A new genus of bamboo coral (Octocorallia: Scleralcyonacea: Keratoisididae) from the Whittard Canyon, Ireland, Northeast Atlantic
Declan Morrissey A B * , A. Louise Allcock B and Andrea M. Quattrini A
A B * , A. Louise Allcock B and Andrea M. Quattrini A
A
B
Abstract
Deep-sea corals are rarely identified to species due to a lack of taxonomic expertise and paucity of sampling. Herein we describe a new genus from the family Keratoisididae collected from the Northeast Atlantic. Using both nuclear (2010 conserved element loci) and complete mitogenome phylogenies, we found this genus to be closely related to the genera Dokidisis and Jasonisis. In the nuclear phylogeny, each genus occupied a distinct well-supported clade. All three genera lack thorned- or double-star sclerites in the pharynx; instead they have flattened rods, a potential unifying feature of the keratoisidid group J3 of Watling et al. (2022). The newly described genus Explorisis gen. nov. has a unique sclerome including spindles and tapered rods that differentiates it from its sister genera. Explorisis katharina sp. nov. is characterised by volcano to cylindrical shaped polyps, striated rods and spindles in the polyp body, and elongated flattened rods in the coenenchyme, whereas Explorisis poppyae sp. nov. has heavily granulated spindles and rods in both the polyp body and coenenchyme. Genetic variation within the mitogenomes across both Explorisis gen. nov. species is limited with mutations in just 3 of 14 protein coding regions.
ZooBank: urn:lsid:zoobank.org:pub:141BD76E-8C83-43BE-8E1E-B8C53CD7CEF7
Keywords: Anthozoa, canyons, deep sea, mitogenomes, new species, systematics, taxonomy, ultraconserved elements.
Introduction
Octocorals are important benthic fauna capable of forming dense aggregations referred to as Marine Animal Forests (Rossi et al. 2017, 2022; Orejas et al. 2022). These forests support a highly diverse invertebrate fauna, including polychaetes, squat lobsters and serpent stars (Parimbelli 2020; Maxwell et al. 2022). Colonies can also act as nurseries for a range of taxa, including elasmobranchs (Etnoyer and Warrenchuk 2007; Morrissey et al. 2023a), cephalopods (Vecchione 2019) and basket stars (Neves et al. 2020). Despite their ecological importance, the taxonomy of octocorals, particularly in the deep sea, is poorly developed, in contrast to other taxonomic groups. Recently, however, there have been major taxonomic revisions within coral taxonomy at or above the family level (McFadden et al. 2022) and new species and genera are still being described, even in well-studied oceans (e.g. Altuna and López-González 2023).
Keratoisididae is a globally distributed and exclusively deep-sea (Watling et al. 2011) group of octocorals that is easily distinguishable from other articulated corals by the sclerome, which comprises needles, spindles, rods and scales, and by differences in the composition of the axis (Heestand Saucier et al. 2021). Branching pattern has been the sole diagnostic trait for genera within the family, with nodal branching in one plane characteristic of the genus Isidella and nodal branching in multiple planes characteristic of Acanella. Species are assigned to Keratoisis when branching occurs from the internodes and unbranched colonies were assigned to Lepidisis. However, this group has been the subject of major taxonomic revisions (e.g. Watling and France 2021; Watling et al. 2022) because multiple molecular phylogenies have revealed that branching pattern is labile (France 2007; Dueñas et al. 2014) and not characteristic of any one genus resulting in polyphyly across the family. New genera have been described to resolve some of the observed polyphyly (Bayer 1990; Watling and France 2011; Alderslade and McFadden 2012; Lapointe and Watling 2015; Watling 2015; France and Watling 2024), but there are more that await description (Lapointe and Watling 2022).
A two-locus phylogeny (mtMutS-5′, mtMutS-3′ and partial 18S) identified 13 distinct groups within the family, with varying levels of support (France 2007; Watling et al. 2022), most of which have at least one genus that is typical of the group: A1 (Acanella), B1 (Adinisis), C1 (Tanyostea), G1 (Bathygorgia), H1 (Onkoisis), I4 (Tridentisis), M1 (Orstomisis) and S1 (Cladarisis). Other groups have two nominal genera such as D2 (Keratoisis and Eknomisis), I1 (Lepidisis and Isidella) and J3 (Dokidisis and Jasonisis), whereas D1 and F1 do not have any formally described representative genera. The phylogeny in Watling et al. (2022) did not have sufficient resolution to elucidate relationships among these predefined groups. A phylogeny of Keratoisididae generated from hundreds of nuclear loci (Morrissey et al. 2023b) resolved the relationships among most keratoisidid groups and found that some groups were not monophyletic and that the division between others would need to be reconsidered. Furthermore, comparing phylogenies constructed from conserved elements and complete mitogenomes revealed mitonuclear discordance within the family (Morrissey et al. 2024).
Keratoisidid group J3 is globally distributed with specimens found in all ocean basins, and is prolific offshore Ireland (Morrissey et al. 2022). Currently, J3 is typified by two genera, Dokidisis and Jasonisis, yet specimens in the J3 clade collected from Ireland were found to be morphologically distinct from these two genera. This study aimed to (1) describe a new genus of keratoisidid collected offshore of Ireland and (2) investigate the phylogenetic position of this genus to other J3 keratoisidids based on both mitogenomes and conserved elements, particularly with respect to Jasonisis and Dokidisis.
Methods
Sample collection
During two cruises aboard RV Celtic Explorer in June 2014 and August 2021, four bamboo corals were collected: one from the Whittard Canyon in the south of Ireland at 2669 m and three from the Fangorn Bank to the northwest of Ireland between 1248 and 1393 m. All specimens were deposited in the Smithsonian Institution National Museum of Natural History (NMNH; Supplementary Table S1). In addition, tissue from the holotype of Dokidisis australis (TMAG-K3834) was obtained from the Tasmanian Museum and Art Gallery (TMAG).
DNA extraction and library preparation
DNA was extracted from the newly gathered specimens and the holotype of Dokidisis australis using a DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions. DNA was quantified using a Qubit Fluorometer using a broad-range assay kit. Libraries were prepared as outlined in Quattrini et al. (2018, 2020) using ~100 ng of DNA that was sheared to 400–800 bp using sonication. Libraries were then prepared using a Kapa Hyper Prep Kit (Roche), indexed using custom iTru dual-indexed primers (Glenn et al. 2019), and amplified with the following thermocycler conditions; 98°C for 45 s, followed by 12 cycles of 98°C for 15 s, 60°C for 30 s, and 72°C for 30 s, followed by a final extension at 72°C for 1 min. The library of D. australis was pooled into a larger set of eight individuals and enriched with the octocoral-specific RNA bait set (octocoral-v2, Erickson et al. 2021). All libraries were sequenced on a NOVASeq platform (150 bp, paired-end reads) by the Oklahoma Medical Research Foundation NGS Core (Oklahoma City, OK, USA).
Bioinformatics
In addition to the five newly generated libraries, raw reads from 124 keratoisidid specimens with data publicly available in Sequence Read Archives (Quattrini et al. 2018, 2020; McFadden et al. 2022; Morrissey et al. 2023b, 2024) were downloaded, including from the type specimen for Jasonisis thresheri (TMAG-K3879). A list of J3 and I4 specimens (the known sister group to J3, Morrissey et al. 2023a) used in this study are presented in Supplementary Table S1. Extraction of conserved elements followed the established methodology of Quattrini et al. (2018) using the ‘octocoral-v2 bait set’ (Erickson et al. 2021). Briefly, raw reads were processed using PhylUCE (ver. 1.7.1, see https://github.com/faircloth-lab/phyluce; Faircloth 2016) following tutorial one. Reads were trimmed using the Illumiprocessor wrapper (ver. 2.0.9, B. C. Faircloth, see https://github.com/faircloth-lab/illumiprocessor; Bolger et al. 2014) with default values and assembled using SPAdes (ver. 3.14, see https://github.com/ablab/spades; Bankevich et al. 2012) with the careful option (--careful) enabled, which performs a mismatch correction. Baits targeting conserved elements were matched to the assembled contigs (70% identity, 70% coverage) using phyluce_assembly_match_contigs_to_probes to locate the targeted loci, which were then extracted using phyluce_assembly_get_match_counts and phyluce_assembly_get_fastas_from_match_counts, exported into separate FASTA files and aligned with default parameters using phyluce_align_seqcap_align, which implements MAFFT (ver. 5, see https://mafft.cbrc.jp/alignment/software/; Katoh and Standley 2013) and trims the edges of loci. A data matrix of locus alignments was created using phyluce_align_get_only_loci_with_min_taxa in which each locus had 50 or 75% taxon occupancy.
Mitochondrial genome assembly and gene annotation
Mitogenomes were assembled from Illumina reads using NOVOPlasty (ver. 4.3.1, see https://github.com/ndierckx/NOVOPlasty; Dierckxsens et al. 2017) using mtMutS from a closely related individual (GenBank: ON971030.1, Morrissey et al. 2022) as a seed or by extracting the gene from contigs generated by SPAdes. Gene annotations were performed in Geneious Prime (ver. 2023.1.1, see https://www.geneious.com) using available keratoisidid mitogenomes. The Open Reading Frame (ORF) finder implemented in Geneious Prime was used to edit annotations, encompassing the largest ORF using known cnidarian start (ATG, GTG) and stop codons (TAA, TAG).
Phylogenomics
Maximum likelihood (ML) reconstructions were built from concatenated alignments of loci from the 50 and 75% taxon-occupancy matrices using IQ-TREE (ver. 2.1.3, see http://www.iqtree.org/; Minh et al. 2020). Partitions were initially by locus, with the best model of evolution selected by ModelFinder (see http://www.iqtree.org/ModelFinder/; Kalyaanamoorthy et al. 2017) within IQ-TREE implementing the greedy strategy (--m MFP + MERGE). This method merges partitions until the model fit does not increase. Node support was determined using 10,000 ultrafast bootstraps (-B 10,000, ufbs).
Species trees were constructed from the individual trees of each locus in each taxon-occupancy matrix. First, each locus underwent phylogenetic reconstruction in IQ-TREE with the best model of evolution for each locus determined by ModelFinder (Kalyaanamoorthy et al. 2017) and node support determined from 1000 ufbs. Nodes with support less than 30% were collapsed from every tree using Newick Utilities (ver. 1.6, see https://github.com/tjunier/newick_utils; Junier and Zdobnov 2010) and long branches were pruned using TreeShrink (ver. 1.2.1, see https://github.com/uym2/TreeShrink; Mai and Mirarab 2018). ASTRAL III (ver. 5.7.8, see https://github.com/smirarab/ASTRAL; Mirarab et al. 2014), a multispecies coalescent species tree method (Zhang et al. 2018), was used to construct the species trees with node support generated from the Local Posterior Probability (LPP) reflecting the probability of a branch being a true branch based on the given set of gene trees. LPP values were then assigned to the nodes of the ML trees.
Twenty-seven additional complete mitogenomes were added to the dataset (Supplementary Table S2, Brugler and France 2008; van der Ham et al. 2009; Muthye et al. 2022; Morrissey et al. 2024) and all individual Protein coding genes (PCGs) were aligned separately in Geneious Prime using MAFFT and translation align settings implemented in Geneious Prime. Genes were then concatenated using Concatenator (ver. 0.2.1, see https://github.com/iTaxoTools/ConcatenatorGui; Vences et al. 2022).
A Maximum likelihood (ML) phylogenomic reconstruction was carried out in IQ-TREE (with partitions initially set by gene and codon position within each gene, with same settings as the conserved elements reconstruction (--m MFP + MEGRE, -B 10,000). Bayesian inference (BI) was executed in MrBayes (ver. 3.2.6, see https://github.com/NBISweden/MrBayes/; Ronquist and Huelsenbeck 2003) with a default general time reversible model with settings and model parameters as follows: lset nst = 6, rates = invgamma, with two Markov Chain Monte Carlo (MCMC) runs for 10,000,000 generations, tree sampling every 1000 generations and a burn-in of 25% applied. Output was checked for stationarity in Tracer (ver. 1.7.1, see https://github.com/beast-dev/tracer/releases/tag/v1.7.1; Rambaut et al. 2018). Bayesian Posterior Probability (BPP) values were transposed onto the ML trees.
Morphology
In situ images were taken using Kongsberg OE14-208 Digital Still Camera attached to Holland I and colonies were pictured again on board using a Nikon P900. Ex situ images were taken with a Cannon 5EOSDR at the NMNH. Images were stacked using CombineZP (ver. 1.0.0, see http://micropics.org.uk/). Polyp morphology was examined using light microscopy. In addition, nine other closely related specimens, previously collected from Irish waters and housed at the Smithsonian Institution National Museum of Natural History, were re-examined.
Sclerites were prepared using the same methods outlined in Morrissey et al. (2022). Briefly, coral polyps were dissected to separate the body wall, tentacles, pharynx and coenenchyme. The tissue from each part was dissolved in 5% sodium hypochlorite (i.e. household bleach), followed by hydrogen peroxide and then washed first in distilled water then in ethanol. Sclerites were then allowed to dry overnight. Sclerites were examined with an OLYMPUS DSX100 where measurements were taken directly using the inbuilt software.
Using a 5 × 0 detail paintbrush, sclerites were mounted onto a double-sided carbon adhesive tab fixed to a metal stub. Sclerites were coated in an 80:20 high purity gold-palladium alloy using a LEICA EM ACE600 achieving a coat thickness of 8–10 nm and were imaged using a Thermofisher APEREO Field Emission Scanning Electron Microscope at the Centre for Imaging, National Museum of Natural History. Descriptions of the sclerite shape and texture were described using the nomenclature established by Bayer et al. (1983) and that which is commonplace in keratoisidid taxonomy.
Micro Computed Tomography (Micro-CT)
One polyp from USNM1516893 was carefully removed and placed into a 200-µL pipette tip, filled with 95% ethanol and sealed, ensuring there was no air trapped inside. This specimen was scanned using the GE Phoenix v|tome|x M 240/180kV Dual Tube Micro CT machine at the Smithsonian Institution National Museum of Natural History Micro Computed Tomography Imaging Center with a 3.529-µm voxel size. Raw CT data were then reconstructed using Phoenix datos|x (see https://www.bakerhughes.com/waygate-technologies/ndt-software/phoenix-datosx-industrial-ct-acquisition-reconstruction-software). Slices were exported using VG Studio Max (ver. 3.2, see https://www.volumegraphics.com/). The resulting scans were analysed using Dragonfly (ver. 2022.2, see https://dragonfly.comet.tech/). We manually segmented a mesh to produce a 3-D model that highlights the sclerites. The resulting model was uploaded to Morphosource (https://doi.org/10.17602/M2/M606264 and https://doi.org/10.17602/M2/M606282).
Taxonomy
Explorisis gen. nov.
ZooBank: urn:lsid:zoobank.org:act:F0184C63-F31C-4950-97EA-6093BF475E31
Colony branches dichotomously or trichotomously at the nodes. Main axis is slender and is covered in a thin coenenchyme. Polyps are volcano, barrel, or trumpet shaped. Longitudinally striated or granulated spindles are rods found in the polyp body. Sclerites can project past the base of the tentacles but not always following the mesenteries. Tapered rods and spindles similar to the polyp body, lightly scalloped flattened rods and clubs can be present in coenenchyme. Flattened rods with irregular borders are found in both tentacles and pharynx. Pharyngeal sclerites can sometimes be wider in the middle than at the ends.
The genus name incorporates the name of the Irish research vessel, RV Celtic Explorer, aboard which all Explorisis specimens used in this study were collected. The root is then combined with isis, as is customary for genera in this family. The gender of this genus is feminine.
Group J3 is now typified by three nominal genera, Explorisis, Dokidisis and Jasonisis. All three genera lack double or thorned stars in the pharynx; instead the pharynx contains flat rods with irregular borders. Explorisis has a distinct sclerome with tapered rods and spindles in the polyp body. In comparison, Dokidisis is diagnosed by the presence of blunt rods in the polyp body and Jasonisis contains densely packed scales with fluted margins. Living tissue of all Explorisis specimens turned dark brown after presentation in alcohol. This has been observed in specimens of other J3 species including Jasonisis thresheri and Dokidisis australis. All individuals were found growing on living Solenosmilia variabilis framework (Supplementary Figs S1 and S2) suggesting a strong association with this species.
Explorisis katharina sp. nov.
(Fig. 1–5, Table 1, Supplementary Fig. S1.)
ZooBank: urn:lsid:zoobank.org:act:D03E8E96-A5C7-4FA5-B1FA-9DEC67BCF65B
Holotype: USNM1516893, Whittard Canyon, Ireland, Northeast Atlantic, collected on 14 June 2014, 48.4672°, −10.3994°, 2669 m, RV Celtic Explorer cruise CE14009.
Paratypes: USNM1704356, Fangorn Bank, Northeast Atlantic, collected 7 August 2021, 55.3661°, −19.8412°, 1286 m, RV Celtic Explorer cruise CE21010. USNM1704369, Fangorn Bank, Northeast Atlantic, collected 9 August 2021, 55.2173°, −19.8798°, 1393 m, RV Celtic Explorer cruise CE21010. USNM1704370, Fangorn Bank, Northeast Atlantic, collected 9 August 2021, 55.2242°, −19.8673°, 1241 m, RV Celtic Explorer cruise CE21010.
USNM1593518, Whittard Canyon, Ireland, Northeast Atlantic, collected on 1 June 2017, −10.681°, 48.615°, 1678 m, RV Celtic Explorer cruise CE17008. USNM1593520, Whittard Canyon, Ireland, Northeast Atlantic, collected on 1 June 2017, −10.681°, 48.615°, 1695 m, RV Celtic Explorer cruise CE17008. USNM1593521, Whittard Canyon, Ireland, Northeast Atlantic, collected on 1 June 2017, −10.681°, 48.615°, 1599 m, RV Celtic Explorer cruise CE17008. USNM1593522, Whittard Canyon, Ireland, Northeast Atlantic, collected on 1 June 2017, −10.681°, 48.615°, 1613 m, RV Celtic Explorer cruise CE17008. USNM1593523, Irish Continental Slope, Ireland, Northeast Atlantic, collected on 14 August 2018, −12.551°, 54.0611°, 1361 m, RV Celtic Explorer cruise CE18012. USNM1593524, Irish Continental Slope, Ireland, Northeast Atlantic, collected on 21 August 2018, −12.403°, 54.0602°, 1397 m, RV Celtic Explorer cruise CE18012. USNM1593525, Irish Continental Slope, Ireland, Northeast Atlantic, collected on 22 August 2018, −12.555°, 54.0643°, 1374 m, RV Celtic Explorer cruise CE18012. USNM1593526, Whittard Canyon, Ireland, Northeast Atlantic, collected on 1 June 2017, −10.68°, 54.0643°, 1374 m, RV Celtic Explorer cruise CE17008. USNM1691939, Irish Continental Slope, Ireland, Northeast Atlantic, collected June 2013, ~54.0552°, −12.5457°, ~1361 m, RV Celtic Explorer cruise CE13008.
Whittard Canyon, Ireland, Northeast Atlantic, 2669 m.
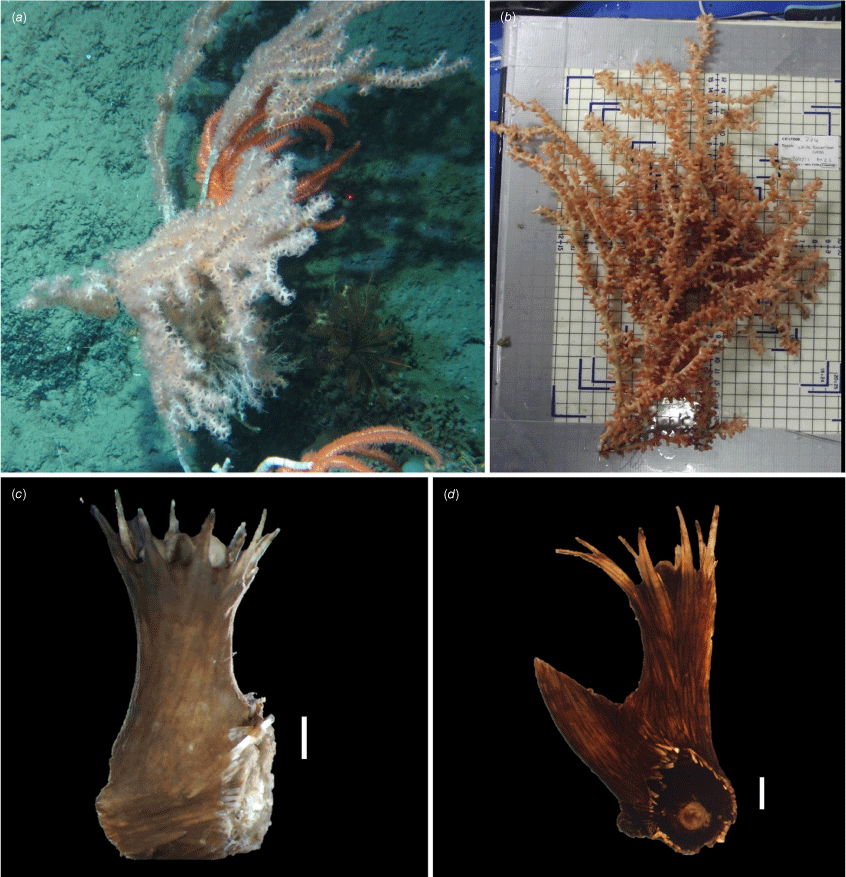
Explorisis katharina gen. nov., sp. nov. (a–c) In situ imagery of USNM1516893 taken by the ROV Holland I aboard RV Celtic Explorer cruise CE140009. (d) Ex situ image taken on deck. The arrow points to USNM1516893.

Explorisis katharina gen. nov., sp. nov. (a) Full image of preserved holotype USNM1516893. (b, c) Close up of polyps.

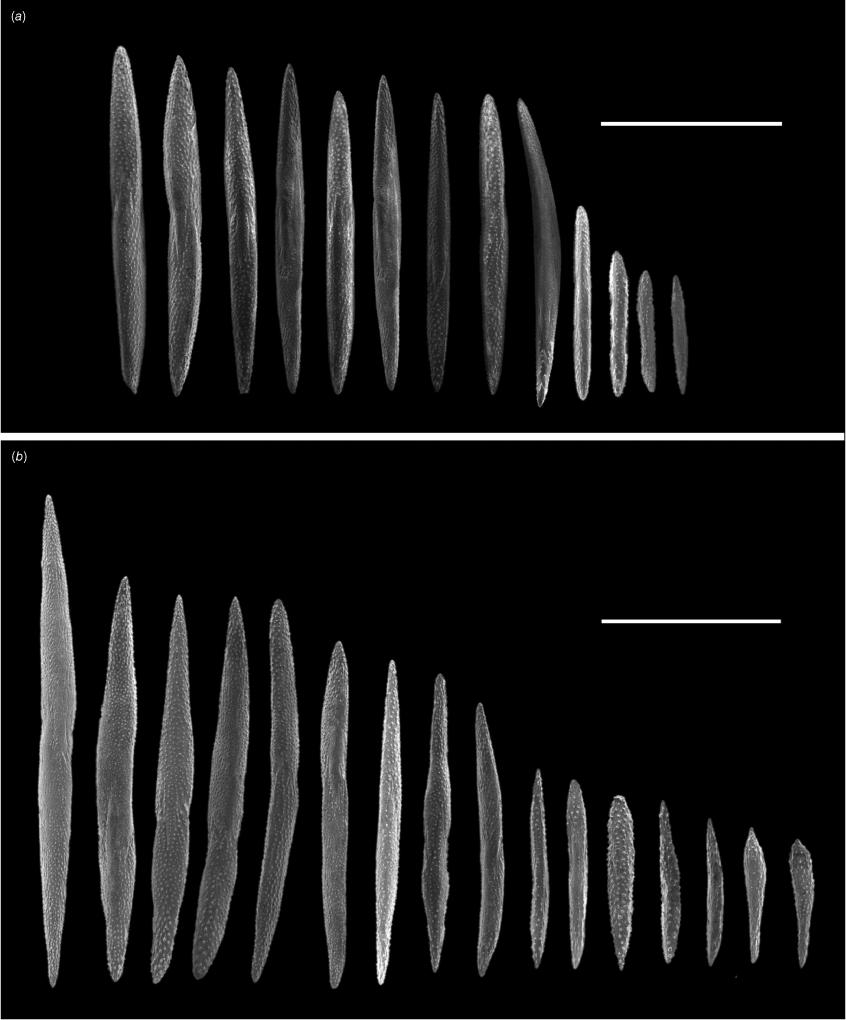
Explorisis katharina gen. nov., sp. nov. holotype USNM1516893, sclerites from the (a) polyp body and (b) coenenchyme. Scale bar: 1 mm (a); 200 µm (b).
Micro-CT reconstruction of a polyp from Explorisis katharina gen. nov., sp. nov. holotype USNM1516893. This figure is available as a 3-D model on Morphosource (see https://doi.org/10.17602/M2/M606264 and https://doi.org/10.17602/M2/M606282).

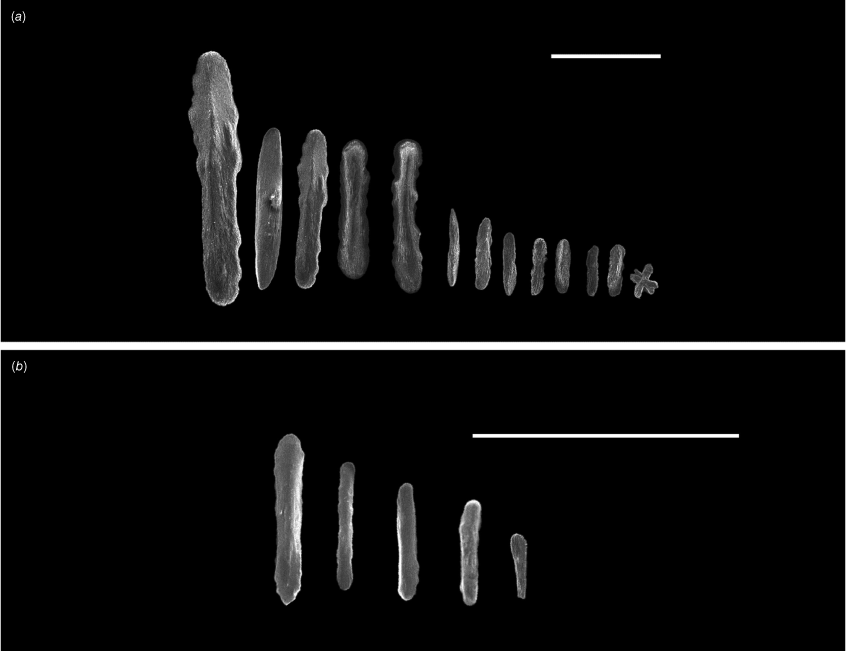
Explorisis katharina gen. nov., sp. nov. holotype USNM1516893, sclerites from the (a) tentacles and (b) pharynx. Scale bar: 100 µm (a); 50 µm (b).

| Species | Gross Morphology | Branching | Polyps | Body Sclerites | Tentacle Sclerites | Pharynx Sclerites | Coenenchyme Sclerites | |
|---|---|---|---|---|---|---|---|---|
| Explorisis katharina | Planar | Nodal | Volcano to cylindrical | Tapered rods and spindles, striated | Elongated flattened rods | Flat rods, irregular borders | Flat rods, irregular borders | |
| Explorisis poppyae | Planar | Nodal | Volcano, cylindrical or trumpet | Tapered rods and spindles, granulated | Tapered rods and spindles, granulated | Flat rods, irregular borders | Flat rods, irregular borders | |
| Dokidisis australis | Unbranched or forks once | Nodal | Cylindrical | Blunt rods, clubs | Blunt rods, clubs | Flat rods | Elongated flat rods | |
| Dokidisis krishanani | Unknown | Unknown | Volcano to cylindrical | Short blunt rods | Short blunt rods | Reported absent | Elongated flat rods | |
| Jasonisis thresheri | Planar | Nodal | Tall & narrow | Scales | Scales | Flat rods | scales |
Species in bold are the type species of the genus.
Colony branches dichotomously or trichotomously at the nodes. Main axis is slender and is covered in a thin coenenchyme. The polyp body contains needles and spindles, which can be longitudinally striated and project past the base of the tentacles but do not always follow the mesenteries. Flattened rods with irregular borders are found in the coenenchyme and pharynx.
Holotype: the holotype consists of two branches, each ~20 cm in length, with longitudinal striated internodes and nodes that are light brown (Fig. 2). Nodes are ~1.7 mm whereas the length of the internodes varies from 6 to 12 mm, with an average of 10.1 mm. Polyps vary from a cylindrical to volcano shape (Fig. 2b, c), are orange in colour in situ, and are primarily arranged on three sides of the coral axis. Coenenchyme is transparent to orange in situ and dark brown upon preservation.
The colony is planar and branches dichotomously and trichotomously from the nodes (Fig. 1). There are no anastomosing branches observed.
These arise primarily on three sides of the coenenchyme and are wider than they are tall. Both cylindrical and volcano polyps are on average 5 mm tall and between 2.6 and 2.8 mm in width when measured from the base. When the polyps are contracted, the tentacles fold over the top of the oral area, although this can be difficult to observe when the polyps are volcano shaped.
Spindles and needles (Fig. 3) are arranged primarily longitudinally along the polyp body (Fig. 4), at a slight angle in the proximal part of the polyp where they protrude slightly into the coenenchyme. Sclerites sometimes project past the base of the tentacles following an intertentacular pattern. Sclerite size ranges from 0.288 to 2.284 mm in length and 0.037 to 0.164 mm in width. Sclerites are usually obstructed from view by the darkened living tissue. Flattened rods and rodlets are arranged longitudinally along both the tentacles and pinnules ranging in size to 0.095–0.213 mm in length and 0.015–0.026 mm in width (Fig. 5). Flattened rods are lightly scalloped. Flattened rods are also present in the coenenchyme (Fig. 3) and are 0.123–0.466 mm in length and 0.026–0.067 mm wide arranged longitudinally along the colony axis, although they become irregular as coenenchyme transitions to polyp. Rodlets and diamond shaped rods are found in the pharynx (Fig. 5) and are between 0.078 and 0.087 mm in length and 0.013 to 0.022 mm in width.
Named after D. Morrissey’s maternal grandmother Kathleen O’Boyle with ‘katharina’ (treated as a noun in apposition) being the Latin origin of Kathleen. Morrissey attributes a lifetime of passion for both science and marine life to trips he made with his family, including his grandmother, to the Pollock Holes, Co. Clare, Ireland.
Explorisis poppyae sp. nov.
(Fig. 6–8, Table 1, Supplementary Fig S2.)
ZooBank: urn:lsid:zoobank.org:act:269A8534-BC11-4F98-A34A-4DA2EC28FC66
Holotype: USNM1593518, Whittard Canyon, Ireland, Northeast Atlantic, collected on 1 June 2017, −10.681 48.615, 1678 m, RV Celtic Explorer cruise CE17008.
Paratypes: USNM1593520, Whittard Canyon, Ireland, Northeast Atlantic, collected on 1 June 2017, −10.681°, 48.615°, 1695 m, RV Celtic Explorer cruise CE17008. USNM1593521, Whittard Canyon, Ireland, Northeast Atlantic, collected on 1 June 2017, −10.681°, 48.615°, 1599 m, RV Celtic Explorer cruise CE17008. USNM1593522, Whittard Canyon, Ireland, Northeast Atlantic, collected on 1 June 2017, −10.681°, 48.615°, 1613 m, RV Celtic Explorer cruise CE17008. USNM1593523, Irish Continental Slope, Ireland, Northeast Atlantic, collected on 14 August 2018, −12.551°, 54.0611°, 1361 m, RV Celtic Explorer cruise CE18012. USNM1593524, Irish Continental Slope, Ireland, Northeast Atlantic, collected on 21 August 2018, −12.403°, 54.0602°, 1397 m, RV Celtic Explorer cruise CE18012. USNM1593525, Irish Continental Slope, Ireland, Northeast Atlantic, collected on 22 August 2018, −12.555°, 54.0643°, 1374 m, RV Celtic Explorer cruise CE18012. USNM1593526, Whittard Canyon, Ireland, Northeast Atlantic, collected on 1 June 2017, −10.68°, 54.0643°, 1374 m, RV Celtic Explorer cruise CE17008. USNM1691939, Irish Continental Slope, Ireland, Northeast Atlantic, collected June 2013, ~54.0552°, −12.5457°, ~1361 m, RV Celtic Explorer cruise CE13008.
USNM1516893, Whittard Canyon, Ireland, Northeast Atlantic, collected on 14 June 2014, 48.4672°, −10.3994°, 2669 m, RV Celtic Explorer cruise CE14009. USNM1704356, Fangorn Bank, Northeast Atlantic, collected 7 August 20201, 55.3661°, −19.8412°, 1286 m, RV Celtic Explorer cruise CE21010. USNM1704369, Fangorn Bank, Northeast Atlantic, collected 9 August 2021, 55.2173°, −19.8798°, 1393 m, RV Celtic Explorer cruise CE21010. USNM1704370, Fangorn Bank, Northeast Atlantic, collected 9 August 20201, 55.2242°, −19.8673°, 1241 m, RV Celtic Explorer cruise CE21010.
Whittard Canyon, Ireland, Northeast Atlantic, 1678 m.
Explorisis poppyae sp. nov. (a) In situ imagery of USNM1593518 taken by the ROV Holland I aboard RV Celtic Explorer cruise CE17008. (b) Ex situ image taken on deck. (c) Light microscopy of a polyp and (d) polyp cleared in clove oil. Imagery adapted from Morrissey et al. (2022). Scale bar: 1 mm.
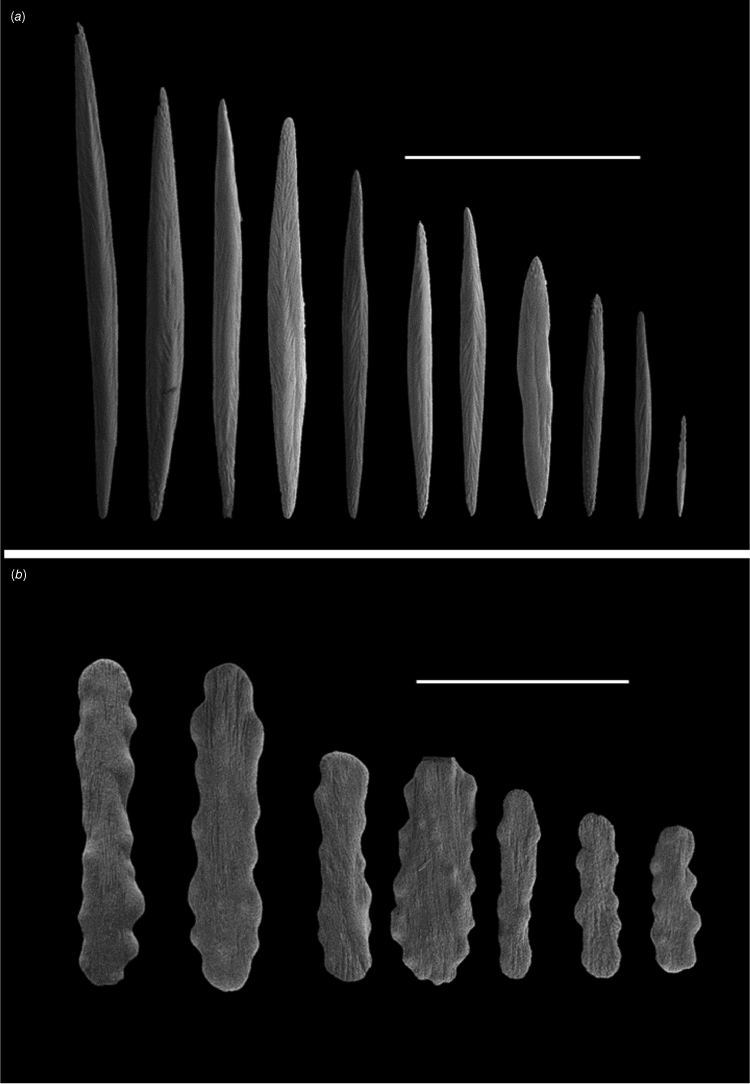
Colony branches dichotomously or trichotomously fromthe nodes. Main skeletal axis varies in thickness and is covered in either a thin or thick coenenchyme. The polyp body contains tapered rods and spindles, which are heavily granulated and project past the base of the tentacles but do not always follow the mesenteries. Rods and spindles, similar to the polyp body, and clubs are present in the coenenchyme. Flattened rods with irregular borders are present in the pharynx.
Holotype: The accessioned voucher is only a 3-cm single fragment of the colony. Tissue is brown.
The colony is planar (Fig. 6), over 30 cm tall, branching dichotomously forming a fan, although no anastomosing of branches was observed. Branching occurs at the nodes. Node and internode lengths are unknown. Polyps trumpet shaped ~5 mm long, orange to pink in colour, and originate from all sides of the coenenchyme (Fig. 6). Tissue turned brown upon preservation.
Arise primarily on three sides of the axis and are ~5 mm long and 3 mm wide. Tentacles fold over the top of the oral area.
Heavily granulated spindles and rods (Fig. 7) are arranged longitudinally in the body and range in size from 2 to 0.63 mm. Single or fusiform sclerites project past the base of the tentacles following the mesenteries. Heavily granulated spindles and clubs, 2.8–0.73 mm, are found in the coenenchyme (Fig. 7). Sclerites are usually obstructed from view by the darkened tissue. Flattened rods present in both the tentacles, 0.23–0.045 mm, and pharynx ~0.3 mm.
From in situ and ex situ photography, there is morphological variation among the paratypes. Colony morphologies are all planar but vary in size and shape, perhaps representing different growth stages of the species (Supplementary Fig. S2). Furthermore, paratypes of E. poppyae found within the Whittard Canyon system exhibit a more similar colony morphology with larger colonies, increased polyp density, and thicker coenenchyme. Those found outside the canyons along the slope are more sparsely branched, smaller and have fewer polyps that are more volcano-like in shape.
Named after A. L. Allcock’s daughter, Poppy Johnson, who is as beautiful as both the flower after which she is named, and this coral that now bears her name.
Explorisis poppyae exhibits a range of colony morphologies suggesting that differences in gross colony morphology may be due to different growth stages of the species or due to environmental drivers. Drivers such as increased food availability within canyon systems could promote growth leading to larger colonies with more polyps and thicker coenenchyme when compared to open slope environments.
Results
Mitogenome composition
The complete mitogenomes of four individuals of E. katharina were assembled (Supplementary Table S2). Mitogenomes were between 19,384 and 19,398 bp in size, had the expected 14 PCGs (ATP6, ATP8, cob, cox1-3, nad1-6, nad4l and mtMutS), 2 rRNAs (rns and rnl) and 1 transfer RNA (tRNA – Met). Genes were arranged as previously reported for the family (Octocoral gene order B, Brugler and France 2008). There are two previously sequenced mitogenomes from the paratypes of E. poppyae (USNM1593420: OR326692, USNM1691939: OR326698; Morrissey et al. 2024) publicly available. The mitogenome of USNM1593420 was 19,398 bp, whereas USNM1691939 was 19,419 bp.
Among sequenced individuals of Explorisis, only five nucleotide substitutions were observed across all PCGs: 1 bp in nad1 and 2 bp in both mtMutS and nad4. In addition, five other differences were present due to the presence of ambiguities. There was also a 21-bp insertion in mtMutS of just USNM1691939. All individuals shared identical sequences between the cob and nad6 spacer (igr4), which is 622 bp long. This region has been previously identified as being highly variable among bamboo corals (van der Ham et al. 2009), however Morrissey et al. (2024) determined the presence of a conserved ORF with unknown function within this region. This Unknown ORF, ORF1, was also present (537 bp, 179 aa) in both species. The mitogenome of one paratype of E. poppyae (USNM1593520) was identical to the holotype (USNM1516893) and a paratype (USNM1704369) of E. katharina. The mitogenome of Dokidisis australis was not recovered.
Phylogenomic position of Explorisis
A total of 2874 conserved elements were recovered with 2010 and 778 loci present within the 50 and 75% taxon-occupancy matrices respectively that consisted of all individuals (Supplementary Table S3). As expected, Explorisis was recovered within group J3 of Watling et al. (2022) within both the conserved elements and complete mitogenome phylogenies. Dokidisis, Explorisis and Jasonisis each occupied distinct, highly supported clades (Fig. 9). As the relationships among keratoisidid groups were identical to previous inferences that used conserved elements (Morrissey et al. 2023b, 2024), we only present the phylogenomic relationships, generated from the 50% taxon-occupancy matrix, among group J3 bamboo corals with I4 as the known sister group.
Maximum likelihood trees constructed from (a) the 50% conserved elements taxon occupancy matrix (2010 loci) with just keratoisidid group J3 and sister group I4 visualised and (b) the complete mitogenomes (14 Protein Coding Genes). All nodes represent 100% ultrafast bootstrap support unless otherwise stated. The colour of each node represents the transposed ASTRAL local posterior probability/MrBayes Bayesian Posterior Probability values. Pink indicates maximum LPP/BPP support (1), green is ≥0.95 and <1 LPP/BPP, blue is <0.95 and ≥0.75 LPP/BPP, and yellow is <0.75 LPP/BPP. No colour node means an unsupported relationship. The dashed line in (b) represents a branch that was manually shortened for visualisation purposes. A1, B1, C1, D1, D2, F1, I1, I4 and J3, refer to the keratoisidid groups defined by Watling et al. (2022). Specimens in bold represent sequences derived from type specimens.
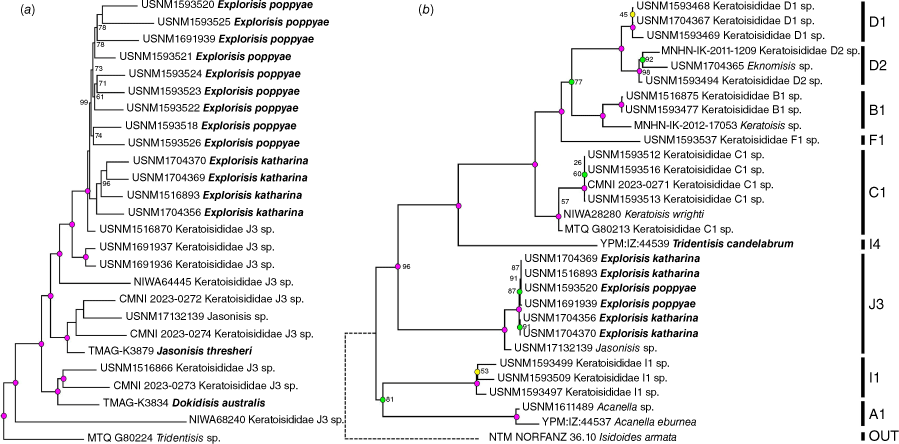
Phylogenomic relationships among Explorisis species
In both the 50 and 75% taxon-occupancy ML phylogenies, all individuals of E. katharina and E. poppyae were recovered as reciprocally monophyletic, highly supported clades. Within the ASTRAL phylogenies, however, E. katharina was not recovered as monophyletic, instead it was recovered as a series of sequential sisters to a monophyletic E. poppyae. Explorisis poppyae was recovered monophyletic in both ASTRAL phylogenies with maximum support in the 50% taxon-occupancy matrix tree and lower support in the 75% (LPP 0.64) (Supplementary Fig. S3). Neither species was recovered as monophyletic in the phylogeny constructed from the complete mitogenomes where the relationships among all Explorisis individuals were unresolved.
Discussion
Due to the large scale polyphyly at both species and genus level within keratoisidids, and the presence of numerous monotypic genera, true genus level diagnostic characters have been difficult to define. Although Jasonisis remains monotypic, the shared similarities in the sclerome among the species of Explorisis and among those of Dokidisis suggests that overall sclerome composition is a diagnostic trait of a genus within Keratoisididae whereas subtle differences in composition, sclerite texture, and microstructures can be used to diagnose species. Previously, sclerite composition, shapes, and micro ornamentations have previously been put forward as diagnostic traits for molecular clade level designations in keratoisidids (Morrissey et al. 2023b) and this study suggests that further elucidation of these characters can be used to form and revise genera. Dokidisis, Explorisis and Jasonisis have overall distinct scleromes comprising different types of body and coenenchyme sclerites, yet all three genera share similar pharyngeal sclerites: irregular shaped rods instead of thorn- and double-stars as present in other keratoisidid genera (Lapointe and Watling 2022; Morrissey et al. 2022; Watling et al. 2022). Among all current nominal species within group J3 of Watling et al. (2022), only flattened rods have been recovered from the pharynx. This trait may be diagnostic of the complete J3 group, but more species need to be examined to confirm this. The variation of colony morphologies exhibited by E. poppyae further emphasises the caution needed when describing new species on just changes in their gross morphology, exemplified by the species Anthothela grandiflora where colonies may grow as a thin membrane with no medulla or as a more arborescent shape with branches and an axial medulla (Moore et al. 2017).
The Micro-CT scan produced a highly accurate 3-D model of the polyp of E. katharina. However, at the scanned resolution, it is impossible to examine sclerite microstructures or texture, only the orientation of larger sclerites. This limits the usefulness of Micro-CT in octocoral taxonomy. Further scans of coral polyps with higher resolution may produce models in which this information is visible. If better models are produced, it may be possible to substitute SEM imagery with 3-D models, reducing a labour-intensive process involved in identifying and describing species. This model will act as a Cybertype, complimenting the physical specimen by allowing researchers to examine the sclerome of the holotype in three dimensions, without the need to travel or request the specimen. Cybertypes have been generated for many other marine invertebrate groups such as annelids (Faulwetter et al. 2013) and sponges (Karcz et al. 2015; Shilov et al. 2023). The generation of Cybertypes enables equitable and inclusive taxonomic research, removing barriers to morphological information that have always been present for researchers from lower-income countries or those without formal affiliations to academic institutes.
Phylogenomics using conserved elements has been able to guide species discovery (Morrissey et al. 2023b) by confirming and disputing hypotheses put forward by previous phylogenies constructed from few genes (Watling et al. 2022). Although there are many undescribed genera and species within the family Keratoisididae (Watling and France 2011; Lapointe and Watling 2022; Morrissey et al. 2022; Watling et al. 2022), phylogenomics can also help guide revisions of existing genera. For example, nominal species of Lepidisis and Keratoisis, the two genera that currently contain most of the nominal species of the Keratoisididae, occur in numerous molecular clades. Ongoing revisions of those genera have determined that true Lepidisis is within clade I1 and that most of the described species of this genus need reassignment (Watling and France 2021). However, attempts to reassign these species has been hampered by a lack of genomic resolution on their true position. Keratoisis grayi, the type of the genus Keratoisis, and Eknomisis dalioi, the type of the genus Eknomisis, are both found within D2, yet mitochondrial based phylogenies have been unable to untangle these genera (Dueñas et al. 2014; Morrissey et al. 2022; Watling et al. 2022), while there is a clear divide using conserved elements. The sequencing of additional type material is fundamental to support additional taxonomic revisions across the family.
The nuclear-based phylogenies showed clear separation of E. katharina and E.poppyae into two highly supported clades within Explorisis whereas phylogenies constructed from complete mitogenomes did not return each species as monophyletic. The limited usefulness of single- and multi-gene mitochondrial markers within Octocorallia has been well documented (McFadden et al. 2011) e.g. among species of Chrysogorgia (Pante et al. 2015), Paramuricea (Radice et al. 2016; Quattrini et al. 2022) and Paragorgia (Herrera and Shank 2016). Mutation rates in the octocoral mitogenome are incredibly low, in part due to the presence of the mismatch repair gene mtMutS (Bilewitch and Degnan 2011; Muthye et al. 2022). Among all individuals of Explorisis, 11 of the 14 PCGS exhibited no variation, and only 0.255–0.76, 0.04–0.2 and 0.01–0.06% variation was observed in mtMutS, nad4 and nad1 respectively. The space between cob and nad6, which has previously been used for delimiting species due to its high variability (van der Ham et al. 2009), was also identical across both species of Explorisis. Low sequence variation of the same magnitude has been reported within the species Paramuricea grayi (Coelho et al. 2022) and among species of Eunicella (Hooper et al. 2021) highlighting the mitochondrial genomes limited use in delineating boundaries.
Conclusion
This study described a new genus and two species belonging to the family Keratoisididae. The genus Explorisis is closely related to both Dokidisis and Jasonisis and has a unique sclerome that distinguishes it from both genera. The sclerome of Explorisis is primarily dominated by tapered rods and spindles whereas Dokidisis and Jasonisis are distinct by having primarily blunt rods and scales. Phylogenies constructed from complete mitogenomes did not return either species of Explorisis as monophyletic, rather the relationship among all individuals of Explorisis was unresolved. However, both species were reciprocally monophyletic in phylogenies constructed from hundreds of nuclear loci.
Data availability
Newly generated raw read data are available in NCBI Genbank SRA archive BioProject PRJNA1073973 and complete mitogenomes were uploaded to GenBank. See Supplementary Tables S1 and S2.
Declaration of funding
Declan Morrissey was funded by an Irish Research Council Postgraduate Scholarship (GOIPG/2019/3682), and a Kenneth Jay Boss Fellowship in the Department of Invertebrate Zoology from The Smithsonian Institution National Museum of Natural History. Corals were collected during research cruise CE13008 aboard RV Celtic Explorer using ROV Holland I, which was funded by the Marine Institute under the Irish National Ship Time Programme awarded to A. L. Allcock, CE14009 aboard RV Celtic Explorer using ROV Holland I, which was funded by the Marine Institute under the Irish National Ship Time Programme awarded to Martin White (University of Galway), CE21010 aboard RV Celtic Explorer using ROV Holland I, which was funded by the Marine Institute under the Irish National Ship Time Programme awarded to A. L. Allcock. Expeditions CE17008 and CE18012 aboard RV Celtic Explorer using ROV Holland I were funded by Science Foundation Ireland and the Marine Institute under Investigators Programme Grant SFI/15/IA/3100 and co-funded under the European Regional Development Fund 2014–2020 awarded to A. L. Allcock. The author payment charges for this publication were funded with the support of the Marine Institute under the Marine Research Programme with the support of the Irish Government.
Acknowledgements
Molecular work on new specimens was undertaken in part in the Laboratories of Analytical Biology (LAB) at the NMNH. The computations in this paper were conducted on the Smithsonian High-Performance Cluster (SI/HPC), Smithsonian Institution (see https://doi.org/10.25572/SIHPC). The authors are grateful to Nina I. Ramos (FIU/SI NMNH) for her help photographing the Explorisis katharina holotype; Scott Whittaker for assisting in the Scanning Electron Microscopy; and Freya Goetz for scanning and processing the Micro-CT. All imaging was conducted at the Scientific Imaging Lab at the National Museum of Natural History, Smithsonian Institution. D. Morrissey also thanks Jamie Maxwell for identifying the sea spider. The authors are grateful to the Tasmanian Museum and Art Gallery for facilitating the loan of TMAG-K3834. We thank the crew and scientists of cruises CE13008, CE14009, CE17008, CE18012 and CE21010 aboard RV Celtic Explorer and the crew operating ROV Holland I for all their help.
References
Alderslade P, McFadden CS (2012) A new genus and species of the family Isididae (Coelenterata: Octocorallia) from a CMAR Biodiversity study, and a discussion on the subfamilial placement of some nominal isidid genera. Zootaxa 3154, 21-39.
| Crossref | Google Scholar |
Altuna Á, López-González PJ (2023) A new genus and species of deep-sea Coralliidae (Octocorallia) from the northeastern Atlantic, the first polymorphic octocoral genus with a membranous-stolonate colony growth form. Deep-Sea Research – I. Oceanographic Research Papers 201, 104157.
| Crossref | Google Scholar |
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19, 455-477.
| Crossref | Google Scholar | PubMed |
Bayer FM (1990) A new isidid octocoral (Anthozoa: Gorgonacea) from New Caledonia, with descriptions of other new species from elsewhere in the Pacific Ocean. Proceedings of the Biological Society of Washington 103, 205-228.
| Google Scholar |
Bilewitch JP, Degnan SM (2011) A unique horizontal gene transfer event has provided the octocoral mitochondrial genome with an active mismatch repair gene that has potential for an unusual self-contained function. BMC Evolutionary Biology 11, 228.
| Crossref | Google Scholar | PubMed |
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30, 2114-2120.
| Crossref | Google Scholar | PubMed |
Brugler MR, France SC (2008) The mitochondrial genome of a deep-sea bamboo coral (Cnidaria, Anthozoa, Octocorallia, Isididae): genome structure and putative origins of replication are not conserved among octocorals. Journal of Molecular Evolution 67, 125-136.
| Crossref | Google Scholar | PubMed |
Coelho M, Ledoux J-B, Boavida J, Paulo D, Gómez-Gras D, Bensoussan N, López-Sendino P, Cerrano C, Kipson S, Bakran-Petricioli T, Garrabou J, Serrão EA, Pearson GA (2022) Complete mitochondrial genome of the branching octocoral Paramuricea grayi (Johnson, 1861), phylogenetic relationships and divergence analysis. Mitochondrial DNA. Part B, Resources 7, 1985-1988.
| Crossref | Google Scholar | PubMed |
Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Research 45, e18.
| Crossref | Google Scholar | PubMed |
Dueñas LF, Alderslade P, Sánchez JA (2014) Molecular systematics of the deep-sea bamboo corals (Octocorallia: Isididae: Keratoisidinae) from New Zealand with descriptions of two new species of Keratoisis. Molecular Phylogenetics and Evolution 74, 15-28.
| Crossref | Google Scholar |
Erickson KL, Pentico A, Quattrini AM, McFadden CS (2021) New approaches to species delimitation and population structure of anthozoans: Two case studies of octocorals using ultraconserved elements and exons. Molecular Ecology Resources 21, 78-92.
| Crossref | Google Scholar | PubMed |
Etnoyer P, Warrenchuk J (2007) A catshark nursery in a deep gorgonian field in the Mississippi Canyon, Gulf of Mexico. Bulletin of Marine Science 81, 553-559.
| Google Scholar |
Faircloth BC (2016) PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 32, 786-788.
| Crossref | Google Scholar | PubMed |
Faulwetter S, Vasileiadou A, Kouratoras M, Thanos D, Arvanitidis C (2013) Micro-computed tomography: Introducing new dimensions to taxonomy. ZooKeys 263, 1-45.
| Crossref | Google Scholar | PubMed |
France SC (2007) Genetic analysis of bamboo corals (Cnidaria: Octocorallia: Isididae): Does lack of colony branching distinguish Lepidisis from Keratoisis? Bulletin of Marine Science 81, 323-333.
| Google Scholar |
France SC, Watling L (2024) Toward a revision of the bamboo corals: part 6, illuminating a new candelabrum genus (Octocorallia: Keratoisididae). Zootaxa 5497(4), 505-519.
| Crossref | Google Scholar |
Glenn TC, Nilsen RA, Kieran TJ, Sanders JG, Bayona-Vásquez NJ, Finger JW, Pierson TW, Bentley KE, Hoffberg SL, Louha S, Garcia-De Leon FJ, Del Rio Portilla MA, Reed KD, Anderson JL, Meece JK, Aggrey SE, Rekaya R, Alabady M, Belanger M, Winker K, Faircloth BC (2019) Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially indexed Illumina libraries (iTru & iNext). PeerJ 7, e7755.
| Crossref | Google Scholar | PubMed |
Heestand Saucier E, France SC, Watling L (2021) Toward a revision of the bamboo corals: part 3, deconstructing the Family Isididae. Zootaxa 5047, 247-272.
| Crossref | Google Scholar | PubMed |
Herrera S, Shank TM (2016) RAD sequencing enables unprecedented phylogenetic resolution and objective species delimitation in recalcitrant divergent taxa. Molecular Phylogenetics and Evolution 100, 70-79.
| Crossref | Google Scholar | PubMed |
Hooper L, Jenkins TL, Griffiths AM, Moore KA, Stevens JR (2021) The complete mitochondrial genome of the pink sea fan, Eunicella verrucosa (Pallas, 1766). Mitochondrial DNA Part B 6, 3309-3311.
| Crossref | Google Scholar | PubMed |
Junier T, Zdobnov EM (2010) The Newick utilities: high-throughput phylogenetic tree processing in the Unix shell. Bioinformatics 26, 1669-1670.
| Crossref | Google Scholar | PubMed |
Kalyaanamoorthy S, Minh BQ, Wong TFK, Von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587-589.
| Crossref | Google Scholar | PubMed |
Karcz J, Woznica A, Binkowski M, Klonowska-Olejnik M, Bernas T, Karczewski J, Migula P (2015) SEM-EDS and X-ray micro computed tomography studies of skeletal surface pattern and body structure in the freshwater sponge Spongilla lacustris collected from Goczalkowice Reservoir habit (Southern Poland). Folia Histochemica et Cytobiologica 53, 88-95.
| Crossref | Google Scholar | PubMed |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30, 772-780.
| Crossref | Google Scholar | PubMed |
Lapointe A, Watling L (2015) Bamboo corals from the abyssal Pacific: Bathygorgia. Proceedings of the Biological Society of Washington 128, 125-136.
| Crossref | Google Scholar |
Lapointe A, Watling L (2022) Towards a revision of the bamboo corals (Octocorallia): part 5, new genera and species of Keratoisididae from the Tasmanian deep sea. Zootaxa 5168, 137-157.
| Crossref | Google Scholar | PubMed |
McFadden CS, Benayahu Y, Pante E, Thoma JN, Nevarez PA, France SC (2011) Limitations of mitochondrial gene barcoding in Octocorallia. Molecular Ecology Resources 11, 19-31.
| Crossref | Google Scholar | PubMed |
McFadden CS, van Ofwegen LP, Quattrini AM (2022) Revisionary systematics of Octocorallia (Cnidaria: Anthozoa) guided by phylogenomics. Bulletin of the Society of Systematic Biologists 1, 8735.
| Crossref | Google Scholar |
Mai U, Mirarab S (2018) TreeShrink: fast and accurate detection of outlier long branches in collections of phylogenetic trees. BMC Genomics 19, 272.
| Crossref | Google Scholar | PubMed |
Maxwell J, Taboada S, Taylor ML (2022) Gorgoniapolynoe caeciliae revisited: the discovery of new species and molecular connectivity in deep-sea commensal polynoids from the Central Atlantic. Deep-Sea Research – I. Oceanographic Research Papers 185, 103804.
| Crossref | Google Scholar |
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37, 1530-1534.
| Crossref | Google Scholar | PubMed |
Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T (2014) ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics (Oxford, England) 30, i541-i548.
| Crossref | Google Scholar | PubMed |
Moore KM, Alderslade P, Miller KJ (2017) A taxonomic revision of Anthothela (Octocorallia: Scleraxonia: Anthothelidae) and related genera, with the addition of new taxa, using morphological and molecular data. Zootaxa 4304, 1-212.
| Crossref | Google Scholar |
Morrissey D, Untiedt CB, Croke K, Robinson A, Turley E, Allcock AL (2022) The biodiversity of calcaxonian octocorals from the Irish Continental Slope inferred from multilocus mitochondrial barcoding. Diversity 14, 576.
| Crossref | Google Scholar |
Morrissey D, Lim A, Howell KL, White M, Wheeler AJ, Allcock AL (2023a) The north-east Atlantic margin: a review of the geology, geography, oceanography, and vulnerable megabenthic ecosystems of the continental slope of Ireland and the United Kingdom. Oceanography and MArine Biology: An Annual Review 61, 219-292.
| Crossref | Google Scholar |
Morrissey D, Gordon JD, Saso E, Bilewitch JP, Taylor ML, Hayes V, McFadden CS, Quattrini AM, Allcock AL (2023b) Bamboozled! Resolving deep evolutionary nodes within the phylogeny of bamboo corals (Octocorallia: Scleralcyonacea: Keratoisididae). Molecular Phylogenetics and Evolution 188, 107910.
| Crossref | Google Scholar |
Morrissey D, Quattrini AM, Allcock AL (2024) Analysis of mitogenomes from the family Keratoisididae reveals mitonuclear discordance and the presence of unknown open reading frames. Research Square [Preprint, version 1, posted 12 March 2024].
| Crossref | Google Scholar |
Muthye V, Mackereth CD, Stewart JB, Lavrov DV (2022) Large dataset of octocoral mitochondrial genomes provides new insights into mt-mutS evolution and function. DNA Repair 110, 103273.
| Crossref | Google Scholar | PubMed |
Neves BM, Wareham Hayes V, Herder E, Hedges K, Grant C, Archambault P (2020) Cold-water soft corals (Cnidaria: Nephtheidae) as habitat for juvenile basket stars (Echinodermata: Gorgonocephalidae). Frontiers in Marine Science 7, 547896.
| Crossref | Google Scholar |
Orejas C, Carreiro-Silva M, Mohn C, Reimer J, Samaai T, Allcock AL, Rossi S (2022) Marine animal forests of the world: definition and characteristics. Research Ideas and Outcomes 8, e96274.
| Crossref | Google Scholar |
Pante E, Abdelkrim J, Viricel A, Gey D, France SC, Boisselier MC, Samadi S (2015) Use of RAD sequencing for delimiting species. Heredity 114, 450-459.
| Crossref | Google Scholar | PubMed |
Quattrini AM, Faircloth BC, Dueñas LF, Bridge TCL, Brugler MR, Calixto-Botía IF, DeLeo DM, Forêt S, Herrera S, Lee SMY, Miller DJ, Prada C, Rádis-Baptista G, Ramírez-Portilla C, Sánchez JA, Rodríguez E, McFadden CS (2018) Universal target-enrichment baits for anthozoan (Cnidaria) phylogenomics: new approaches to long-standing problems. Molecular Ecology Resources 18, 281-295.
| Crossref | Google Scholar | PubMed |
Quattrini AM, Rodríguez E, Faircloth BC, Cowman PF, Brugler MR, Farfan GA, Hellberg ME, Kitahara MV, Morrison CL, Paz-García DA, Reimer JD, McFadden CS (2020) Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nature Ecology & Evolution 4, 1531-1538.
| Crossref | Google Scholar | PubMed |
Quattrini AM, Herrera S, Adams JM, Grinyó J, Allcock AL, Shuler A, Wirshing HH, Cordes EE, McFadden CS (2022) Phylogeography of Paramuricea: the role of depth and water mass in the evolution and distribution of deep-sea corals. Frontiers in Marine Science 9, 849402.
| Crossref | Google Scholar |
Radice VZ, Quattrini AM, Wareham VE, Edinger EN, Cordes EE (2016) Vertical water mass structure in the North Atlantic influences the bathymetric distribution of species in the deep-sea coral genus Paramuricea. Deep-Sea Research – I. Oceanographic Research Papers 116, 253-263.
| Crossref | Google Scholar |
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67, 901-904.
| Crossref | Google Scholar | PubMed |
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574.
| Crossref | Google Scholar | PubMed |
Rossi S, Bramanti L, Gori A, Orejas C (2017). Animal forests of the world: an overview. In ‘Marine Animal Forests: The Ecology of Benthic Biodiversity Hotspots’. (Eds S Rossi, L Bramanti, A Gori, C Orejas) pp. 1–28. (Springer) 10.1007/978-3-319-21012-4_1
Rossi S, Bramanti L, Horta P, Allcock L, Carreiro-Silva M, Coppari M, Denis V, Hadjioannou L, Isla E, Jimenez C, Johnson M, Mohn C, Orejas C, Ramšak A, Reimer J, Rinkevich B, Rizzo L, Salomidi M, Samaai T, Schubert N, Soares M, Thurstan RH, Vassallo P, Ziveri P, Zorrilla-Pujana J (2022) Protecting global marine animal forests. Science 376, 929.
| Crossref | Google Scholar | PubMed |
Shilov VA, Kamenev YO, Semenchenko AA, Kiyashko SI, Mordukhovich VV (2023) New sponge species of the family Vulcanellidae (Demospongiae: Tetractinellida) from the Piip submarine volcano and adjacent areas (Bering Sea, NW Pacific). Deep-Sea Research – II. Topical Studies in Oceanography 208, 105229.
| Crossref | Google Scholar |
van der Ham JL, Brugler MR, France SC (2009) Exploring the utility of an indel-rich, mitochondrial intergenic region as a molecular barcode for bamboo corals (Octocorallia: Isididae). Marine Genomics 2, 183-192.
| Crossref | Google Scholar |
Vecchione M (2019) ROV observations on reproduction by deep-sea cephalopods in the central Pacific Ocean. Frontiers in Marine Science 6, 403.
| Crossref | Google Scholar |
Vences M, Patmanidis S, Kharchev V, Renner SS (2022) Concatenator, a user-friendly program to concatenate DNA sequences, implementing graphical user interfaces for MAFFT and FastTree. Bioinformatics Advances 2, vbac050.
| Crossref | Google Scholar | PubMed |
Watling L (2015) A new genus of bamboo coral (Octocorallia: Isididae) from the Bahamas. Zootaxa 3918, 239-249.
| Crossref | Google Scholar | PubMed |
Watling L, France SC (2011) A new genus and species of bamboo coral (Octocorallia: Isididae: Keratoisidinae) from the New England seamounts. Bulletin of the Peabody Museum of Natural History 52, 209-221.
| Crossref | Google Scholar |
Watling L, France SC (2021) Toward a revision of the bamboo corals: part 2, untangling the genus Lepidisis (Octocorallia: Isididae). Bulletin of the Peabody Museum of Natural History 62, 97-110.
| Crossref | Google Scholar |
Watling L, France SC, Pante E, Simpson A (2011) Biology of deep-water octocorals. In ‘Advances in Marine Biology’. (Ed. MB Lesser) pp. 41–122. (Academic Press) 10.1016/B978-0-12-385529-9.00002-0
Watling L, Saucier EH, France SC (2022) Towards a revision of the bamboo corals (Octocorallia): part 4, delineating the family Keratoisididae. Zootaxa 5093, 337-375.
| Crossref | Google Scholar | PubMed |
Zhang C, Rabiee M, Sayyari E, Mirarab S (2018) ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19, 153.
| Crossref | Google Scholar | PubMed |