonization status increases the empirical use of glycopeptides in MRSA bacteraemia and may decrease mortality. Clin. Microbiol. Infect. , .| Knowing prior methicillin-resistant Staphylococcus aureus (MRSA) infection or colonization status increases the empirical use of glycopeptides in MRSA bacteraemia and may decrease mortality.Crossref | GoogleScholarGoogle Scholar | 24224545PubMed |
[2] Harris, A.D. et al. (2006) What infection control interventions should be undertaken to control multidrug-resistant gram-negative bacteria? Clin. Infect. Dis. 43, S57–S61.
| 16894516PubMed |
[3] Schechner, V. et al. (2013) Asymptomatic rectal carriage of blaKPC producing carbapenem-resistant Enterobacteriaceae: who is prone to become clinically infected? Clin. Microbiol. Infect. 19, 451–456.
| Asymptomatic rectal carriage of blaKPC producing carbapenem-resistant Enterobacteriaceae: who is prone to become clinically infected?Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC38nhtVSrsw%3D%3D&md5=57c1ac761db5a20308e833b5f4789a59CAS | 22563800PubMed |
[4] Espedido, B. et al. (2005) Wide dissemination of a carbapenemase plasmid among gram-negative bacteria: implications of the variable phenotype. J. Clin. Microbiol. 43, 4918–4919.
| Wide dissemination of a carbapenemase plasmid among gram-negative bacteria: implications of the variable phenotype.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVOrtb%2FK&md5=37d0b707f2009ec8273efc749d1a0555CAS | 16145178PubMed |
[5] van Hal, S.J. et al. (2009) Immediate appearance of plasmid-mediated resistance to multiple antibiotics upon antibiotic selection: an argument for systematic resistance epidemiology. J. Clin. Microbiol. 47, 2325–2327.
| Immediate appearance of plasmid-mediated resistance to multiple antibiotics upon antibiotic selection: an argument for systematic resistance epidemiology.Crossref | GoogleScholarGoogle Scholar | 19420178PubMed |
[6] Weintrob, A.C. et al. (2010) Natural history of colonization with gram-negative multidrug-resistant organisms among hospitalized patients. Infect. Control Hosp. Epidemiol. 31, 330–337.
| 20175687PubMed |
[7] D’Agata, E.M. et al. (2002) High rate of false-negative results of the rectal swab culture method in detection of gastrointestinal colonization with vancomycin-resistant enterococci. Clin. Infect. Dis. 34, 167–172.
| 11740703PubMed |
[8] Funk, J.A. et al. (2000) The effect of fecal sample weight on detection of Salmonella enterica in swine feces. J. Vet. Diagn. Invest. 12, 412–418.
| The effect of fecal sample weight on detection of Salmonella enterica in swine feces.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3cvmsFGjsg%3D%3D&md5=6393013551088a66eb6747039d223073CAS | 11021427PubMed |
[9] Lautenbach, E. et al. (2005) Test characteristics of perirectal and rectal swab compared to stool sample for detection of fluoroquinolone-resistant Escherichia coli in the gastrointestinal tract. Antimicrob. Agents Chemother. 49, 798–800.
| Test characteristics of perirectal and rectal swab compared to stool sample for detection of fluoroquinolone-resistant Escherichia coli in the gastrointestinal tract.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXht12nurY%3D&md5=3479688b5e7849e08959783059657979CAS | 15673772PubMed |
[10] Perez, F. et al. (2011) Effect of antibiotic treatment on establishment and elimination of intestinal colonization by KPC-producing Klebsiella pneumoniae in mice. Antimicrob. Agents Chemother. 55, 2585–2589.
| Effect of antibiotic treatment on establishment and elimination of intestinal colonization by KPC-producing Klebsiella pneumoniae in mice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXnt1Kit7s%3D&md5=ea9fd055ee7a988aa8b815854e393066CAS | 21422202PubMed |
[11] Turnidge, J. et al. (2012) Gram-negative survey: 2011 antimicrobial susceptibility report, Commonwealth of Australia, Department of Health and Ageing. March.
[12] Fernando, G.A. et al. (2010) A risk for returned travellers: the ‘post-antibiotic era’. Med. J. Aust. 193, 59.
| 20618119PubMed |
[13] Espedido, B.A. et al. (2013) Whole genome sequence analysis of the first Australian OXA-48-producing outbreak-associated Klebsiella pneumoniae isolates: the resistome and in vivo evolution. PLoS ONE 8, e59920.
| Whole genome sequence analysis of the first Australian OXA-48-producing outbreak-associated Klebsiella pneumoniae isolates: the resistome and in vivo evolution.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXmtFWrsb4%3D&md5=e75fde614a9a009d2c49f1998b735196CAS | 23555831PubMed |
[14] Peleg, A.Y. et al. (2005) Dissemination of the metallo-beta-lactamase gene blaIMP-4 among gram-negative pathogens in a clinical setting in Australia. Clin. Infect. Dis. 41, 1549–1556.
| 1:CAS:528:DC%2BD2MXhtlahsb%2FI&md5=8949ac9baa85cb32c9e818eea1824223CAS | 16267725PubMed |
[15] Recommendations for the control of multi-drug resistant Gram-negatives: carbapenem resistant Enterobacteriacea. http://www.safetyandquality.gov.au/publications/mrgn-guidelines-enterobacteriacea/
[16] Centers for Disease Control and Prevention (CDC) (2009) Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb. Mortal. Wkly Rep. 58, 256–260.
| 19300408PubMed |
[17] Savard, P. et al. (2013) The challenges of carbapenemase-producing Enterobacteriaceae and infection prevention: protecting patients in the chaos. Infect. Control Hosp. Epidemiol. 34, 730–739.
| The challenges of carbapenemase-producing Enterobacteriaceae and infection prevention: protecting patients in the chaos.Crossref | GoogleScholarGoogle Scholar | 23739078PubMed |
[18] Nordmann, P. et al. (2012) Detection of carbapenemase producers in Enterobacteriaceae by use of a novel screening medium. J. Clin. Microbiol. 50, 2761–2766.
| Detection of carbapenemase producers in Enterobacteriaceae by use of a novel screening medium.Crossref | GoogleScholarGoogle Scholar | 22357501PubMed |
[19] Carrer, A. et al. (2010) Use of ChromID extended-spectrum beta-lactamase medium for detecting carbapenemase-producing Enterobacteriaceae. J. Clin. Microbiol. 48, 1913–1914.
| Use of ChromID extended-spectrum beta-lactamase medium for detecting carbapenemase-producing Enterobacteriaceae.Crossref | GoogleScholarGoogle Scholar | 20237104PubMed |
[20] Cohen Stuart, J. and Leverstein-Van Hall, M.A. (2010) Guideline for phenotypic screening and confirmation of carbapenemases in Enterobacteriaceae. Int. J. Antimicrob. Agents 36, 205–210.
| Guideline for phenotypic screening and confirmation of carbapenemases in Enterobacteriaceae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXptVSrs7w%3D&md5=54452b335f55970b9ff7ac042faed7dfCAS | 20598859PubMed |
[21] Walther-Rasmussen, J. and Hoiby, N. (2006) OXA-type carbapenemases. J. Antimicrob. Chemother. 57, 373–383.
| OXA-type carbapenemases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtlOgtLc%3D&md5=f0be06449ac60853154cdc127a5fb669CAS | 16446375PubMed |
[22] Poirel, L. et al. (2012) OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67, 1597–1606.
| OXA-48-like carbapenemases: the phantom menace.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xotleqtrk%3D&md5=2507cb4b1921aaec18d306d7177b1f04CAS | 22499996PubMed |
[23] Huang, T.D. et al. (2014) Temocillin and piperacillin/tazobactam resistance by disc diffusion as antimicrobial surrogate markers for the detection of carbapenemase-producing Enterobacteriaceae in geographical areas with a high prevalence of OXA-48 producers. J. Antimicrob. Chemother. 69, 445–450.
| Temocillin and piperacillin/tazobactam resistance by disc diffusion as antimicrobial surrogate markers for the detection of carbapenemase-producing Enterobacteriaceae in geographical areas with a high prevalence of OXA-48 producers.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXlvFOktg%3D%3D&md5=28b5fceba59cdf459efef8aa89c9e448CAS | 24055766PubMed |
[24] Nordmann, P. et al. (2012) Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 18, 1503–1507.
| Rapid detection of carbapenemase-producing Enterobacteriaceae.Crossref | GoogleScholarGoogle Scholar | 22932472PubMed |
[25] Ginn, A.N. et al. (2013) Limited diversity in the gene pool allows prediction of third-generation cephalosporin and aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae. Int. J. Antimicrob. Agents 42, 19–26.
| Limited diversity in the gene pool allows prediction of third-generation cephalosporin and aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXot1CjtLg%3D&md5=ada1ef59ef7fd25ab16ec83e7749e23eCAS | 23706544PubMed |
[26] Richter, S.N. et al. (2012) Ultrarapid detection of blaKPC(1)/(2)-(1)(2) from perirectal and nasal swabs by use of real-time PCR. J. Clin. Microbiol. 50, 1718–1720.
| Ultrarapid detection of blaKPC(1)/(2)-(1)(2) from perirectal and nasal swabs by use of real-time PCR.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XnsF2lur8%3D&md5=d4c69c9978f71e30e005cc158a64bf33CAS | 22378915PubMed |
[27] Liu, W. et al. (2012) Sensitive and rapid detection of the New Delhi metallo-beta-lactamase gene by loop-mediated isothermal amplification. J. Clin. Microbiol. 50, 1580–1585.
| Sensitive and rapid detection of the New Delhi metallo-beta-lactamase gene by loop-mediated isothermal amplification.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XnsF2lt7g%3D&md5=88f9aad5859d900f530971d12b76faacCAS | 22357496PubMed |
[28] Naas, T. et al. (2013) Real-time PCR for detection of blaOXA-48 genes from stools. J. Antimicrob. Chemother. 68, 101–104.
| Real-time PCR for detection of blaOXA-48 genes from stools.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhvVChtr7N&md5=d63c0ef2ef7eac8a694207a6c8f36bcaCAS | 22969079PubMed |
[29] Peter, H. et al. (2012) Direct detection and genotyping of Klebsiella pneumoniae carbapenemases from urine by use of a new DNA microarray test. J. Clin. Microbiol. 50, 3990–3998.
| Direct detection and genotyping of Klebsiella pneumoniae carbapenemases from urine by use of a new DNA microarray test.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXktlSktQ%3D%3D&md5=692bffd4b988db69cff844bde204a90eCAS | 23035190PubMed |
[30] Avlami, A. et al. (2010) Detection of metallo-beta-lactamase genes in clinical specimens by a commercial multiplex PCR system. J. Microbiol. Methods 83, 185–187.
| Detection of metallo-beta-lactamase genes in clinical specimens by a commercial multiplex PCR system.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtlWjsrvI&md5=35176b35c126c21bff84d6d0c08ded56CAS | 20807554PubMed |
Biographies
Dr Paul Ingram graduated from the University of Western Australia in 1999. He is both an Infectious Diseases Physician and Clinical Microbiologist working at Royal Perth Hospital, Western Australia. He completed an Infectious Diseases Fellowship at the National University of Singapore and is currently Clinical Senior Lecturer at the University of Western Australia. His interests include beta-lactamases in gram-negative bacteria, the epidemiology and infection control responses to antibiotic resistance and antimicrobial stewardship.
Jon Iredell is an Infectious Disease Physician and Microbiologist who divides his time between Westmead Hospital in a combined Infectious Diseases and Microbiology Department and his research, which is supported by the NHMRC at the University of Sydney. His major interests are in critical infection, including the study of bacterial septic shock, and in bacterial genetics and ecology. He is Director, NHMRC Centre of Research Excellence in Critical Infection and conjoint Professor of Medicine and Microbiology at Sydney Medical School/Westmead Millennium Institute and the Marie Bashir Institute and Director of Infectious Diseases, Western Sydney and Westmead Hospital.


