A molecular assessment of species boundaries and relationships in the Australian brine shrimp Parartemia (Anostraca: Parartemiidae)
Md Aminul Islam A B * , Jennifer Chaplin A , Angus D’Arcy Lawrie A , Mahabubur Rahman A C and Adrian Pinder B
A B * , Jennifer Chaplin A , Angus D’Arcy Lawrie A , Mahabubur Rahman A C and Adrian Pinder B
A
B
C
Abstract
Australian salt lakes contain a diverse range of endemic invertebrates. The brine shrimp Parartemia is among the most speciose and salt-tolerant of these invertebrates. The morphotaxonomy of Parartemia is well established but there has only been limited molecular assessment of the phylogenetic relationships and boundaries of the morphospecies. We used multiple genetic markers (nuclear 28S and mitochondrial 16S and COI) and tree-building methods (Bayesian inference and maximum likelihood) to investigate the phylogeny of Parartemia. We also used species delimitation methods to test the validity of morphological species designations. The data set included all but 2 of the 18 described Parartemia morphospecies, collected from a total of 93 sites from across southern Australia plus some sequences from GenBank. The results identified large amounts of molecular divergence (e.g. COI P-values of up to 25.23%), some groups of closely related species (which also usually shared some morphological similarities) and some distinctive species, although the relationships among divergent lineages were generally not well resolved. The most conservative set of results from the species delimitation analyses suggests that the morphotaxonomy is largely accurate, although many morphospecies comprised divergent genetic lineages separated by COI P-values of up to 17.02%. Two putative new morphospecies, three cryptic species and one synonymy were identified. Our findings improve the knowledge of Parartemia taxonomy and will facilitate the development of future studies and conservation of this taxon.
Keywords: anostraca, cryptic species, extremophiles, high mitochondrial DNA divergence, integrative taxonomy, invertebrate conservation, morphological and molecular species congruence, salt lakes, species delimitation.
Introduction
Most biological disciplines depend on adequate taxonomy, particularly accurate species determination (Bortolus 2008; Jackson et al. 2022), which ideally should be supported by multiple lines of evidence (Padial et al. 2010). Nevertheless, only ~1.2 million–1.9 million of an estimated 5 million–15 million species have been described (Stork 1997; Mora et al. 2011; Costello et al. 2013; Jackson et al. 2022). Furthermore, taxonomic effort has not been evenly spread across taxa or ecosystems (Donaldson et al. 2016; Di Marco et al. 2017; Troudet et al. 2017). The biodiversity of inland saline waters is one area that requires better documentation (Lawrie et al. 2021; Saccò et al. 2021).
Salt lakes are exceedingly common in Australia, where they are usually ephemeral and show variable and often very high salinities (Bayly and Williams 1966; De Deckker 1983; Lawrie et al. 2021). Some invertebrate taxa have undergone substantial diversification in these environments or their precursors (De Deckker 1983; Remigio et al. 2001; Lawrie et al. 2021; Lawrie et al. 2023; Rahman et al. 2023). The endemic brine shrimp Parartemia, the only genus in the Parartemiidae (Weekers et al. 2002), is probably the best example of this. This taxon and its closest known relative Artemia are often regarded as extremophiles due to their occurrence in hypersaline lakes (Timms 2014). Divergence between these two lineages is ancient, dating back to around the breakup of Gondwana (Coleman et al. 1998; Anufriieva and Shadrin 2013). Native Artemia species occur on all continents except Australia and Antarctica (Ruebhart et al. 2008; Muñoz and Pacios 2010); however, Parartemia is endemic to Australia. Although much more widespread, Artemia with nine sexually reproducing species plus a range of parthenogens (Rogers 2013; Asem et al. 2023a, 2023b) appears to have far fewer species than Parartemia, which has eighteen described morphospecies (Timms 2014).
The first described Parartemia species was P. zietziana by Sayce (1903), which occurs in South Australia, Victoria and Tasmania (Timms et al. 2009). Prior to this, all Parartemia specimens were probably misidentified as Artemia (Timms 2014). Parartemia zietziana was considered to be the sole representative of the genus until Linder (1941) described six species from Western Australia: P. contracta, P. cylindrifera, P. extracta, P. informis, P. longicaudata and P. serventyi. Another species, P. minuta, was described by Geddes (1973). More recently, Timms and Hudson (2009) added four morphospecies (P. acidiphila, P. auriciforma, P. triquetra and P. yarleensis) from South Australia and Timms (2010) added another six (P. bicorna, P. boomeranga, P. laticaudata, P. mouritzi, P. purpurea and P. veronicae) from Western Australia, giving a total of eighteen described morphospecies. A detailed identification guide for Parartemia morphospecies has been provided by Timms (2012). Three male morphological characters – the ventral process, medial process, and the space between the second antenna basal antennomeres – are important in species diagnoses (Timms 2012). Although Parartemia morphotaxonomy is well established, it has been subject to only limited molecular testing (see below). More intensive sampling is also likely to reveal more species (Timms 2010).
Remigio et al. (2001) conducted the only previous study on the molecular phylogeny of Parartemia. The results were mainly based on 341 bp of the mitochondrial 16S rRNA gene and a small number of specimens per species (mostly just one) from eight known species and two putative new species (corresponding to their samples POP4 and POP5). The results suggested that Parartemia morphospecies are genetically distinct, with high levels of genetic divergence between species. The study identified four main clades (labelled A–D), two of which contained the haplotypes of multiple species whereas the other two contained haplotypes of single distinctive species. Their molecular data supported some previous hypotheses of species relationships based on limited morphological traits but not others. Parartemia minuta was the most genetically distinctive species in their dataset. A comprehensive analysis, with a more complete set of species and additional genetic loci, is required to fully understand Parartemia systematics.
Salt lake environments in Australia (and elsewhere) are deteriorating due to the effects of global climate change (e.g. mainly increasing aridity) and other anthropogenic effects (Williams 2002; Timms 2005; Jellison et al. 2008). This has led to general concerns about the fate of the invertebrate inhabitants of these lakes (Timms 2005; Pinder et al. 2009; Timms et al. 2009; Lawrie et al. 2021). Some species of Parartemia have been singled out as vulnerable to extinction, although in some cases this appears to be based on incomplete data. For example, P. contracta currently has ‘Vulnerable’ status on the IUCN Red List based on a 1996 assessment, but since that assessment more populations have been discovered and the listing may be unnecessary (Timms et al. 2009). On the other hand, Timms et al. (2009) suggested that P. boomeranga (formerly Parartemia sp. ‘c’) and P. extracta should be assessed for inclusion in the IUCN Red List as ‘Vulnerable’ because distributional records indicate that the ranges of these species are contracting. In a separate study, Timms (2012) suggested that P. boomeranga was extremely rare or possibly even extinct. To manage their conservation, a thorough understanding of the phylogeny, taxonomy and distributions of Parartemia species is needed (see Rogers and Aguilar 2020).
We used multiple molecular markers to investigate the phylogenetic relationships among almost all described Parartemia morphospecies, typically with multiple representatives from multiple populations of each morphospecies. We also used single mitochondrial DNA loci (16S and COI) and species delimitation methods to assess the validity of 16 described Parartemia morphospecies that were included in this study as well as of two putative new morphospecies found in our collections. Finally, we compared our results to those of Remigio et al. (2001) to test the validity of their findings regarding the phylogeny of Parartemia, which were based on a more limited data set.
Materials and methods
Specimen collection and preservation
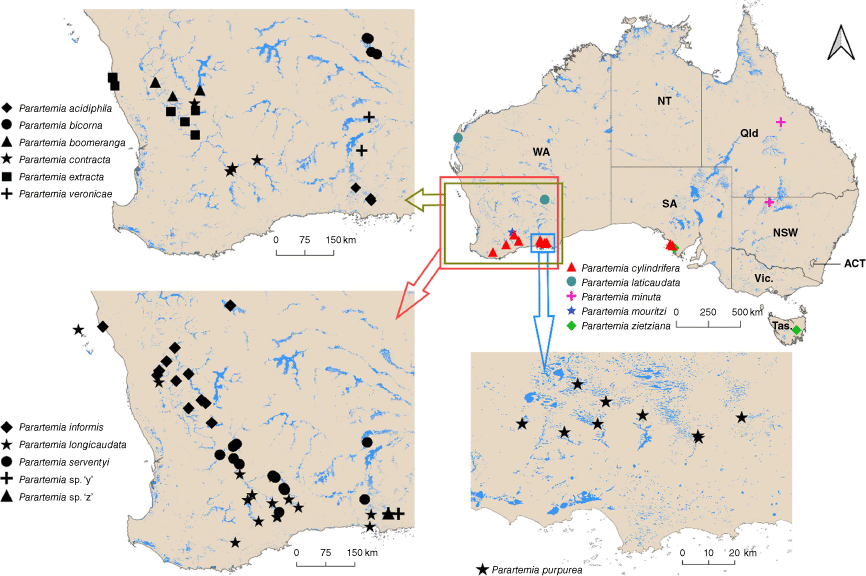
We collected Parartemia specimens from 84 salt lakes in Australia (mainly Western Australia) between September 2017 and August 2022 (Fig. 1 and Supplementary Table S1). Specimens were collected using a dip net and then euthanised by freezing and preserved in 100% ethanol. Specimens from a site in Tasmania (provided by the Tasmanian Museum and Art Gallery) and eight sites in Western Australia (from the Department of Biodiversity, Conservation and Attractions, the Western Australian Museum and the Stantec Australia Pty Ltd) were also used (details in Supplementary Table S1).
Approximate locations of Parartemia collection sites. Sites in close proximity may not always be distinguishable in the figure (see detailed information in Supplementary Table S1). The hydrological information, shown in blue, is sourced from the national surface water database of Geoscience Australia (www.ga.gov.au). The full Australian map includes the state and territory boundaries, WA, Western Australia; SA, South Australia; NT, Northern Territory; Qld, Queensland; NSW, New South Wales; ACT, Australian Capital Territory; Vic., Victoria; and Tas., Tasmania. No specimens of P. minuta were collected in this study but GenBank sequences for this species from Lake Buchanan in Qld and an unspecified site in the Paroo area in NSW were included (details in Supplementary Table S1).
Identification of morphospecies
All Parartemia specimens were identified to morphospecies using the morphological criteria of Timms (2012). All known morphospecies were represented in the samples except for P. auriciforma, P. yarleensis and P. minuta. Parartemia auriciforma is only known from one site in central Australia, P. yarleensis is only known from inland South Australia and P. minuta occurs in inland areas in South Australia, Victoria, New South Wales and Queensland (Timms et al. 2009). Our attempts to obtain either fresh or ethanol-preserved specimens of these three species were unsuccessful. However, for P. minuta, we included 28S and 16S sequences from GenBank (see below) and morphological data from Timms (2012). We collected two groups of specimens whose morphology did not match those of any described species (see ‘Results’) – herein these groups are referred to as Parartemia sp. ‘y’ and Parartemia sp. ‘z’.
DNA extractions, PCR amplification and sequencing
Genomic DNA was extracted from thoracic segments V through VII or thoracopods III through VII using a Masterpure Complete DNA and RNA Purification Kit (Epicentre) following the manufacturer’s instructions. Taxonomically significant characters (head, genital segments and abdomen) were left intact for later cross-checking with molecular findings if required. Negative controls, i.e. assays with reagents but no added tissue or DNA, were included in every extraction to check for contamination.
One nuclear genetic region (28S) and two mitochondrial genetic regions (16S and COI) were used. Details of the PCR primers and amplicon lengths are given in Table 1. Except for the universal primers LCO1490 and HCO2198 (Folmer et al. 1994), all primers were designed specifically for Parartemia because it was not possible to amplify the target region from all species or all populations of a species with preexisting primers (see Table 1, Supplementary Tables S2 and S3).
| Name | Forward primers | Name | Reverse primers | |
|---|---|---|---|---|
| 28S | ||||
| 28S11 | ACAAGTACCGCGAGGGAAAGT | 28S32 | CGCCAGTTCTGCTTACCAAAA | |
| 28S71 | TGGTAAACTCCATCTAAGGCTAA | |||
| 16S | ||||
| 16SarPara | CGCCTGTTTAACAAAAACATAGC | 16SbrPara | TGAACTCAGATCACGTAGGG | |
| COI | ||||
| LCO1490 | GGTCAACAAATCATAAAGATATTGG | HCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | |
| LCOPara | CAATCACAAAGATATTGGAACCC | HCOPara | ACTTCAGGGTGACCAAAAAATCAG | |
| COI101 | GCACCTATTATCGGCCACTTT | COI102 | TGGTGGGCTCAGACAACAAA | |
| Facid-1 | TCTACGAACCATAGGGACATTG | Rboom-2 | TTCTGGGTGACCAAAAAACCAG | |
| Fboom-2 | ACTCTACAAACCATAAGGACATTG | Rinfo-1 | CCTCTGGATGGCCGAAAAATC | |
| Fext-2 | ATTCTACGAATCACAAGGATATTGG | Rlong-1 | CTTCTGGGTGACCAAAAAACCA | |
| Finfo-1 | TATGCAACGCTGACTATATTCTAC | |||
| Finfo-3 | TATGCAACGCTGGCTGTACTC | |||
| Flong-1 | ACTCTACAAATCATAAGGACATCG | |||
| Flong-2 | ACTCTACAAATCATAAGGACATTGG | |||
| Fser-1 | ACTCTACAAACCATAAGGACATCG | |||
All primers were designed in this study, except for the universal primers LCO1490 and HCO2198 (Folmer et al. 1994). Details of primers used for each species are in Supplementary Tables S2 and S3.
PCR reaction volumes were 25 μL containing 5 µL of GoTaq Reaction Buffer (Promega), 0.5 μL of dNTPs (10 mM per nucleotide), 0.25 μL of each of forward and reverse primers (10 µM), 2 μL of MgCl2 (25 mM), 0.35 μL of bovine serum albumin (10 µg µL–1), 0.125 μL of GoTaq G2 Hot Start Taq Polymerase, 1 μL of DNA and adjusted to the final volume using PCR grade water. PCR reactions for all three markers were (i) 95°C initial denaturation temperature for 5 min; (ii) 40 cycles of denaturation at 94°C for 1 min, annealing at 48°C for 1 min and extension at 72°C for 45 s; and (iii) a final extension at 72°C for 7 min. PCR products were purified using Exo-SAP purification (Dugan et al. 2002) and sequenced in the forward and reverse directions in an automatic ABI 3700 sequencer (Applied Biosystems) by Macrogen Inc. (South Korea).
Sequence data
The sequencing chromatograms were visualised in Chromas (ver. 2.6.5, Technelysium Pty Ltd, Australia). Forward and reverse sequences were compared, and any ambiguities were corrected. The consensus 16S sequences were aligned using MAFFT online version (see http://mafft.cbrc.jp/alignment/server/) with the G-INS-i strategy, whereas the consensus COI sequences were aligned using MUSCLE (ver. 5, see http://www.drive5.com/muscle/; Edgar 2004) in MEGA X (ver. 10.2.6, see https://www.megasoftware.net; Kumar et al. 2018). The 28S sequences were aligned as described for 16S except that we used the Q-INS-i strategy and made manual adjustments by eye in regions containing indels. We found no evidence that nuclear copies of COI had inadvertently been included in our Parartemia dataset as, for example, there were few amino acid substitutions, and no indels or stop codons, in the translated sequences (see Raupach and Radulovici 2015). Haplotypes in the 28S, 16S and COI datasets were identified using DnaSP (ver. 6.12.03, see http://www.ub.edu/dnasp/; Rozas et al. 2017). All new haplotypes have been deposited in GenBank with accession numbers listed in Supplementary Table S1. This study also used GenBank sequences for Parartemia (including one 28S and two 16S haplotypes of P. minuta) and a range of outgroup taxa (see Supplementary Tables S1 and S4). A multigene (28S, 16S and COI) concatenated dataset was assembled in MEGA X (Kumar et al. 2018), using individuals for which data from at least two of the three targeted genetic regions were available.
Phylogenetic analysis
Phylogenetic analyses were conducted using Bayesian inference (BI) and maximum likelihood (ML) frameworks on the concatenated dataset and on single locus 28S, 16S and COI datasets. The former is widely recognised as the most reliable approach for phylogenetic analysis (Wiens and Moen 2008). The concatenated dataset was used to investigate species relationships within Parartemia. The 16S and COI markers were also used to investigate the validity of Parartemia morphospecies, as they are effective in identifying crustacean species (Remigio et al. 2001; Costa et al. 2007; Raupach and Radulovici 2015).
The best nucleotide substitution models (GTR + I + G for 28S and TrN + G + I for 16S and COI) were selected using jModelTest 2 (ver. 2.1.9, see https://github.com/ddarriba/jmodeltest2; Darriba et al. 2012) based on the Bayesian information criterion (BIC). Maximum likelihood molecular clock tests conducted in MEGA X (Kumar et al. 2018) revealed that the partitioned concatenated dataset and the single locus datasets did not adhere to the assumptions of a strict clock. BI analysis for each dataset was separately conducted in BEAST (ver. 1.10.4, see https://beast.community/beast; Suchard et al. 2018), employing the identified substitution models (see above) with the uncorrelated relaxed clock and coalescent constant population size as tree priors. The analysis was run for 50 million generations, and the estimated sample size (ESS) was checked by looking at the log-output file in Tracer (ver. 1.7.2, see https://github.com/beast-dev/tracer/releases/tag/v1.7.2; Rambaut et al. 2018). A burn in of 25% of the initial trees was discarded, and the final tree was produced in TreeAnnotator (a BEAST-distributed program) and visualised in FigTree (ver. 1.4.4, A. Rambaut, see http://tree.bio.ed.ac.uk/software/figtree/). The analysis was also performed for each dataset using the uncorrelated relaxed clock and Yule process as tree priors, but the ESS was low (<200), even after increasing the MCMC chain length to 100 million generations, and thus the results were discarded.
The ML phylogenetic analysis was performed separately for the concatenated, 28S, 16S and COI datasets on the IQ-TREE web server (see http://iqtree.cibiv.univie.ac.at; Trifinopoulos et al. 2016) using 5000 ultrafast bootstrap replicates and the above-mentioned substitution models.
Validity of Parartemia morphospecies and pairwise distances
Three species delimitation analyses were employed to assess the validity of Parartemia morphospecies using the 16S and COI datasets (without outgroups). Firstly, the Assemble Species by Automatic Partitioning (ASAP), which relies on pairwise genetic distances (Puillandre et al. 2021), was conducted on the ASAP website (see https://bioinfo.mnhn.fr/abi/public/asap) using the Kimura (K80) substitution model (Kimura 1980) and default settings. Of the 10 best ASAP partition schemes included in the results, the one with the lowest ASAP score and another one with the smallest number of partitions were chosen. Secondly, the maximum likelihood implementation of the multi-rate Poisson tree processes (mPTP) (Kapli et al. 2017) was performed on the mPTP webserver (see https://mptp.h-its.org) with default settings using a BI phylogenetic tree generated by BEAST (Suchard et al. 2018) based on the same parameters as outlined in the ‘Phylogenetic analysis’ section (see above). Lastly, the General Mixed Yule Coalescent (GMYC) analysis (Fujisawa and Barraclough 2013) was performed in R (ver. 4.3.3, R Foundation for Statistical Computing, Vienna, Austria, see https://www.r-project.org/) using the same BI phylogenetic tree as used for mPTP (see Michonneau 2016 for further details).
MEGA X (Kumar et al. 2018) was used to compute uncorrected p-distances and Kimura two-parameter (K2P) (Kimura 1980) distances between Parartemia haplotypes in the 28S, 16S and COI datasets.
Results
General information
After aligning and trimming, a total of 28 28S (674 bp excluding gaps), 100 16S (474 bp excluding gaps) and 161 COI (658 bp) haplotypes of Parartemia were identified in sequences from 50, 113 and 232 individuals respectively.
Substantial genetic divergence was present in the 16S (e.g. p-distance of up to 20.34%) and especially the COI (e.g. p-distance of up to 25.23%) genetic regions in Parartemia. As expected, divergence in the 28S region was more limited (e.g. p-distance of up to 2.8%), with few differences between most species (Supplementary Table S5). Accordingly, the relationships among Parartemia species were poorly resolved in 28S phylogeny (Supplementary Fig. S1), although the 28S data did provide strong support for the monophyly of Parartemia (see below).
Monophyly of Parartemia and species relationships
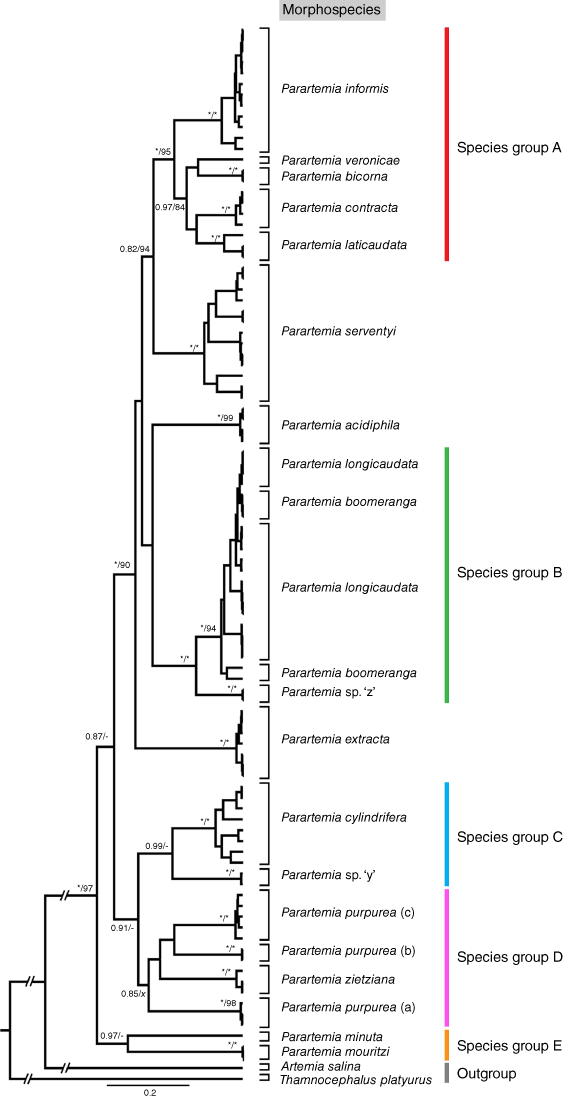
All Parartemia haplotypes formed a well-supported monophyletic group in both BI and ML concatenated phylogenetic trees (BPP = 1 and 97% bootstrap value; Fig. 2 and Supplementary Fig. S2), as well as in the BI and ML 28S (Supplementary Fig. S1) and 16S trees (see Fig. 3 and Supplementary Fig. S3) and the ML COI tree (see Supplementary Fig. S4). In the BI COI tree, all Parartemia haplotypes formed a monophyletic group, but the node support was low (see Fig. 4).
Bayesian inference (BI) phylogenetic tree for the Parartemia concatenated dataset (COI, 16S and 28S). Maximum likelihood (ML) phylogenetic tree is available in Supplementary Fig. S2. Bayesian Posterior Probability (BPP, when ≥0.80) and bootstrap values from the ML tree (when ≥80%) are indicated at nodes (BPP/bootstrap). For nodes where one value was above the threshold and the other was below, the latter is indicated by hyphen (-). BPP values of 1 and bootstraps of 100% are indicated by asterisks (*). Node with bootstrap value (x) indicates that the node-level species composition is not supported by the ML phylogenetic tree. Species names are given based on Parartemia morphotaxonomy (morphospecies). Coloured bars to the right of the tree indicate species groups identified within Parartemia in this study, named alphabetically (A–E).
Bayesian inference (BI) phylogenetic tree of Parartemia 16S haplotypes. Maximum likelihood (ML) phylogenetic tree is available in Supplementary Fig. S3. Bayesian Posterior Probability (BPP, when ≥0.80) and bootstrap values from the ML tree (when ≥80%) are indicated at nodes (BPP/bootstrap). For nodes where one value was above the threshold and the other was below, the latter is indicated by hyphen (-). BPP values of 1 and bootstraps of 100% are indicated by asterisks (*). Each haplotype is denoted by its GenBank accession number followed by its morphospecies name. The green, blue, magenta and orange vertical rectangles indicate recovered species partitions based on Assemble Species by Automatic Partitioning (ASAP) lowest score scheme (ASAP1), ASAP least partition scheme (ASAP2), multi-rate Poisson Tree Processes (mPTP) and Generalised Mixed Yule Coalescent (GMYC) analyses respectively. Species hypotheses (SH) determined by this study are indicated by black bars under the SH heading. Grey-coloured bars to the right of the coloured rectangles indicate species groups (SG) identified within Parartemia in this study, named alphabetically (A–E).
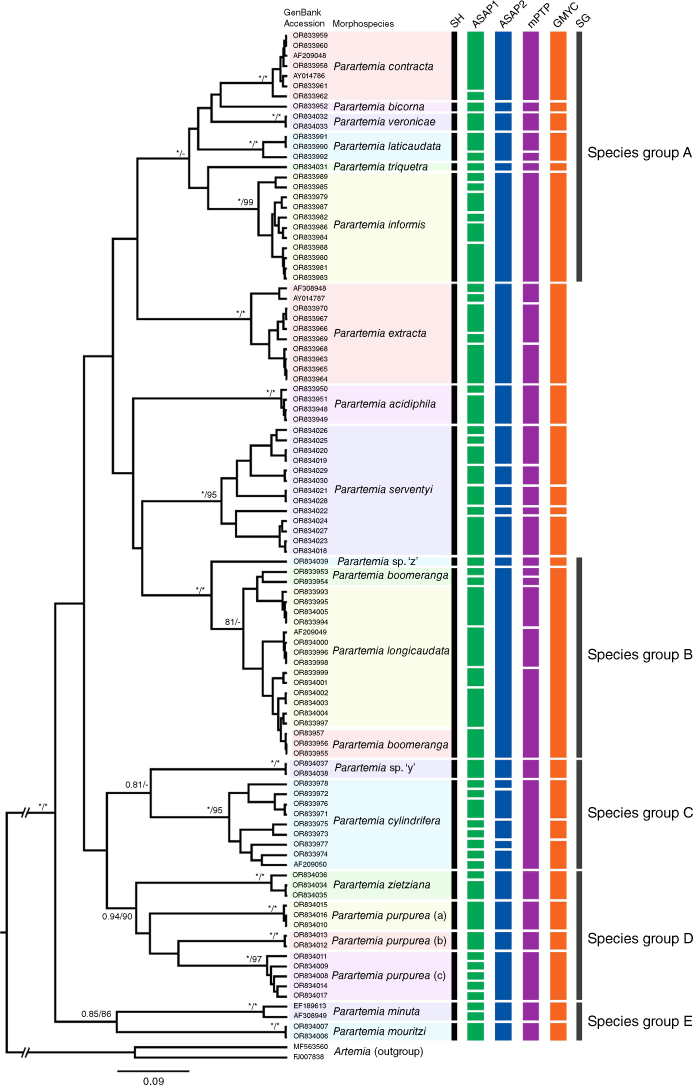
Bayesian inference (BI) phylogenetic tree of Parartemia COI haplotypes. Maximum likelihood (ML) phylogenetic tree is available in Supplementary Fig. S4. Bayesian Posterior Probability (BPP, when ≥0.80) and bootstrap values from the ML tree (when ≥80%) are indicated at nodes (BPP/bootstrap). For nodes where one value was above the threshold and the other was below, the latter is indicated by a hyphen (-). BPP values of 1 and bootstraps of 100% are indicated by asterisks (*). Each haplotype is denoted by its GenBank accession number followed by its morphospecies name. The green, blue, magenta and orange vertical rectangles indicate recovered species partitions based on Assemble Species by Automatic Partitioning (ASAP) lowest score scheme (ASAP1), ASAP least partition scheme (ASAP2), multi-rate Poisson Tree Processes (mPTP) and Generalised Mixed Yule Coalescent (GMYC) analyses respectively. Species hypotheses (SH) determined by this study are indicated by black bars under the SH heading. Grey-coloured bars to the right of the coloured rectangles indicate species groups (SG) identified within Parartemia in this study, named alphabetically (A–E; species group E is not shown due to the absence of COI data for P. minuta).
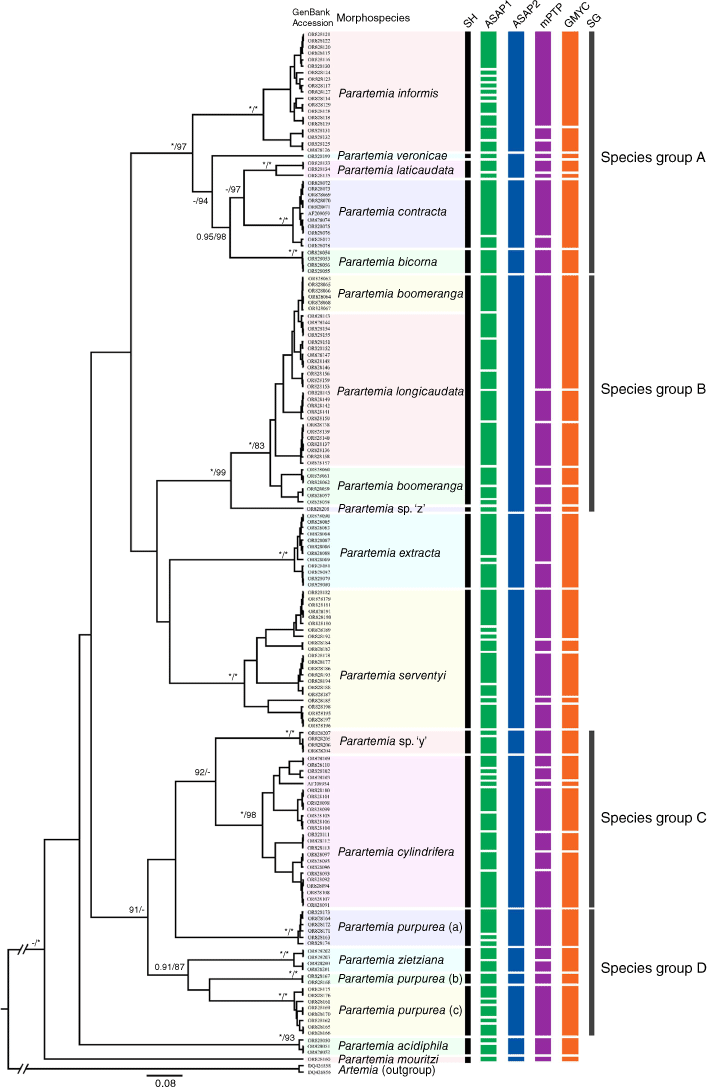
Although some aspects of the relationships among Parartemia species were not well resolved, particularly for deeper divergences, some species groups and divergent species were evident. The haplotypes of five species (P. bicorna, P. contracta, P. informis, P. laticaudata and P. veronicae) were invariably grouped together in a single usually well-supported clade in the BI and ML concatenated, 16S and COI trees (species group A in Fig. 2–4). A sixth species, P. triquetra, for which there were no 28S or COI data, was also included in this group in the 16S trees (Fig. 3 and Supplementary Fig. S3). Similarly, Parartemia sp. ‘z’ and the P. boomeranga and P. longicaudata morphotypes always formed a single group (species group B in Fig. 2–4). Parartemia cylindrifera and Parartemia sp. ‘y’ also consistently grouped together, forming a well-supported clade in the BI trees; this clade was present but less strongly supported in the ML trees (species group C in Fig. 2–4). Parartemia zietziana and the different lines of P. purpurea (see below) also usually grouped together but the clade was not always well supported (species group D in Fig. 2–4). The concatenated and 16S BI trees also indicated that, although P. minuta and P. mouritzi showed considerable divergence from each other, they formed a base group in the phylogeny (species group E in Fig. 2 and 3; no COI data are available for the former species). Three species (P. serventyi, P. extracta and P. acidiphila) were each distinctive (see Fig. 2–4).
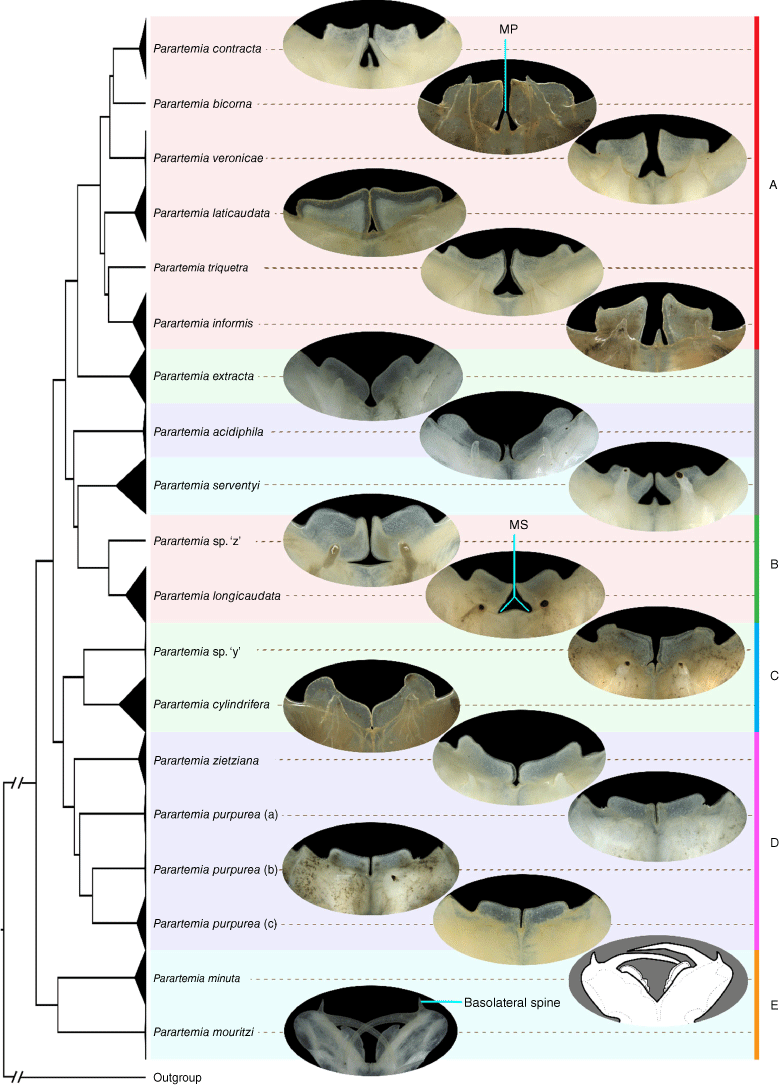
The morphology of the medial process (MP) or medial space (MS) in the heads of males in species that occurred in the same species groups (A–E) in the molecular phylogenies were usually similar (Table 2, Fig. 5). For example, the six species in group A all had an undivided medial process (Fig. 5). Apart from P. yarleensis, which was not included in the molecular phylogeny, P. serventyi was the only other species to show this feature (Table 2) and, although the P. serventyi haplotype clade was distinctive, it was closest to species group A clade in the concatenated trees (Fig. 2 and Supplementary Fig. S2). Another example is group C, which was formed by P. cylindrifera and Parartemia sp. ‘y’, the only Parartemia species that have a small bifid medial process (Table 2). Parartemia minuta and P. mouritzi in species group E had different MS structures but are the only known Parartemia species possessing basolateral spines (see Fig. 5).
| Morphospecies | Description | |
|---|---|---|
| Parartemia acidiphila Timms & Hudson, 2009 | MP present (large with small bifid apex) | |
| Parartemia cylindrifera Linder, 1941 | MP present (small bifid structure) | |
| Parartemia sp. ‘y’ (this study) | ||
| Parartemia bicorna Timms, 2010 | MP present (small, medium or large; no bifid apex) | |
| Parartemia contracta Linder, 1941 | ||
| Parartemia informis Linder, 1941 | ||
| Parartemia laticaudata Timms, 2010 | ||
| Parartemia serventyi Linder, 1941 | ||
| Parartemia triquetra Timms & Hudson, 2009 | ||
| Parartemia veronicae Timms, 2010 | ||
| Parartemia yarleensis Timms & Hudson, 2009 A | ||
| Parartemia auriciforma Timms & Hudson, 2009 A | MP absent; MS broad (flat, concave or convex) | |
| Parartemia boomeranga Timms, 2010 | ||
| Parartemia longicaudata Linder, 1941 | ||
| Parartemia sp. ‘z’ (this study) | ||
| Parartemia extracta Linder, 1941 | MP absent; MS round | |
| Parartemia minuta Geddes, 1973 A | MP absent; MS open V-shaped | |
| Parartemia mouritzi Timms, 2010 | MP absent; MS flat with a small V-shaped central notch | |
| Parartemia purpurea Timms, 2010 | MP absent; MS either absent or an almost closed diamond-shaped | |
| Parartemia zietziana Sayce, 1903 |
Photographs of these morphological features are in Fig. 5.
Photographs showing the morphology of the medial process (MP) or medial space (MS) in the heads of male Parartemia from 19 species confirmed in this study. The male head of P. minuta has been redrawn from Timms (2014). Coloured bars indicate five species groups (A–E) in Parartemia identified in the molecular phylogenies. The grey-coloured bar indicates the three molecularly distinctive species. The tree topology is derived from the collapsed 16S phylogenetic tree (Fig. 3).
Species delimitation
The three species delimitation methods identified different numbers of partitions among the Parartemia morphospecies. For the 16S dataset, the ASAP scheme with the lowest score (ASAP1; threshold distance 0.01) gave 56 partitions and the ASAP least partition scheme (ASAP2; threshold distance 0.06) and the mPTP and GMYC methods yielded fairly similar results of 27, 30 and 24 partitions respectively (Fig. 3 and Table 3). For the COI dataset, the GMYC (31) and mPTP (38) methods and especially ASAP1 (62; threshold distance 0.01) gave more partitions than ASAP2 (15; threshold distance 0.16) (Fig. 4 and Table 3). As is described below, the ASAP2 scheme showed the closest match with the Parartemia morphospecies. The other methods indicate the presence of one or more partitions within most morphospecies (Table 3). Our species hypotheses mainly conform with the ASAP2 scheme (see Table 3 and below).
| Morphospecies | 16S | COI | Number of species | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p | n | h | ASAP1 | ASAP2 | mPTP | GMYC | p | n | h | ASAP1 | ASAP2 | mPTP | GMYC | |||
| Parartemia acidiphila | 4 | 4 | 4 | 2 | 1 | 1 | 1 | 4 | 8 | 3 | 2 | 1 | 1 | 1 | 1 | |
| Parartemia bicorna | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | 4 | 1 | 1 | 1 | 1 | 1 | |
| Parartemia contracta | 4 | 7 | 7 | 2 | 1 | 1 | 1 | 4 | 13 | 11 | 2 | 1 | 2 | 1 | 1 | |
| Parartemia cylindrifera | 8 | 9 | 9 | 8 | 5 | 1 | 3 | 12 | 29 | 24 | 9 | 1 | 7 | 5 | 1 | |
| Parartemia sp. ‘y’ | 2 | 4 | 2 | 1 | 1 | 1 | 1 | 2 | 6 | 4 | 2 | 1 | 1 | 1 | 1 A | |
| Parartemia extracta | 7 | 10 | 10 | 5 | 1 | 3 | 1 | 7 | 17 | 12 | 3 | 1 | 2 | 1 | 1 | |
| Parartemia informis | 12 | 13 | 11 | 6 | 1 | 1 | 1 | 12 | 29 | 19 | 10 | 1 | 3 | 2 | 1 | |
| Parartemia boomeranga | 5 | 5 | 5 | 7 | 1 | 5 | 1 | 5 | 13 | 12 | 9 | 1 | 6 | 5 | 1 B | |
| Parartemia longicaudata | 15 | 18 | 14 | ″ | ″ | ″ | ″ | 15 | 36 | 24 | ″ | ″ | ″ | ″ | ″ B | |
| Parartemia sp. ‘z’ | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | ″ | 1 | 1 | 1 A | |
| Parartemia minuta | – | 2 | 2 | 2 | 1 | 1 | 1 | – | – | – | – | – | – | – | 1 | |
| Parartemia mouritzi | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Parartemia purpurea | 9 | 10 | 10 | 7 | 3 | 3 | 3 | 9 | 18 | 16 | 9 | 3 | 3 | 3 | 3 C | |
| Parartemia serventyi | 13 | 14 | 13 | 7 | 5 | 5 | 4 | 13 | 32 | 22 | 8 | 1 | 5 | 5 | 1 | |
| Parartemia triquetra | 1 | 1 | 1 | 1 | 1 | 1 | 1 | – | – | – | – | – | – | – | 1 | |
| Parartemia laticaudata | 2 | 3 | 3 | 2 | 1 | 2 | 1 | 2 | 10 | 3 | 2 | 1 | 2 | 2 | 1 | |
| Parartemia veronicae | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ″ | 1 | 1 | 1 | |
| Parartemia zietziana | 2 | 3 | 3 | 2 | 1 | 1 | 1 | 2 | 6 | 4 | 2 | 1 | 2 | 1 | 1 | |
| Total | 88 | 113 | 100 | 56 | 27 | 30 | 24 | 91 | 232 | 161 | 62 | 15 | 38 | 31 | 19 | |
Where the results of species delimitation analyses are shared across multiple species the partitions below are indicated by a double prime symbol (″). The number of species represented by each morphospecies, based on the results of this study, is given in the final column. ASAP1, Assemble Species by Automatic Partitioning lowest score scheme; ASAP2, Assemble Species by Automatic Partitioning least partition scheme; mPTP, multi-rate Poisson Tree Processes; GMYC, Generalised Mixed Yule Coalescent method.
For both the 16S and COI genetic regions, the maximum amount of intraspecific genetic distance varied among species and tended to be larger in species that were sampled from a greater number of sites (see Table 4 and Supplementary Table S1). For any one species, the maximum intraspecific genetic distance was always less than its minimum genetic distance from another species, but overall there was a limited amount of overlap between minimum interspecific and maximum intraspecific distances for both the COI and 16S datasets (Table 4, Supplementary Tables S6 and S7).
| Species | 16S rRNA | COI mtDNA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | K2P-distance | p-distance | N | K2P-distance | p-distance | ||||||
| M. Intra. | Inter. | M. Intra. | Inter. | M. Intra. | Inter. | M. Intra. | Inter. | ||||
| P. acidiphila | 4 | 1.49 | 13.97–23.06 | 1.47 | 12.58–19.54 | 3 | 1.86 | 20.19–27.70 | 1.82 | 17.33–22.49 | |
| P. bicorna | 1 | – | 10.69–18.70 | – | 9.85–16.35 | 4 | 0.46 | 18.20–27.10 | 0.46 | 15.81–22.04 | |
| P. contracta | 7 | 3.02 | 10.38–21.21 | 2.93 | 9.62–18.03 | 11 | 4.42 | 16.86–29.47 | 4.26 | 14.74–23.71 | |
| P. cylindrifera | 9 | 10.73 | 12.62–23.03 | 9.83 | 11.51–19.50 | 24 | 14.08 | 17.88–29.30 | 12.46 | 15.65–23.71 | |
| P. extracta | 10 | 8.08 | 13.30–21.19 | 7.53 | 12.13–18.24 | 12 | 3.79 | 20.95–28.51 | 3.65 | 17.93–22.95 | |
| P. informis | 11 | 6.67 | 8.97–22.75 | 6.28 | 8.39–19.29 | 19 | 13.75 | 17.64–28.51 | 12.16 | 15.35–22.95 | |
| P. laticaudata | 3 | 4.56 | 9.95–20.52 | 4.40 | 9.24–17.65 | 3 | 11.66 | 16.43–29.98 | 10.49 | 14.44–24.01 | |
| P. longicaudata–P. boomeranga | 19 | 8.06 | 10.14–23.44 | 7.52 | 9.39–19.71 | 36 | 13.12 | 16.06–28.05 | 11.70 | 13.98–22.80 | |
| P. minuta | 2 | 4.58 | 15.88–21.85 | 4.39 | 14.23–18.62 | – | – | – | – | – | |
| P. mouritzi | 2 | 0.21 | 16.38–23.40 | 0.21 | 14.68–19.75 | 1 | – | 21.95–27.92 | – | 18.84–22.80 | |
| P. purpurea (a) | 3 | 0.63 | 14.76–24.59 | 0.63 | 13.21–20.34 | 6 | 2.81 | 20.43–29.47 | 2.74 | 17.63–23.71 | |
| P. purpurea (b) | 2 | 0.42 | 15.26–23.06 | 0.42 | 13.60–19.50 | 2 | 0.46 | 20.99–30.38 | 0.46 | 17.93–24.32 | |
| P. purpurea (c) | 5 | 4.34 | 12.85–22.62 | 4.18 | 11.74–19.08 | 8 | 3.93 | 20.67–32.02 | 3.80 | 17.78–25.23 | |
| P. serventyi | 13 | 10.49 | 11.36–24.59 | 9.60 | 10.46–20.34 | 22 | 20.21 | 18.25–32.02 | 17.02 | 15.96–25.23 | |
| P. triquetra | 1 | – | 9.85–21.85 | – | 9.21–18.62 | – | – | – | – | – | |
| P. veronicae | 2 | 0.21 | 8.97–20.52 | 0.21 | 8.39–17.61 | 1 | – | 16.43–26.48 | – | 14.44–21.73 | |
| P. zietziana | 3 | 3.47 | 13.48–22.27 | 3.35 | 12.16–18.87 | 4 | 4.28 | 20.45–27.20 | 4.10 | 17.63–22.34 | |
| Parartemia sp. ‘y’ | 2 | 0.21 | 13.56–19.80 | 0.21 | 12.16–17.19 | 4 | 1.38 | 17.88–27.70 | 1.37 | 15.65–22.49 | |
| Parartemia sp. ‘z’ | 1 | – | 10.14–21.12 | – | 9.39–18.20 | 1 | – | 16.06–27.42 | – | 13.98–22.34 | |
N, number of haplotypes per species; M. Intra., maximum intraspecific distance; and Inter., range in interspecific distance. Further details are in Supplementary Tables S6 and S7.
In total, 8 of the 18 Parartemia morphospecies (16 described and 2 newly identified undescribed species) were each represented by a single well-supported clade (or a distinct haplotype when the species was represented by a single haplotype) that exactly corresponded to a single ASAP2 partition in both 16S and COI datasets (Fig. 3 and 4; also see Table 3). Another two species, P. minuta and P. triquetra (respectively represented by two haplotypes and a single haplotype), each corresponded to a single ASAP2 partition in the 16S dataset but were not included in the COI dataset (Table 3). For this group, genetic distances were highest in P. extracta, which was collected from seven sites plus two 16S sequences from GenBank and had maximum p-distances of 7.53% for 16S and 3.65% for COI. They were also high in P. informis, which was collected from 12 sites and had maximum p-distances of 6.28% for 16S and 12.16% for COI (Table 4 and Supplementary Table S1). The geographic distributions of the divergent 16S lineages within each of these species did not overlap (Supplementary Fig. S5 and S6). This species group also included the putative new morphospecies Parartemia sp. ‘y’, which was collected from two sites and represented by two 16S and four COI haplotypes (Table 3), with maximum p-distances of 0.21 and 1.37% respectively (Table 4). The minimum p-distance between the haplotypes of Parartemia sp. ‘y’ and those of any other species (P. cylindrifera) was 12.16% for 16S and 15.65% for COI (Supplementary Table S6).
The remaining eight morphospecies fell into four categories: (i) morphospecies that formed single ASAP2 partitions in the COI but not the 16S dataset, (ii) morphospecies that formed single ASAP2 partitions in the 16S but not the COI dataset, (iii) multiple morphospecies that were combined into a single ASAP2 partition in both the 16S and COI datasets, and (iv) a single morphospecies that was spilt into multiple ASAP2 partitions in both the 16S and COI datasets.
Category (i) comprised P. cylindrifera and P. serventyi, each of which formed multiple (5) partitions in 16S ASAP2 analysis (Table 3), despite each being represented by a single well-supported clade in the phylogenetic trees (see Fig. 2–4) and only a single partition in the COI ASAP2 analysis. These two species, which were sampled from respectively 12 (plus one COI and one 16S sequence from GenBank) and 13 sites (Supplementary Table S1), had the highest 16S and COI genetic distances of all sampled species (Table 4). Within each species, the distributions of the divergent 16S lineages did not overlap, although those of lineages C and E in P. serventyi bordered each other (see Supplementary Fig. S7 and S8).
Category (ii) included P. laticaudata and P. veronicae, which were combined into a single partition in the COI ASAP2 analysis (Table 3). This was surprising because these two species did not form a monophyletic group in any of the phylogenetic analyses (see Fig. 2–4) and were partitioned via the more conserved 16S region. Although the minimum genetic distances between the haplotypes of these two species, e.g. p-distances of 9.24% for 16S and 14.44% for COI, were among the lowest found in the Parartemia species (see Supplementary Table S6), that between the 16S haplotypes of P. verconicae and P. informis was only 8.39% (Supplementary Table S6). Category (ii) also included the putative new species Parartemia sp. ‘z’, which was combined into a single partition with P. boomeranga and P. longicaudata in the COI analysis; it also grouped with these species in the ML 16S tree (Supplementary Fig. S3). Parartemia sp. ‘z’ was represented by single 16S and COI haplotypes. The minimum p-distances between these haplotypes and those of the P. longicaudata–P. boomeranga morphotypes, at 9.39% for 16S and 13.98% for COI, were low for interspecific comparisons (Supplementary Table S6). Nevertheless, the haplotypes of Parartemia sp. ‘z’ were separated from those of P. longicaudata–P.boomeranga in most of the phylogenetic analyses (see Fig. 2–4) and formed their own partition in all species delimitation analyses except COI ASAP2 (Table 3).
Category (iii) comprised P. boomeranga and P. longicaudata, which in combination corresponded to a single ASAP2 partition in both the 16S (a single partition was also suggested by the 16S GMYC analysis; Fig. 3 and Table 3) and (along with Parartemia sp. ‘z’) in the COI datasets (see above; also see Fig. 4 and Table 3). Together, these species formed a single clade in the phylogenetic trees (which also included Parartemia sp. ‘z’ in the ML 16S tree; Supplementary Fig. S3), but neither morphotype formed an exclusive monophyletic group within this broader clade (see Fig. 2–4). Similarly, although the other species delimitation methods identified multiple partitions within the broader P. boomeranga–P. longicaudata clade, none of these partitions exactly corresponded to haplotypes representing one or the other morphotype (Fig. 3 and 4). The amount of divergence (p-distance) within the P. longicaudata–P. boomeranga complex ranged up to 7.52% for 16S and 11.7% for COI, which was less than that in some other species (Table 4). The P. longicaudata morphotype was common and widely distributed whereas we only found the P. boomeranga morphotype at five sites in the northern Wheatbelt within 125 km of each other (Supplementary Table S1 and Supplementary Fig. S9). The P. boomeranga–P. longicaudata complex included a widespread lineage (E) whose distribution overlapped or bordered that of the other four lineages (A–D) whose distributions did not overlap (see Supplementary Fig. S9).
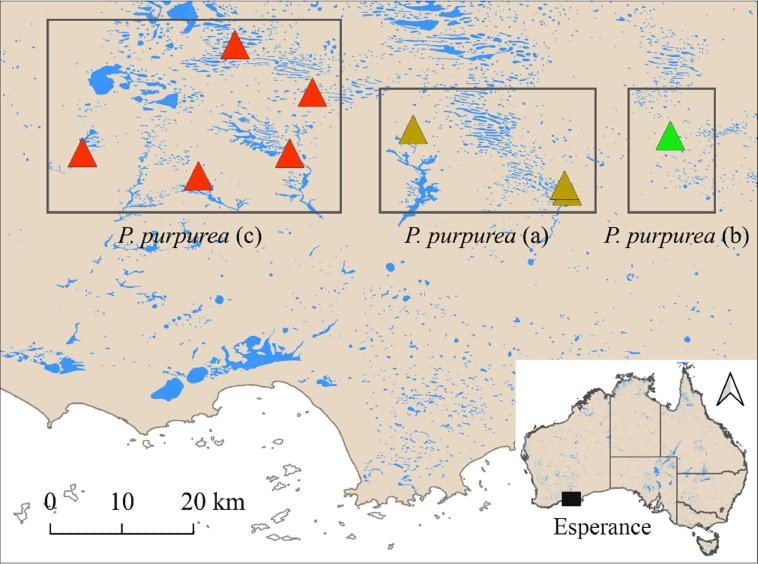
Parartemia purpurea was the only morphospecies in category (iv). Representatives of this taxon fell into three well-supported clades (a, b and c) in the phylogenetic trees (see Fig. 2–4). Some details of the relationship among these clades varied between datasets and tree-building methods (Fig. 2–4). Regardless, each of the three clades corresponded to separate ASAP2 partitions (also for the mPTP and GMYC methods) in both the 16S and COI datasets (Fig. 3 and 4) and differed from each other by p-distances of at least 13.6 and 18.24% for the 16S and COI data respectively (see details in Supplementary Table S6), which are greater than the maximum intraspecific distance recorded in any other morphospecies (Table 4). On this basis, we propose that ‘P. purpurea’ comprises three cryptic species that herein are called P. purpurea (a), P. purpurea (b) and P. purpurea (c) (see Fig. 2–4). The morphology of some specimens of each species were checked but no characteristic differences were found. All three species were found within 85 km of each other in the Esperance hinterland, but their geographical distributions did not overlap (see Fig. 6).
Morphology of new species
The overall morphology of the putative new species Parartemia sp. ‘y’ was like that of P. cylindrifera (with which it also had the most molecular similarities). However, the ventral processes in the males of Parartemia sp. ‘y’ featured nearly rectangular distolateral outgrowths and obtuse distomesial corners, whereas those of other species including P. cylindrifera had round distolateral corners protruding ventrally and round distomesial corners (Fig. 7).
Photographs of putative new species of Parartemia showing distinctive morphological features relative to morphologically similar species. Comparison of male head of Parartemia sp. ‘y’ (a) with that of P. cylindrifera (b). Comparison of male head and posterior thoracic and anterior abdominal segments of Parartemia sp. ‘z’ (c, d) with those of the P. boomeranga (e, f) and P. longicaudata (g, h) morphotypes. Morphological features mentioned in the text are indicated.
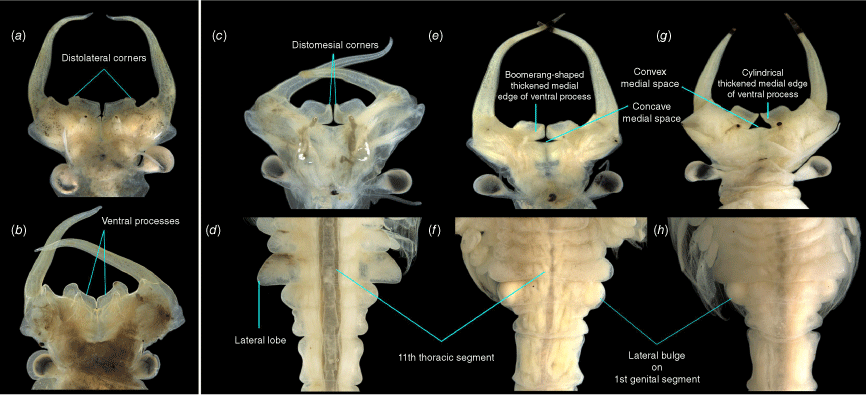
The overall morphology of Parartemia sp. ‘z’ was similar to the P. boomeranga and P. longicaudata morphotypes (with which it also had the most molecular similarities) but the thickened medial edges of the ventral processes in the males were strongly concave (boomerang-shape) contrasting with their weakly concave (boomerang-shape) or cylindrical shape in P. boomeranga–P. longicaudata (Fig. 7). Parartemia sp. ‘z’ was also easily discernible from P. boomeranga–P. longicaudata by the substantially larger lateral lobes on the eleventh thoracic segment in males, which were more than double the size of lateral bulges on the first genital segment (Fig. 7).
Parartemia species POP4 and POP5
To determine the species identity of POP4 and POP5 from Remigio et al. (2001), we trimmed our 16S sequences and compared them to two short 16S sequences (335 and 337 bp) from POP4 and POP5 on GenBank (data not presented). The sequence for POP4 (accession number AY014794) was identical to the corresponding region in one of three P. laticaudata haplotypes. The sequence for POP5 (accession number AY014795) was similar (minimum and maximum p-distances of 2.2 and 4.3% respectively) to the corresponding region in one of five haplotypes found in P. purpurea (c), but highly divergent from the next closest taxa based on 16S, which were P. purpurea (b) and P. purpurea (a) (minimum p-distances of 16.1 and 17.1% respectively). These findings indicate that POP4 corresponds to P. laticaudata and POP5 to P. purpurea (c).
Discussion
This study provides the first molecular phylogeny of Parartemia based on an almost complete suite of species and broad geographic sampling. The findings generally support suggestions that this taxon is monophyletic, includes large amounts of molecular divergence, generally exhibits congruent patterns of genetic and morphological divergence and contains a large number of species (see Remigio et al. 2001; Timms 2014; Lawrie et al. 2021). It also provides evidence of two new morphospecies and of cryptic speciation.
Molecular divergence
The amount of molecular divergence found in Parartemia was large, as was also noted by Remigio et al. (2001). Using COI p-distances as an example, the maximum divergence in Parartemia (25.23%) was higher than that reported for a range of other branchiopod genera, such as Triops (15.6%; Meusel and Schwentner 2017), Limnadopsis (18.6%; Schwentner et al. 2011), Eocyzicus (19.0%; Schwentner et al. 2014), Ozestheria (21.2%; Schwentner et al. 2015) and Artemia (21.8%; Muñoz et al. 2008). It was also greater than that detected in other invertebrate genera that have a long history in Australian salt lakes or their precursors, for example, Coxiella gastropods (18.9%; Lawrie et al. 2023) or Australocypris giant ostracods (18.9%; Rahman 2024). The extreme conditions in salt lakes may accelerate the rate of molecular evolution in halophilic cladocerans and anostracans, like Parartemia (Hebert et al. 2002). Such rate heterogeneity would help to explain why divergence levels in Parartemia are high relative to branchiopods from fresh or low salinity water but not necessarily in comparison with Artemia or other genera of halophilic crustaceans. It may also be that some Parartemia lineages are particularly old or that habitat specialisation has driven divergence among some lineages (see below).
Phylogenetic relationships
Some aspects of the phylogeny of Parartemia, particularly deeper divergences, were not fully resolved in this study. This may be because the selected genetic markers were not suitable for higher-level taxonomic resolution in Parartemia or because these deeper divergences may be associated with rapid cladogenesis (e.g. see Kang et al. 2008; Whitfield and Kjer 2008; Pinceel et al. 2013). However, some groups of related species and some distinctive species were evident. The fact that species thought to be closely related based on the molecular data usually also showed similarities in the structures of their medial processes or medial space in the males adds evidence that these groups are real. Like Remigio et al. (2001), we found that P. minuta (which occurs in central and eastern Australia) was usually positioned at the base of the phylogeny and, in our study, usually grouped with P. mouritzi (which occurs in Western Australia and was not sampled by Remigio et al. 2001). These are the only two Parartemia species known to possess basolateral spines (see Timms 2012), providing further evidence that they are sister species. Like Remigio et al. (2001), we found that P. extracta was a distinctive species. Remigio et al. (2001) found two new species of Parartemia, which they called POP4 and POP5 and that we have identified as P. laticaudata and P. purpurea (c) respectively. The species groups that Remigio et al. (2001) called clades A and B were not apparent in our samples. For example, that study placed P. longicaudata (which included their POP1, POP3 and POP6 samples), P. cylindrifera, P. purpurea (c) (then POP5) and P. zietziana in clade A whereas our study usually placed P. purpurea (c) and P. zietziana in the same clade but P. longicaudata in a different clade and P. cylindrifera in yet another one. The discrepancies between the results of this study and those of Remigio et al. (2001) can be attributed to the results of the latter study being based on only a short 16S fragment and only a small subset of species (see Introduction). Increased taxonomic representation has been shown to alter perceptions of species relationships in a range of taxa, including Branchinella fairy shrimps in Australia (Pinceel et al. 2013).
On the basis that new anostracans species are expected to arise from widespread species in peripheral habitats, Rogers (2015) predicted that anostracan genera will comprise a series of small clades, each containing a basal widespread species and a derived species with a narrow distribution at the periphery of the widespread one (see also Rogers and Aguilar 2020). Our phylogenies provide several examples of widespread and narrowly distributed sister species including (with the widespread species listed first) – P. cylindrifera and Parartemia sp. ‘y’, P. longicaudata–P. boomeranga and Parartemia sp. ‘z’, P. informis and P. triquerta and P. minuta and P. mouritzi (see Timms et al. 2009 and below for distributional information). However, other aspects of the relationships between these sister species do not fit with the above prediction. For example, relative to their widespread counterparts, the narrow range species were usually ancestral in the phylogenies or had either overlapping (e.g. P. cylindrifera and Parartemia sp. ‘y’) or disjunct distributions (e.g. P. informis and P. triquerta). Intraspecific divergence within some widespread Parartemia species may more closely conform with the above prediction and will be examined in detail in a future study on phylogeographic patterns in Parartemia.
Species delimitation
The most recent previous estimate of the number of Parartemia morphospecies was 18 (Timms 2010). Our study suggests that Parartemia consists of at least 21 species. These species comprise: (i) 13 described morphospecies that have been confirmed using the molecular data in this study; (ii) 2 putative new morphospecies; (iii) 3 cryptic species of P. purpurea; (iv) 1 species encompassing the P. longicaudata and P. boomeranga morphotypes; and (v) 2 described morphospecies that were not included in this study. These data suggest that Parartemia is the most speciose genus of halophilic invertebrates found in Australia, notwithstanding that the taxonomy of some of the other invertebrate groups is rudimentary (see Lawrie et al. 2021). Based on the currently available data, the next most diverse genus is Coxiella (a gastropod) with at least 15 species, although recent molecular evidence indicates that these species are spread over several unrecognised genera (four clades in Lawrie et al. 2023). Australocypris with 10 species is the most species-rich genus in Mytilocypridinae giant ostracods (Rahman 2024). Parartemia is much more species-rich than Artemia, which only has nine recognised species even though Artemia is essentially globally distributed (Rogers 2013; Asem et al. 2023b). Compared to Artemia, speciation in Parartemia is likely facilitated by their heavy, sinking resting eggs, which tend to retard dispersal (McMaster et al. 2007; Timms et al. 2009). Australia also has a diverse range of Branchinella fairy shrimps in fresh or low-salinity water (Pinceel et al. 2013; Rogers and Timms 2014). This diversity and that in Parartemia is probably linked to a long history of aridity and persistent ephemeral water bodies in the Australian landscape (Rogers and Timms 2014). Rogers (2015) and references therein have argued that species diversity in anostracans will be enhanced in an older landscape like Australia where more occupied habitats will favour speciation by habitat specialisation over colonisation of vacant habitats. This fits with the high levels of ecological specialisation apparent among Parartemia species, e.g. with regard to pH or substrate geochemistry (see Timms et al. 2009; Timms 2012).
Our species hypotheses for Parartemia are based on the most conservative results from the species delimitation analyses (i.e. the ASAP scheme with the least number of partitions), which tended to match morphological species boundaries. This fits with the suggestion that morphological diagnosis of anostracan species is generally straightforward (Rogers and Aguilar 2020), although this is not always the case (e.g. see Ketmaier et al. 2008; Pinceel et al. 2013; Rogers and Aguilar 2020). Many Parartemia morphospecies comprised multiple divergent lineages that were partitioned in some of the species delimitation analyses. Other than for the three lineages of P. purpurea (discussed below), we have interpreted the divergent lineages within morphospecies as examples of intraspecific variation rather than as cryptic species for the following reasons. (1) The presence of morphologically similar but genetically divergent lineages may be linked to an accelerated rate of molecular evolution in Parartemia (see above). (2) With the exception of P. longicaudata, the geographic distributions of the divergent lineages of morphospecies were essentially nonoverlapping. Also, conspecific divergent lineages never co-occurred in the same water body (see Supplementary Fig. S5–S9). (3) Species delimitation methods are sensitive to the population structure of a species and sometimes delimit genetically divergent populations within a species (Luo et al. 2018; Gaytán et al. 2020). (4) Defining species based on molecular data alone is challenging (Jörger and Schrödl 2013; Fišer et al. 2018). (5) We wanted to avoid the pitfalls of taxonomic inflation (see Padial and De la Riva 2006).
We found two putative new morphospecies of Parartemia – Parartemia sp. ‘y’ and Parartemia sp. ‘z’ – in our samples. These taxa exhibited both distinctive morphological and genetic characteristics relative to other described species. The evidence is stronger for Parartemia sp. ‘y’, which was found at two sites, than for Parartemia sp. ‘z’, which was only found at a single site. The results of this study provide the first evidence of cryptic species in Parartemia, namely P. purpurea (a), P. purpurea (b) and P. purpurea (c), although cryptic species have been reported in a range of anostracans (e.g. Pinceel et al. 2013; Rogers 2014) and other branchiopods (e.g. Schwentner et al. 2013; Meusel and Schwentner 2017). The three P. purpurea lineages were consistently separated from each other in the species delimitation analyses and did not always form a single well-supported clade in the phylogenetic trees. No morphological differences were detected among individuals from these lineages, including in relation to characters important in species diagnosis in Parartemia (see ‘Introduction’), although it is possible that further scrutiny could reveal some subtle differences. Ecological or physiological specialisation may have been crucial in driving cryptic speciation in P. purpurea (e.g. Rogers 2014).
The two new Parartemia morphospecies as well as the three cryptic species of P. purpurea were found in only the Esperance hinterland region of Western Australia. This brings the total number of Parartemia species found in this region to 9 (out of 21), 5 of which have only been found in this region (this study and Timms et al. 2009). In general, the region is well known for the diverse range of invertebrates found in its salt lakes (see Timms 2009). Coxiella gastropods (Lawrie et al. 2023) and Australocypris ostracods (Rahman et al. 2023) also have high species richness and multiple endemic species in this region. The region hosts a large number and variety of salt lakes (see Timms 2009 for details), which are probably important factors driving divergence and diversity. It also seems likely that this region has served as a refugium in evolutionary time, limiting extinctions, possibly because of favourable hydrological or climatic conditions (e.g. see Jansson 2003; Davis et al. 2013).
Our results suggest that the P. boomeranga and P. longicaudata morphotypes comprise a single monophyletic lineage. These results are based on a comprehensive sampling of these morphotypes. Parartemia boomeranga was sampled across its documented range, including three lakes near the type locality (unnamed lake east of Gunyidi, −30.12, 116.24; see Timms 2010). Parartemia longicaudata was also sampled across its entire known geographical range, from Esperance to the Houtman Abrolhos Islands, including the neotype locality (Pink Lake in Esperance; see Supplementary Fig. S9 and Timms 2010). Morphological differences between P. boomeranga and P. longicaudata are minor and confined to the thickened medial edges of the ventral processes in males (boomerang-shaped in the former and cylindrical in the latter) and the shape of the medial space (concave in the former and convex in the latter) (see Fig. 7 and Timms 2010). Regardless, the P. boomeranga and P. longicaudata morphotypes did not form reciprocally monophyletic groups in the molecular phylogeny, so each morphotype must have evolved more than once or may be environmentally induced. Following the guidelines set by the International Code of Zoological Nomenclature (International Trust for Zoological Nomenclature 1999), P. longicaudata being named first has nomenclatural priority over P. boomeranga.
Conservation implications
Timms et al. (2009) provided a detailed assessment of the conservation status of Parartemia species. This assessment can be updated using the improved taxonomic and associated distributional information on Parartemia generated by this study.
Timms et al. (2009) proposed that P. boomeranga (then Parartemia sp. ‘c’) should be assessed as potentially vulnerable. Timms (2012) later advised that this species was extremely rare and at risk of extinction. However, we encountered this species at five sites within the reported range of this species, including Lake Moore (in the northern wheatbelt region in Western Australia) from which this taxon has previously been recorded (Timms et al. 2009) and three sites that are near the type locality (see above). Furthermore, the molecular results suggest that the P. boomeranga morphotype is not a valid species but is synonymous with the common and widely distributed P. longicaudata, which is not regarded as threatened (see Timms et al. 2009).
The two new species and the three cryptic species of P. purpurea discovered in this study all have narrow geographic distributions. Except for P. purpurea (c), these species are known from between only one and three sites in the Esperance hinterland region. All these species have sites in the Kau Rock Nature Reserve or the nearby Beaumont Nature Reserve and are therefore offered some protection but are nonetheless somewhat vulnerable given their rarity and restricted distributions. Parartemia purpurea (c) is known from five or six sites (depending on whether we resampled Remigio et al.’s (2001) POP5 collection site, the exact location of which was not reported) but all are in the Esperance hinterland and none are in nature reserves. The above distributional information does not consider 20 other sites in the Esperance hinterland that were not sampled in this study but are reported to contain the P. purpurea morphotype (see Timms et al. 2009; Timms 2010; Rogers and Timms 2014). Regardless, as an area of evolutionary importance with high species diversity (see above and Timms et al. 2009), the Esperance hinterland should be a priority site for Parartemia conservation (see Moritz 2002; Davis et al. 2013).
Limitations and future work
Future phylogenetic studies on Parartemia should aim to include P. auriciforma and P. yarleensis, which were missing from this study, as well as more samples from salt lakes in remote areas, which have typically been poorly studied and may harbour a high proportion of undiscovered species (Timms 2010). It is also important to formally describe and name the new species discovered in this study so they can be taken into account in conservation planning and legislation (e.g. see Mace 2004; Padial and De la Riva 2006). A better understanding of the evolutionary significance of the divergent lineages within morphospecies is also needed. This could be facilitated via assessments of phylogeographic structures of these species (e.g. see Seidel et al. 2009).
The phylogenetic results of this study are based on a total of three different genetic markers, which were not sufficient to resolve the relationships among some lineages, particularly those reflecting deeper divergences. Future studies should include more markers and loci to improve the resolution of phylogenetic relationships in Parartemia, although there is no simple answer as to the minimum number of markers or loci needed to produce a robust phylogeny or whether markers should be selected at random or systematically (Gatesy et al. 2007).
Conclusion
Our study indicates that Parartemia comprises at least 21 species, including two putative new morphospecies and three cryptic species. The molecular data revealed five groups of related species that were also largely supported by morphological data. The molecular data were also mainly consistent with morphospecies designations, although many morphospecies contained large amounts of divergence. Overall, the results highlight the importance of using both molecular and morphological data for providing robust species hypotheses in Parartemia. The improved taxonomic information for Parartemia will support the development of future studies and conservation assessments of this taxon.
Supplementary material
Supplementary tables and figures are included with this submission and are available online.
Data availability
Data supporting the findings of this study are available as supplementary material. The newly generated sequences have been deposited in GenBank under accession numbers OR828050–OR828208, OR833948–OR834039 and OR834040–OR834069. Further information will be provided upon request.
Declaration of funding
The corresponding author was supported by a Research Training Program (RTP) PhD scholarship from the Department of Education, Australian Government, through Murdoch University.
Acknowledgements
The authors express their gratitude to the Department of Biodiversity, Conservation and Attractions, the Western Australian Museum and the Stantec Australia Pty Ltd in Western Australia, as well as the Tasmanian Museum and Art Gallery, Hobart, for generously providing some specimens. The authors thank Brian V. Timms for his invaluable suggestions during this study. We also thank two reviewers for their constructive criticisms that have improved the quality of the manuscript.
References
Anufriieva E, Shadrin N (2013) Historical biogeography of Artemia (Anostraca): some intriguing aspects of diversification. In ‘BioSyst.EU 2013 Global systematics!’, 18–22 February 2013, Vienna, Austria. (Eds A Kroh, B Berning, E Haring, M Harzhauser, H Sattmann, J Walochnik, D Zimmermann, M Zuschin) Abstract volume, p. 12. (NOBIS Austria: Vienna, Austria) Available at https://archive.bgbm.org/BGBM/staff/wiss/Lack/PDF/BioSystEU2013_Abstract_volume.pdf
Asem A, Gajardo G, Hontoria F, Yang C, Shen C-Y, Rastegar-Pouyani N, Padhye SM, Sorgeloos P (2023a) The species problem in Artemia Leach, 1819 (Crustacea: Anostraca), a genus with sexual species and obligate parthenogenetic lineages. Zoological Journal of the Linnean Society 202, zlad192.
| Crossref | Google Scholar |
Asem A, Yang C, Eimanifar A, Hontoria F, Varó I, Mahmoudi F, Fu C-Z, Shen C-Y, Rastegar-Pouyani N, Wang P-Z (2023b) Phylogenetic analysis of problematic Asian species of Artemia Leach, 1819 (Crustacea, Anostraca), with the descriptions of two new species. Journal of Crustacean Biology 43(1), ruad002.
| Crossref | Google Scholar |
Bayly I, Williams W (1966) Chemical and biological studies on some saline lakes of south-east Australia. Marine and Freshwater Research 17(2), 177-228.
| Crossref | Google Scholar |
Bortolus A (2008) Error cascades in the biological sciences: the unwanted consequences of using bad taxonomy in ecology. AMBIO: A Journal of the Human Environment 37(2), 114-118.
| Crossref | Google Scholar | PubMed |
Coleman M, Geddes MC, Trotman C (1998) Divergence of Parartemia and Artemia haemoglobin genes. International Journal of Salt Lake Research 7(2), 171-180.
| Crossref | Google Scholar |
Costa FO, DeWaard JR, Boutillier J, Ratnasingham S, Dooh RT, Hajibabaei M, Hebert PD (2007) Biological identifications through DNA barcodes: the case of the Crustacea. Canadian Journal of Fisheries and Aquatic Sciences 64(2), 272-295.
| Crossref | Google Scholar |
Costello MJ, May RM, Stork NE (2013) Can we name Earth’s species before they go extinct? Science 339(6118), 413-416.
| Crossref | Google Scholar | PubMed |
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9, 772.
| Crossref | Google Scholar | PubMed |
Davis J, Pavlova A, Thompson R, Sunnucks P (2013) Evolutionary refugia and ecological refuges: key concepts for conserving Australian arid zone freshwater biodiversity under climate change. Global Change Biology 19(7), 1970-1984.
| Crossref | Google Scholar | PubMed |
De Deckker P (1983) Australian salt lakes: their history, chemistry, and biota – a review. Hydrobiologia 105(1), 231-244.
| Crossref | Google Scholar |
Di Marco M, Chapman S, Althor G, Kearney S, Besancon C, Butt N, Maina JM, Possingham HP, von Bieberstein KR, Venter O (2017) Changing trends and persisting biases in three decades of conservation science. Global Ecology and Conservation 10, 32-42.
| Crossref | Google Scholar |
Donaldson MR, Burnett NJ, Braun DC, Suski CD, Hinch SG, Cooke SJ, Kerr JT (2016) Taxonomic bias and international biodiversity conservation research. Science Applications Forum 1(1), 105-113.
| Crossref | Google Scholar |
Dugan KA, Lawrence HS, Hares DR, Fisher CL, Budowle B (2002) An improved method for post-PCR purification for mtDNA sequence analysis. Journal of Forensic Science 47(4), 1-8.
| Crossref | Google Scholar | PubMed |
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32(5), 1792-1797.
| Crossref | Google Scholar | PubMed |
Fišer C, Robinson CT, Malard F (2018) Cryptic species as a window into the paradigm shift of the species concept. Molecular Ecology 27(3), 613-635.
| Crossref | Google Scholar | PubMed |
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3(5), 294-299.
| Google Scholar | PubMed |
Fujisawa T, Barraclough TG (2013) Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets. Systematic Biology 62(5), 707-724.
| Crossref | Google Scholar | PubMed |
Gatesy J, DeSalle R, Wahlberg N (2007) How many genes should a systematist sample? Conflicting insights from a phylogenomic matrix characterized by replicated incongruence. Systematic Biology 56(2), 355-363.
| Crossref | Google Scholar | PubMed |
Gaytán Á, Bergsten J, Canelo T, Pérez‐Izquierdo C, Santoro M, Bonal R (2020) DNA Barcoding and geographical scale effect: the problems of undersampling genetic diversity hotspots. Ecology and Evolution 10(19), 10754-10772.
| Crossref | Google Scholar | PubMed |
Geddes M (1973) A new species of Parartemia (Anostraca) from Australia. Crustaceana 25(1), 5-12.
| Crossref | Google Scholar |
Hebert PD, Remigio EA, Colbourne JK, Taylor DJ, Wilson CC (2002) Accelerated molecular evolution in halophilic crustaceans. Evolution 56(5), 909-926.
| Crossref | Google Scholar | PubMed |
International Trust for Zoological Nomenclature (1999) ‘International Code of Zoological Nomenclature (ICZN)’, 4th edn. (ITZN, c/o Natural History Museum: London) Available at http://hdl.handle.net/1885/92062
Jackson S, Baker A, Eldridge M, Fisher D, Frankham G, Lavery T, MacDonald AJ, Menkhorst P, Phillips M, Potter S (2022) The importance of appropriate taxonomy in Australian mammalogy. Australian Mammalogy 45, 13-23.
| Crossref | Google Scholar |
Jansson R (2003) Global patterns in endemism explained by past climatic change. Proceedings of the Royal Society of London – B. Biological Sciences 270(1515), 583-590.
| Crossref | Google Scholar | PubMed |
Jellison R, Williams WD, Timms BV, Alcocer J, Aladin NV (2008) Salt lakes: values, threats and future. In ‘Aquatic ecosystems: trends and global prospects’. (Ed. NVC Polunin) pp. 94–110. (Cambridge University Press) 10.1017/CBO9780511751790.010
Jörger KM, Schrödl M (2013) How to describe a cryptic species? Practical challenges of molecular taxonomy. Frontiers in Zoology 10(1), 59.
| Crossref | Google Scholar | PubMed |
Kang S, Sultana T, Loktev VB, Wongratanacheewin S, Sohn W-M, Eom KS, Park J-K (2008) Molecular identification and phylogenetic analysis of nuclear rDNA sequences among three opisthorchid liver fluke species (Opisthorchiidae: Trematoda). Parasitology International 57(2), 191-197.
| Crossref | Google Scholar | PubMed |
Kapli P, Lutteropp S, Zhang J, Kobert K, Pavlidis P, Stamatakis A, Flouri T (2017) Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 33(11), 1630-1638.
| Crossref | Google Scholar | PubMed |
Ketmaier V, Pirollo D, De Matthaeis E, Tiedemann R, Mura G (2008) Large-scale mitochondrial phylogeography in the halophilic fairy shrimp Phallocryptus spinosa (Milne-Edwards, 1840) (Branchiopoda: Anostraca). Aquatic Sciences 70(1), 65-76.
| Crossref | Google Scholar |
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16(2), 111-120.
| Crossref | Google Scholar | PubMed |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35(6), 1547-1549.
| Crossref | Google Scholar | PubMed |
Lawrie ADA, Chaplin J, Pinder A (2021) Corrigendum to: biology and conservation of the unique and diverse halophilic macroinvertebrates of Australian salt lakes. Marine and Freshwater Research 72(11), 1695.
| Crossref | Google Scholar |
Lawrie ADA, Chaplin J, Kirkendale L, Whisson C, Pinder A, Mlambo MC (2023) Phylogenetic assessment of the halophilic Australian gastropod Coxiella and South African Tomichia resolves taxonomic uncertainties, uncovers new species and supports a Gondwanan link. Molecular Phylogenetics and Evolution 184, 107810.
| Crossref | Google Scholar | PubMed |
Linder F (1941) Contributions to the morphology and the taxonomy of the Branchiopoda Anostraca. Zoologiska Bidrag från Uppsala 20, 101-302.
| Google Scholar |
Luo A, Ling C, Ho SY, Zhu C-D (2018) Comparison of methods for molecular species delimitation across a range of speciation scenarios. Systematic Biology 67(5), 830-846.
| Crossref | Google Scholar | PubMed |
Mace GM (2004) The role of taxonomy in species conservation. Philosophical Transactions of the Royal Society of London – B. Biological Sciences. 359(1444), 711-719.
| Crossref | Google Scholar | PubMed |
McMaster K, Savage A, Finston T, Johnson MS, Knott B (2007) The recent spread of Artemia parthenogenetica in Western Australia. Hydrobiologia 576(1), 39-48.
| Crossref | Google Scholar |
Meusel F, Schwentner M (2017) Molecular and morphological delimitation of Australian Triops species (Crustacea: Branchiopoda: Notostraca) – large diversity and little morphological differentiation. Organisms Diversity & Evolution 17(1), 137-156.
| Crossref | Google Scholar |
Michonneau F (2016) Using GMYC for species delineation. (University of Washington: Seattle, WA, USA). Available at https://francoismichonneau.net/gmyc-tutorial
Mora C, Tittensor DP, Adl S, Simpson AG, Worm B (2011) How many species are there on Earth and in the ocean? PLoS Biology 9(8), e1001127.
| Crossref | Google Scholar | PubMed |
Moritz C (2002) Strategies to protect biological diversity and the evolutionary processes that sustain it. Systematic Biology 51(2), 238-254.
| Crossref | Google Scholar | PubMed |
Muñoz J, Pacios F (2010) Global biodiversity and geographical distribution of diapausing aquatic invertebrates: the case of the cosmopolitan brine shrimp, Artemia (Branchiopoda, Anostraca). Crustaceana 83(4), 465-480.
| Crossref | Google Scholar |
Muñoz J, Gómez A, Green AJ, Figuerola J, Amat F, Rico C (2008) Phylogeography and local endemism of the native Mediterranean brine shrimp Artemia salina (Branchiopoda: Anostraca). Molecular Ecology 17(13), 3160-3177.
| Crossref | Google Scholar | PubMed |
Padial JM, De la Riva I (2006) Taxonomic inflation and the stability of species lists: the perils of ostrich’s behavior. Systematic Biology 55(5), 859-867.
| Crossref | Google Scholar | PubMed |
Padial JM, Miralles A, De la Riva I, Vences M (2010) The integrative future of taxonomy. Frontiers in Zoology 7(1), 16.
| Crossref | Google Scholar | PubMed |
Pinceel T, Vanschoenwinkel B, Waterkeyn A, Vanhove MP, Pinder A, Timms BV, Brendonck L (2013) Fairy shrimps in distress: a molecular taxonomic review of the diverse fairy shrimp genus Branchinella (Anostraca: Thamnocephalidae) in Australia in the light of ongoing environmental change. Hydrobiologia 700(1), 313-327.
| Crossref | Google Scholar |
Pinder A, Timms BV, Campagna V (2009) Salt-loving shrimps threatened by salinisation? Information sheet 20/2009, science division. (Department of Environment and Conservation, Western Australia) Available at https://library.dbca.wa.gov.au/static/Journals/080525/080525-20.pdf
Puillandre N, Brouillet S, Achaz G (2021) ASAP: assemble species by automatic partitioning. Molecular Ecology Resources 21(2), 609-620.
| Crossref | Google Scholar | PubMed |
Rahman M, Chaplin J, Pinder A (2023) The biology of giant ostracods (Crustacea, Cyprididae), a review focusing on the Mytilocypridinae from Australian inland waters. Marine and Freshwater Research 74(1), 1-19.
| Crossref | Google Scholar |
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67(5), 901-904.
| Crossref | Google Scholar | PubMed |
Raupach MJ, Radulovici AE (2015) Looking back on a decade of barcoding crustaceans. ZooKeys 539, 53-81.
| Crossref | Google Scholar | PubMed |
Remigio E, Hebert P, Savage A (2001) Phylogenetic relationships and remarkable radiation in Parartemia (Crustacea: Anostraca), the endemic brine shrimp of Australia: evidence from mitochondrial DNA sequences. Biological Journal of the Linnean Society 74(1), 59-71.
| Crossref | Google Scholar |
Rogers DC (2013) Anostraca catalogus (Crustacea: Branchiopoda). Raffles Bulletin of Zoology 61(2), 525-546.
| Google Scholar |
Rogers DC (2014) Two new cryptic anostracan (Branchiopoda: Streptocephalidae, Chirocephalidae) species. Journal of Crustacean Biology 34(6), 862-874.
| Crossref | Google Scholar |
Rogers DC (2015) A conceptual model for anostracan biogeography. Journal of Crustacean Biology 35(5), 686-699.
| Crossref | Google Scholar |
Rogers DC, Aguilar A (2020) Molecular evaluation of the fairy shrimp family Branchinectidae (Crustacea: Anostraca) supports peripatric speciation and complex divergence patterns. Zoological Studies 59, 14.
| Crossref | Google Scholar | PubMed |
Rogers DC, Timms BV (2014) Anostracan (Crustacea: Branchiopoda) zoogeography III. Australian bioregions. Zootaxa 3881(5), 453-487.
| Crossref | Google Scholar | PubMed |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution 34(12), 3299-3302.
| Crossref | Google Scholar | PubMed |
Ruebhart DR, Cock IE, Shaw GR (2008) Invasive character of the brine shrimp Artemia franciscana Kellogg 1906 (Branchiopoda: Anostraca) and its potential impact on Australian inland hypersaline waters. Marine and Freshwater Research 59(7), 587-595.
| Crossref | Google Scholar |
Saccò M, White NE, Harrod C, Salazar G, Aguilar P, Cubillos CF, Meredith K, Baxter BK, Oren A, Anufriieva E (2021) Salt to conserve: a review on the ecology and preservation of hypersaline ecosystems. Biological Reviews 96(6), 2828-2850.
| Crossref | Google Scholar | PubMed |
Sayce O (1903) The Phyllopoda of Australia, including descriptions of some new genera and species. Proceedings of the Royal Society of Victoria 15, 224-261.
| Google Scholar |
Schwentner M, Timms BV, Richter S (2011) An integrative approach to species delineation incorporating different species concepts: a case study of Limnadopsis (Branchiopoda: Spinicaudata). Biological Journal of the Linnean Society 104(3), 575-599.
| Crossref | Google Scholar |
Schwentner M, Clavier S, Fritsch M, Olesen J, Padhye S, Timms BV, Richter S (2013) Cyclestheria hislopi (Crustacea: Branchiopoda): a group of morphologically cryptic species with origins in the Cretaceous. Molecular Phylogenetics and Evolution 66(3), 800-810.
| Crossref | Google Scholar | PubMed |
Schwentner M, Timms BV, Richter S (2014) Evolutionary systematics of the Australian Eocyzicus fauna (Crustacea: Branchiopoda: Spinicaudata) reveals hidden diversity and phylogeographic structure. Journal of Zoological Systematics and Evolutionary Research 52(1), 15-31.
| Crossref | Google Scholar |
Schwentner M, Just F, Richter S (2015) Evolutionary systematics of the Australian Cyzicidae (Crustacea, Branchiopoda, Spinicaudata) with the description of a new genus. Zoological Journal of the Linnean Society 173(2), 271-295.
| Crossref | Google Scholar |
Seidel RA, Lang BK, Berg DJ (2009) Phylogeographic analysis reveals multiple cryptic species of amphipods (Crustacea: Amphipoda) in Chihuahuan Desert springs. Biological Conservation 142(10), 2303-2313.
| Crossref | Google Scholar |
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution 4(1), vey016.
| Crossref | Google Scholar | PubMed |
Timms BV (2005) Salt lakes in Australia: present problems and prognosis for the future. Hydrobiologia 552(1), 1-15.
| Crossref | Google Scholar |
Timms BV (2009) Study of the saline lakes of the Esperance Hinterland, Western Australia, with special reference to the roles of acidity and episodicity. Natural Resources and Environmental Issues 15(1), 215-225.
| Google Scholar |
Timms BV (2010) Six new species of the brine shrimp Parartemia Sayce 1903 (Crustacea: Anostraca: Artemiina) in Western Australia. Zootaxa 2715(1), 1-35.
| Crossref | Google Scholar |
Timms BV (2012) An identification guide to the brine shrimps (Crustacea: Anostraca: Artemiina) of Australia. Museum Victoria Science Reports 16, 1-36.
| Crossref | Google Scholar |
Timms BV (2014) A review of the biology of Australian halophilic anostracans (Branchiopoda: Anostraca). Journal of Biological Research – Thessaloniki 21(1), 21.
| Crossref | Google Scholar | PubMed |
Timms BV, Hudson P (2009) The brine shrimps (Artemia and Parartemia) of South Australia, including descriptions of four new species of Parartemia (Crustacea: Anostraca: Artemiina). Zootaxa 2248(1), 47-68.
| Crossref | Google Scholar |
Timms BV, Pinder AM, Campagna VS (2009) The biogeography and conservation status of the Australian endemic brine shrimp Parartemia (Crustacea, Anostraca, Parartemiidae). Conservation Science Western Australia 7(2), 413-427.
| Google Scholar |
Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44(W1), W232-W235.
| Crossref | Google Scholar | PubMed |
Troudet J, Grandcolas P, Blin A, Vignes-Lebbe R, Legendre F (2017) Taxonomic bias in biodiversity data and societal preferences. Scientific Reports 7(1), 9132.
| Crossref | Google Scholar | PubMed |
Weekers PH, Murugan G, Vanfleteren JR, Belk D, Dumont HJ (2002) Phylogenetic analysis of anostracans (Branchiopoda: Anostraca) inferred from nuclear 18S ribosomal DNA (18S rDNA) sequences. Molecular Phylogenetics and Evolution 25(3), 535-544.
| Crossref | Google Scholar | PubMed |
Whitfield JB, Kjer KM (2008) Ancient rapid radiations of insects: challenges for phylogenetic analysis. Annual Review of Entomology 53, 449-472.
| Crossref | Google Scholar | PubMed |
Wiens JJ, Moen DS (2008) Missing data and the accuracy of Bayesian phylogenetics. Journal of Systematics and Evolution 46(3), 307-314.
| Crossref | Google Scholar |
Williams WD (2002) Environmental threats to salt lakes and the likely status of inland saline ecosystems in 2025. Environmental Conservation 29(2), 154-167.
| Crossref | Google Scholar |