Phylogenomics of endemic Australian Ulopinae (Hemiptera: Cicadomorpha: Cicadellidae)
Olivia Evangelista A * , Nikolai Tatarnic B C and Keith Bayless A D
A * , Nikolai Tatarnic B C and Keith Bayless A D
A
B
C
D
Abstract
Ulopinae is a distinctive subfamily of leafhoppers that is widely distributed across the Afrotropical, Palearctic, Indomalayan and Australasian regions. The ulopine fauna of Australia is entirely endemic and includes two tribes of striking appearance, the Ulopini and Cephalelini. Knowledge of these groups is fragmentary and in many instances, no information is available beyond original descriptions. We assess the monophyly, phylogenetic placement and species-level diversity of the Ulopini genus Austrolopa. Phylogenetic analyses based on sequence data from target nuclear loci (18S, 28S, H2A and H3) and mitochondrial genomes (15 genes) for 23 membracoid taxa yielded congruent topologies. Our results provide strong evidence for the monophyly of Ulopinae and a clade consisting of Ulopini + Cephalelini. However, a non-monophyletic Cephalelini arises from within a polyphyletic Ulopini. Austrolopa was strongly recovered as monophyletic in all analyses, a result also supported by morphological features. The genus currently includes six species, three of which are described based on morphological and molecular data: Austrolopa botanica, sp. nov., Austrolopa rotunda, sp. nov. and Austrolopa sublima, sp. nov. A lectotype designation is provided for Austrolopa kingensis Evans, 1937, sp. reval. Our findings illustrate that the Australian Ulopinae is far more diverse than currently circumscribed and several species of Austrolopa are yet to be recognised.
ZooBank: urn:lsid:zoobank.org:pub:1480285B-8F61-4659-A929-2B1EF3168868
Keywords: Austrolopa, Bush Blitz, bush fires, leafhoppers, Membracoide, narrow-range endemics, species discovery, Ulopini.
Introduction
Leafhoppers in the family Cicadellidae (Hemiptera: Membracoidea) are a megadiverse group of plant-feeding insects occurring in virtually all terrestrial vegetated ecosystems. This group includes over 23 000 described species classified into 19 subfamilies (Dietrich et al. 2022), and is therefore the largest family of hemimetabolous insects, and one of the most diverse groups of herbivores. The abundance and ubiquity likely result from a confluence of several factors, such as the broad range of host plant associations and long evolutionary history. Fossil evidence shows some extant groups to be present since the lower cretaceous (Hamilton 1990) and divergence time estimates indicate that several leafhopper lineages originated between 150 and 120 million years ago, concurrent with the diversification of angiosperm host plants during the late Jurassic and early Cretaceous (Dietrich et al. 2017). Several leafhopper species are also key agricultural pests, causing considerable damage in primary industries through direct feeding or transmission of phytopathogenic agents such as the bacteria Xylella fastidiosa (Krugner et al. 2019).
Despite the outstanding diversity and agricultural significance, major aspects of the evolutionary history of Cicadellidae have only recently been clarified and several key questions remain challenging. Multiple phylogenetic studies strongly support earlier hypotheses that Cicadellidae is paraphyletic with respect to a lineage encompassing the treehoppers (Aetalionidae, Melizoderidae and Membracidae; Hamilton 1983; Dietrich et al. 2001; Cryan and Urban 2012). Over the last decade, the non-monophyly of Cicadellidae was demonstrated in multiple independent studies based on transcriptome (Johnson et al. 2018; Skinner et al. 2019; Hu et al. 2023), anchored hybrid enrichment exome capture (Dietrich et al. 2017) and morphology data (Dietrich et al. 2022). In addition to the non-monophyly of Cicadellidae, these studies also support Ulopinae and Megophthalminae as sister groups to the treehopper families (e.g. Dietrich et al. 2017).
These phylogenetic relationships are consistent with the evidence gathered by Rakitov (1996, 1998; Rakitov and Godoy 2005) on brochosomes, a key evolutionary innovation in leafhoppers that is also considered to have greatly contributed to the diversification. Brochosomes are highly hydrophobic proteinaceous granules secreted in modified segments of the Malpighian tubules that help leafhoppers keep the cuticle dry, preventing these from being encapsulated by water droplets. Leafhoppers exhibit a suite of behaviours and morphological adaptations to coat themselves in brochosomes, such as long setae on the hindlegs that help spread the fluid over the body or powder the brochosomes onto the eggs. Although treehoppers have lost the ability to produce brochosomes, there is still a unique ‘bathing’ behaviour that is shared with ulopine relatives (Rakitov 1996). The physiological loss of brochosome production concomitant with the retention of complex bathing behaviour suggests that these groups are linked by a major evolutionary transition (Dietrich et al. 2001). Information on the evolution and diversity of Ulopinae is therefore fundamental to clarify the classification of Membracoidea and understand the evolution of these unique morphological and behavioural traits.
Although a comprehensive phylogeny of Ulopinae has not been the focus of a previous study, prior research indicates that these insects share a suite of unique morphological traits. In ulopines, the maxillary plate and gena are separated by a distinct cleft, and the mesanepisternum bears a shelf-like projection (Dietrich 2005; World Auchenorrhyncha Database by Dmitriev et al., see https://hoppers.speciesfile.org). The hindlegs have reduced macrosetae, fine structures that are typically used to comb dry brochosomes onto the insects’ integument. Ulopines also have comparatively shorter hindlegs than other membracoids that may be associated with a diminished ability to jump. Many species are either brachypterous or lack hindwings, suggesting that these are slow insects with reduced mobility that rely on crypsis to avoid predation.
Ulopinae currently includes 135 species classified into five tribes. The extant distribution suggests a Gondwanan origin. Species of Ulopini are present worldwide except for the Neotropical and Nearctic regions; Cephalelini shows a disjunct distribution in South Africa, Australia and New Zealand; Coloborrhinini and Mesargini are Afrotropical and Indo-Malayan, respectively; and the monobasic Monteithiini is restricted to montane areas of Papua New Guinea. Australian representatives comprise a considerable share of the putative phylogenetic diversity of Ulopinae, accounting for 12 of the 40 currently recognised genera. The Australian fauna is entirely endemic and represented by two tribes that are strikingly dissimilar. Members of Cephalelini can be remarkably slender due to the strongly projected head and elongated forewings. Some species are potential seed mimics and associated with grassland environments. By contrast, members of Ulopini are relatively small, round cryptic leafhoppers with heavily sclerotised, dark-coloured, densely punctured cuticles.
The taxonomy of these groups has not been properly addressed since Evans (1977), who described most of the Australian fauna. As a result, some diagnostic features do not allow unambiguous identification of the newly collected ulopine specimens, especially at the species level. This study provides the first glimpse into the true diversity of Australian Ulopinae. We assess the monophyly, phylogenetic placement and species-level diversity of Austrolopa. Some of these ulopines appear to be narrow-range endemics (Harvey 2002; Rosauer et al. 2009) in south-eastern Australia, potentially confined to small geographic areas and host plants. We discovered and described three species of Austrolopa within the 200 km between Black Mountain (ACT) and Kosciuszko National Park (NSW). This area has been significantly affected by bushfires, most recently in 2019–20. The extent of the damage from this unprecedented environmental catastrophe cannot be fully determined until the species diversity is more thoroughly catalogued (Marsh et al. 2023). We aim to investigate the phylogeny of Australian Ulopini with a thorough sampling of multiple ulopine lineages, contextualising the evolutionary history and assessing the species-level classification of Austrolopa that includes at-risk species in montane areas of south-eastern Australia.
Materials and methods
Taxon sampling
Fresh samples were obtained from extensive malaise trapping programs at the Australian National Insect Collection and active collecting during various Bush Blitz species discovery expeditions. Other key specimens were sorted from coloured pan traps that were part of a 2-year survey on alpine pollination (Encinas-Viso et al. 2022). The examined material is deposited at the Australian Museum (AM), Australian National Insect Collection (ANIC), Queensland Museum (QM), South Australian Museum (SAMA), Victoria Museum (NMV) and Western Australian Museum (WAM). Taxa included in the phylogenetic sampling are listed in Table 1. Voucher information, gene sampling and NCBI accession numbers are specified in the Supplementary Tables S1 and S2. Specimen metadata were entered into the corresponding institutional databases, and will subsequently be shared with the Australian Faunal Directory and Atlas of Living Australia.
Phylogenomic analyses were performed on the CSIRO Petrichor computing resource. All samples collected by the authors were obtained under appropriate collection permits (ACT permit number PL2017113 and NSW Permit number SL100528, principal licensee David Yeates).
| Family | Subfamily | Tribe | Genus and species | Source | |
|---|---|---|---|---|---|
| Aetalionidae | Aetalioninae | Aetalionini | Aetalion reticulatum | Dietrich et al. (2017) | |
| Aetalionidae | Biturritiinae | Biturritiini | Tropidaspis sp. | Dietrich et al. (2017) | |
| Cicadellidae | Deltocephalinae | Eupelicini | Paradorydium sp. | This study | |
| Cicadellidae | Eurymelinae | Macropsini | Macropsis notata | Wang et al. (2020) | |
| Cicadellidae | Ledrinae | Ledrini | Ledropsis sp. | This study | |
| Cicadellidae | Megophthalminae | Agalliini | Bergallia sp. A | Dietrich et al. (2017) | |
| Cicadellidae | Megophthalminae | Agalliini | Sinoagallia serrata | Dietrich et al. (2017) | |
| Cicadellidae | Ulopinae | Cephalelini | Paracephaleus aff. marginatus | This study | |
| Cicadellidae | Ulopinae | Cephalelini | Paracephaleus marginatus | This study | |
| Cicadellidae | Ulopinae | Cephalelini | Paracephaleus sp. | This study | |
| Cicadellidae | Ulopinae | Cephalelini | Paracephalus dobsonensis | This study | |
| Cicadellidae | Ulopinae | Coloborrhinini | Coloborrhis corticina | Dietrich et al. (2017) | |
| Cicadellidae | Ulopinae | Mesargini | Mesargus hirsutus | Dietrich et al. (2017) | |
| Cicadellidae | Ulopinae | Mesargini | Mesargus serrata | R.-H. Dai and J.-J. Wang (unpub. data) | |
| Cicadellidae | Ulopinae | Ulopini | Austrolopa botanica sp. nov. B | This study | |
| Cicadellidae | Ulopinae | Ulopini | Austrolopa rotunda sp. nov. B | This study | |
| Cicadellidae | Ulopinae | Ulopini | Austrolopa sublima sp. nov. B | This study | |
| Cicadellidae | Ulopinae | Ulopini | Microlopa sp. | This study | |
| Cicadellidae | Ulopinae | Ulopini | Taslopa montana | This study | |
| Cicadellidae | Ulopinae | Ulopini | Ulopa reticulata | Johnson et al. (2018) | |
| Melizoderidae | Melizoderinae | Melizoderini | Melizoderes osborni | Dietrich et al. (2017) | |
| Membracidae | Centrotinae | Terentiini | Acanthuchus sp. | This study | |
| Membracidae | Stegaspidinae | Stegaspidini | Bocydium globulare | Dietrich et al. (2017) |
Taxonomic descriptions
The identity of known species was assessed based on literature and firsthand examination of primary types. Putative new species were compared with several specimens identified by Evans that were used as a basis for multiple studies on Australian leafhoppers (Evans 1937, 1939, 1966, 1977). Our delimitation hypothesis involved an evaluation of morphological features, geographic range and depth of DNA sequence variation. Specimen analysis, verbatim label transcription and dissections followed the methods outlined in Evangelista et al. (2014). Updated information on Membracoidea classification and nomenclature was checked against the World Auchenorrhyncha Database (see https://hoppers.speciesfile.org). Taxonomic descriptions followed the terminology presented by Dmitriev (2010) and Dietrich et al. (2022). Images provided for key taxa were acquired with the BK Visionary Digital Imaging Station that was equipped with Canon EOS 7D/6DII digital cameras connected to various macro lenses (Canon MP-E 65 mm and 200 mm appended to Mitutoyu Microscope Objectives 5×, 10× and 20×). New taxa and nomenclatural acts are attributed to the first author only.
Genomic data acquisition and data mining
The alignment was composed of data originating from four sequencing methods. Newly sequenced shotgun genomes were aligned with (a) genes scraped from the NCBI Nucleotide database, (b) Transcriptome Shotgun Assemblies (TSAs) from NCBI and (c) Anchored Hybrid Enrichment exome capture data, assembled de novo from NCBI Short Read Archive (SRA). For all newly sequenced taxa, DNA was extracted from ethanol-preserved, pinned specimens using the Qiagen DNeasy kit. Non-destructive extractions were performed on the whole body, after detaching the thorax from the abdomen. The quality of each genomic extract was assessed based on the concentration and total amount of DNA obtained with a Qubit fluorometer assay. This was evaluated in conjunction with a DNA fragment size and smear analysis, obtained through the Agilent Fragment Analyser system. DNA extractions were sent to Novogene Singapore for library preparation, indexing, multiplexing and sequencing using an Illumina NovaSeq platform. Samples were sequenced at ~8 GB of raw sequence reads per sample to account for the larger genome sizes in Hemiptera. Raw read quality was checked with FastQC (ver. 0.11.5, A. Andrews, see https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to assess whether further trimming was necessary. Trimmomatic (ver. 0.32, see http://www.usadellab.org/cms/index.php?page=trimmomatic; Bolger et al. 2014) was used to remove adapter contamination and low-quality sequences. Quality filtering was performed using Trimgalore (ver 0.6.7, see https://github.com/FelixKrueger/TrimGalore; Krueger 2015) to remove Nextera primers and low-fidelity reads. The GATB Minia pipeline (ver. 1, 2022 update, see https://github.com/GATB/gatb-minia-pipeline) using kmers 21, 41, 61, 81, 101 and 121 was used to assemble the de novo data including mitochondrial genomes. Previously sequenced AHE data were downloaded from NCBI SRA using the fasterq-dump module on SRAToolkit (ver. 3.0.0, see https://hpc.nih.gov/apps/sratoolkit.html; Leinonen et al. 2011) and quality filtered as described above. These data were assembled de novo by combining the outputs of three assembly strategies: Tadpole, part of BBMap (ver. 38.90, parameters shave rinse pop k = 75, https://github.com/BioInfoTools/BBMap/blob/master/sh/tadpole.sh; Bushnell et al. 2017), Trinity (ver. 2.13.2, default parameters, see https://github.com/trinityrnaseq/trinityrnaseq/wiki; Grabherr et al. 2011) and SPAdes (ver. 3.15.5, kmers 21,33,55,77, see https://github.com/ablab/spades; Bankevich et al. 2012). Each of these assembly strategies resulted in longer contigs for certain gene regions, therefore merging and deduplication of assembly overlap using dedupe.sh (part of BBMap, ver. 38.90) yielded longer, more complete alignments than using the output of any single assembly protocol. This more inclusive assembly strategy was necessary as mitochondrial and ribosomal sequences are not targeted by the AHE probes, however many reads map to mitochondrial and ribosomal genes. Removal of potential cross-contaminated contigs from index mis-specification or laboratory error was performed in BLAST+ (ver. 2.12.0, see https://blast.ncbi.nlm.nih.gov/; Camacho et al. 2009). Representatives of major groups (i.e. Membracidae) were binned together and each assembly was reciprocally blasted with all outside that bin, and any contigs with word size ≥180 and ≥99% identity were removed. All contigs for each assembly were filtered by blasting against a database of common bacterial, plant and vertebrate vector contaminants, with 135-bp wordsize and ≥98 identity.
Two bioinformatic approaches were used to retrieve (a) mitochondrial genes and fully annotated mitochondrial genomes (MitoFinder ver. 1.4.1, see https://github.com/RemiAllio/MitoFinder; Allio et al. 2020) and (b) ribosomal and nuclear protein-coding genes (PhyloHerb, ver. 1.1.0, see https://github.com/lmcai/PhyloHerb; Cai et al. 2022). Both pipelines were implemented using tailored reference sets, including annotated mitochondrial genome data from multiple representatives in Cicadellidae, Melizoderidae, Aetalionidae and Membracidae (Supplementary Table S3). Data mining was performed in PhyloHerb using the default settings to retrieve four nuclear loci: 18S rDNA (18S), 28S rDNA (28S), Histone 2A (H2A) and Histone 3 (H3). Automated assembly of mitochondrial genomes, gene annotation and individual gene extraction were performed in MitoFinder using MetaSpades as the assembling option and MiTFi for the tRNA annotation step. The following parameters were adjusted in MitoFinder depending on the availability of closely related reference data: -min-contig-size, -blast-size, -blast-identity-nucl and -blast-identity-prot. Results were qualitatively examined to assert that sequence data had been successfully obtained and to reduce error and contamination through (a) manual inspection of the alignment, (b) sequence verification using the blastn search tool against the standard nucleotide collection on the NCBI database and (c) neighbour joining analysis for each gene fragment using extensive Membracoidea outgroups obtained from the NCBI nucleotide database.
Phylogenomic analysis
The final alignment consisted of 19 genes: 2 nuclear protein coding genes (H2A and H3), 2 nuclear ribosomal genes (18S and 28S) and 15 mitochondrial genes (2 ribosomal and 13 protein coding genes). The data were assembled into three distinct matrices including (a) all 19 genes, (b) 15 protein coding genes as amino acids and (c) 15 protein coding genes as codons. Phylogenetic reconstruction was performed for the three concatenated matrices in a maximum likelihood framework in IQ-TREE (ver. 2.1.3, see http://www.iqtree.org/; Minh et al. 2020) followed by a minimum of 1000 rapid bootstrap pseudoreplicates. Model selection was performed in ModelFinder (Kalyaanamoorthy et al. 2017) as a module in IQ-TREE (ver. 2.1.3) allowing for free rate models (R). Model choice was also expanded to include free-rate models (i.e. LG4x; Le et al. 2012) in ModelFinder (Kalyaanamoorthy et al. 2017). Gene partitions were merged into metapartitions by ModelFinder as an IQ-TREE module in selected analyses. To search the tree space more thoroughly, the following parameters were set to extend the tree search phase: –ntop 100, –allnni, –nbest 100 and –nstop 1000 –bnni. In addition to bootstrap support, we quantified genealogical concordance by calculating gene concordance factors (GCF) and site concordance factors (SCF) in IQ-TREE (ver. 2.1.3). These are represented in Supplementary Table S6 as gCF_N that indicates the number of gene trees concordant with the branch; and sCF, corresponding to the proportion of informative sites that are concordant with that branch. To calculate gene concordance factors, gene trees were inferred through IQ-TREE with model choice in ModelFinder. Mitochondrial genes were treated separately as gene trees were conflicting due to saturation and to correlate with the partitioning schemes. Although the mitochondrial genes do not recombine and are therefore not independent, gCF as displayed here is a useful correlate to estimation error. Matrices were concatenated with AMAS where necessary (Borowiec 2016).
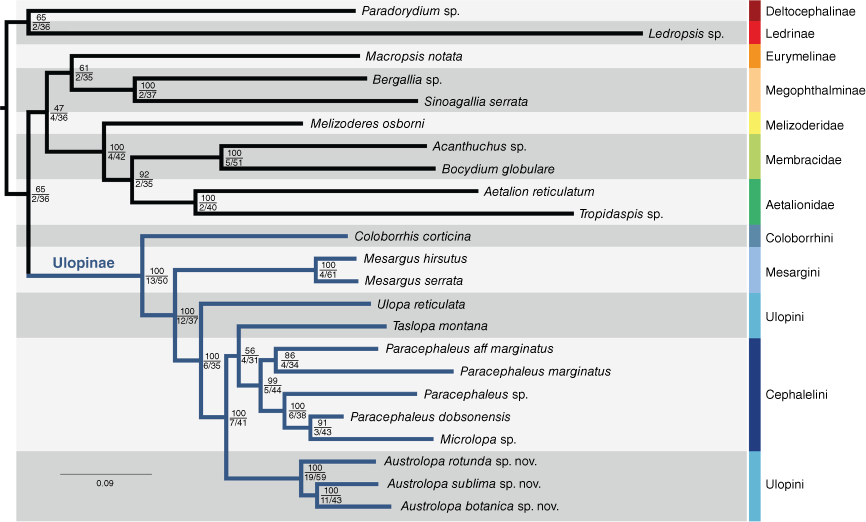
Three matrices analysed were (A) all 19 genes, nucleotides, (B) 15 protein coding genes, translated to amino acids and (C) 15 protein coding genes, analysed under codon aware models. Seven phylogenetic analyses were performed to test the sensitivity of the results to analytical perturbation. The analyses were devised as follows: (1) all genes in a single partition, (2) all genes, partitioned per gene, (3) all genes, partitioned by gene, with partition merging, (4) all genes, protein-coding genes partitioned into codons per gene plus ribosomal genes, (5) as in 4, with partition merging, (6) protein-coding genes only, analysed as amino acids and (7) protein-coding genes only, analysed with codon aware models. Matrix A was used for analyses 1–5; Matrix B was used for analysis 6; and Matrix C was used for analysis 7. Trees were visualised in FigTree (ver. 1.4.4, A. Rambaut, see http://tree.bio.ed.ac.uk/software/figtree/). Fig. 1 was prepared in Adobe Illustrator, rooted a posteriori to Deltocephaline + Ledrinae that are distantly related to Eurymelinae, Megophthalminae, Ulopinae and treehoppers (Dietrich et al. 2017; Skinner et al. 2019).
Maximum likelihood consensus tree inferred from four nuclear (18S, 28S, H2A and H3) and 15 mitochondrial genes, sampled from 23 membracoid taxa. Topologies were estimated in IQ-TREE (ver. 2.1.3) based on a dataset partitioned by gene, using the partition merging option (Matrix A, Analysis 3). Branch support statistics are represented by bootstrap values after 1000 rapid bootstrap pseudoreplicates (top), gene concordance factor out of 19 (gCF_N, bottom left) and site concordance factors (sCF, bottom right). Scale bar represents substitutions per site. Log-likelihood of consensus tree: −156361.937.
Results
The merged assemblies using outputs from Trinity, Tadpole and SpaDES resulted in three matrices with high gene coverage. These included 12 312 and 19 989 nucleotide positions (Matrices A and B respectively) and 4104 amino acid sites (Matrix C). All taxa failed the Chi-Square test for homogeneity of base composition implemented in IQ-TREE (ver. 2.1.3; Schmidt et al. 2002, Minh et al. 2020) except Aetalion reticulatum, Austrolopa sublima sp. nov. and Paracephaleus marginatus. Three taxa were missing more than 50% of the aligned data: Bocydium globulare, Mesargus hirsutus and Paracephaleus dobsonensis. Detailed information on assembled matrices, including gene sampling, parsimony informative sites, GC content and data completeness is provided in the Supplementary Table S4. Specifications of each partition and performance statistics for the seven analytical approaches are listed in Supplementary Table S5. These have yielded well supported relationships within Ulopinae and between Ulopinae and other lineages. Our findings agree with previous hypotheses that three treehopper families are a monophyletic lineage arising from within Cicadellidae (Dietrich et al. 2017; Skinner et al. 2019; Hu et al. 2023). Larger phylogenomic datasets provide strong support for a paraphyletic Cicadellidae with respect to the treehoppers, with Megophthalminae + Ulopinae as sister relatives. Our sensitivity analyses show a different combination of relationships, represented by Megophthalminae as sister to treehoppers in three topologies and Ulopinae as sister to treehoppers in two topologies (Supplementary Table S6). Fig. 1 shows a polyphyletic Cicadellidae, with Ulopinae placed as sister to a clade including a monophyletic Eurymelinae + Megophthalminae as the closest relatives of treehoppers, a phylogenetic arrangement recovered in five out of seven topologies (Supplementary Table S6). The topology in Fig. 1 includes all sampled genes and illustrates the relationships most consistently recovered across the tested parameter space. Taslopa as sister to the Paracephaleus group is a hypothesis with very low support, and 4 of the 19 genes and 31% of the sites were in concordance (Fig. 1). In analyses 5–7, involving codon models (7), amino acids (6) or allowing codons from different genes to merge into metapartitions (5), Austrolopa was sister to the Paracephaleus group instead, though only 2 of the 19 genes were in concordance with the position of Taslopa. Topologies representing the other six phylogenetic analyses are shown in Supplementary Fig. S1–S6.
Our results provide compelling evidence for the monophyly of Ulopinae, with Coloborrhinini and Mesargini as successive sister groups to the remaining Ulopinae and a monophyletic Ulopini + Cephalelini. These two tribes, however, were recovered as non-monophyletic with respect to each other. Cephalelini was rendered non-monophyletic as Microlopa was recovered as sister to an undescribed species of Paracephaleus. Within Ulopinae, the topologies differ regarding the placement of a single taxon, Taslopa montana, either placed as sister to Microlopa + ‘Cephalelini’ (Fig. 1) or as sister to the remaining Australian Ulopini + Cephalelini (Supplementary Fig. S1). Austrolopa was strongly recovered as monophyletic in all analyses. Representatives of this genus can be easily recognised based on external characters of the head and forewings. Constituent species also show a well-marked variation in features of the head and male genitalia that allow accurate delimitation and recognition (discussed below). The three Austrolopa taxa sampled showed deep sequence divergences, demonstrating that these are representative of distinct, novel species-level units.
Taxonomy
Austrolopa Evans, 1937
Austrolopa brunensis Evans, 1937 by original designation.
Austrolopa differs from the other ulopine genera in having a crown distinctly flat, anterior margin arched or somewhat acute in dorsal view; two ocelli, located dorsally on crown; transition to face carinate, often strongly laminate; face long and broad, featuring a median longitudinal carina apically; frontoclypeus enlarged, spatulate and elongate, completely concealing anteclypeus in frontal view; hindwings present or absent.
Small, slender, cryptically coloured leafhoppers, yellow, tan, dark brown or a combination thereof; head, thorax and forewings coarsely and densely punctured. Head with crown flat anteriorly, anterior margin arched or somewhat acute in dorsal view; eyes prominent, ovoid to slightly transverse; two ocelli present, located dorsally on crown; surface of crown slightly concave between ocelli and between ocelli and eyes; transition to face sharp, distinctly carinate; face long, broad, nearly flat, forming ~45° angle with crown in lateral view; coronal suture longitudinally grooved on crown, longitudinally carinate on vertex, most distinctly keeled between anterior margin and ecdysial line, delimiting a longitudinal depression on both sides, from vertex to posterior margin of frontoclypeus; ecdysial line well below transverse carina of crown, shape variable; supra-antennal ledges transverse, nearly perpendicular to ecdysial line, bent apically; frontoclypeus spatulate, broad and elongate, completely concealing anteclypeus in frontal view, projected beyond posterior margin of head, outline of posterior margin variable. Thorax with pronotum flat or humped in profile, rectangular in dorsal view, anterior and posterior margins subparallel. Forewings fully developed, elytra-like, featuring three anteapical cells; one vein s, two r-m and two m-cu present; venation often reticulate, additional crossveins usually in claval area, between CuP and Pcu; membrane densely punctured throughout (except in A. victoriensis); apex round or acute; appendix absent. Hindwings present or absent; when present: RA2 indistinct, MA fused with MP, A1 and Pcu fused basally and diverging preapically; one r-m, one m-cu and jugal lobe present. Abdomen finely punctured, smooth, lacking projections or tubercles. Male pygofer with slender, laminate subgenital plate, lateral plate somewhat triangular in lateral view, partially enclosed dorsally; parameres well developed, shank elongate, apically hooked; aedeagus apically bifid, each branch further divided into two slender projections.
The most important diagnostic feature of Austrolopa is the elongate and spatulate frontoclypeus that is projected into a ledge that entirely conceals the anteclypeus in frontal view. Forewings are elytra-like and hindwings may be present or absent, a trait that does not seem to be subject to intraspecific variation. These leafhoppers are not distinctly sexually dimorphic, except for a typical difference in size (males smaller than females). An examination of the Evans collection at the Australian Museum revealed several curatorial issues including incorrect labelling of primary types that warrants a lectotype designation for A. kingensis.
Tasmania, Australian Capital Territory, New South Wales, Northern Territory, Queensland, Victoria, Western Australia (Fletcher 2012). In this study, representatives from the first three states were examined.
Bossiaea (Fabaceae), Cassinia (Asteraceae), Hakea (Proteaceae), Mirbelia (Fabaceae), Melaleuca (Myrtaceae), Podocarpus (Podocarpaceae), Pultenaea (Fabaceae), Richea (Epacridaceae) and Trymalium (Rhamnaceae) (Moir et al. 2003; Fletcher 2009, 2012).
Anacephaleus minutus Evans, 1937: holotype ♀ (coll.: D.C. Swan), Perth, Western Australia (AM, K290269); Anacephaleus punctatus Evans, 1937 (=Notocephalius hartmeyeri Jacobi, 1909): holotype ♀ Kiata, Victoria, F.E. Wilson col. (AM, K290270); Anacephaleus simplex Evans, 1937 (=Alocephalus ianthe (Kirkaldy, 1906): holotype ♀, Mordialloc, Victoria (NMV, T-14423); Anacephaleus ulopae Evans, 1937, holotype ♀Adelaide, South Australia, A.M. Lea col. (SAMA, 20-017509); Microlopa minuta Evans, 1966, holotype ♂ Waldheim HS, Cradle Mountain, Tasmania, ii.1964, J.W. Evans col. (AM, K290274); Uloprora risdonensis Evans, 1939, Holotype ♀ Risdon, Tasmania, V.V. Hickman col. (AM, K290279); Woodella wanungarae Evans, 1966 (=Doowella wanungarae (Evans, 1966)): holotype ♂ Mt Wanungara, McPherson Ranges, Queensland, 1958, T. Woodward coll. [in moss] (QM). Additional Ulopinae featured in the phylogenetic analyses are listed in Supplementary Table S1. Specimens of Austrolopa are listed in the examined material of the corresponding species.
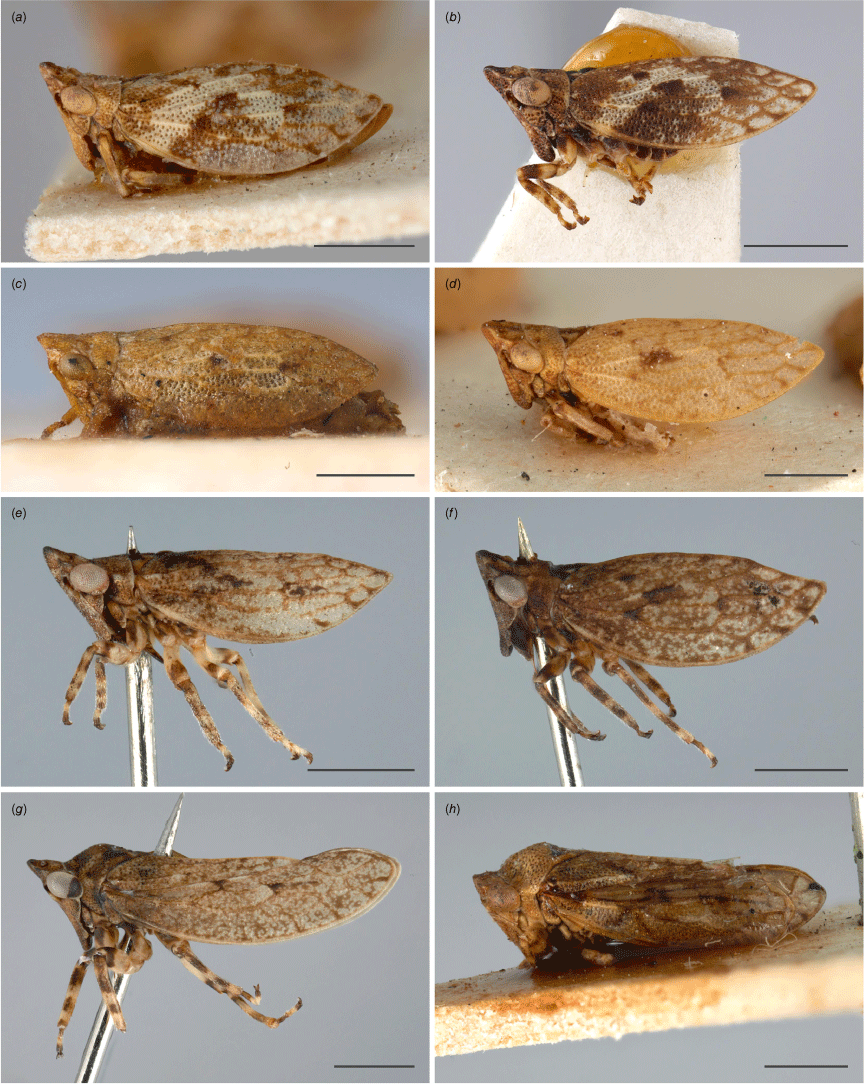
| 1 | Pronotum humped dorsally; hindwings present (Fig. 2g, h) ...2 Pronotum flat dorsally; hindwings absent (Fig. 2a–f) ...3 |
| 2 | Crown narrow, approximately as long as eyes, evenly long throughout (Fig. 3f); forewing apex membranous, amber hyaline (Fig. 2h) ...Austrolopa victoriensis Crown projected forward, somewhat acute and longest in middle (Fig. 3e); forewings coriaceous throughout, spotted dark brown (Fig. 2g) ...Austrolopa sublima, sp. nov. |
| 3 | Anterior margin of vertex broadly arched in anteroventral view; posterior margin of frontoclypeus round to slightly acute, longest in middle (Fig. 4a, c) ...4 Anterior margin of vertex irregularly arched, slightly angled, nearly flat in middle; posterior margin of frontoclypeus somewhat truncate (Fig. 4b, d) ...5 |
| 4 | Anterior margin of vertex and ecdysial line broadly arched in anteroventral view (Fig. 4a); forewings opaque, entirely pigmented yellow and brown, in lateral view: broadly arched in repose, curved toward acute apex (Fig. 2a, b, 3a) ...Austrolopa brunensis Anterior margin of vertex and ecdysial line strongly curved upward towards middle (Fig. 4c); forewings slightly translucent, sparsely pigmented apically; in lateral view feebly arched in repose, nearly flat until near apex (Fig. 2e, 3c) ...Austrolopa botanica, sp. nov. |
| 5 | Body, head and forewings tan to pale yellow (Fig. 2c, d); head weakly produced, vertex very narrow at middle: distance between anterior margin and ecdysial line shorter than width of eyes in anteroventral view (Fig. 4b) ...Austrolopa kingensis, sp. reval. Body, head and forewings dark (Fig. 2f); head produced, vertex broad at middle: distance between anterior margin and ecdysial line distinctly higher than width of eyes in anteroventral view (Fig. 4d) ...Austrolopa rotunda, sp. nov. |
Austrolopa brunensis Evans, 1937
(Fig. 2a, b, 3a, 4a, 5a, 6a–e.)
Austrolopa brunensis Evans, 1937, p. 48.
Austrolopa tasmaniensis Evans, 1947, p. 140.
Austrolpa brunneus Evans, 1951, p. 5 [sic].
Type locality: Bruny Island, Tasmania.
Female (mm). Body length: 3.74; forewing length: 2.75; head width: 1.29; crown width: 0.93; crown length: 0.44; face length: 1.23.
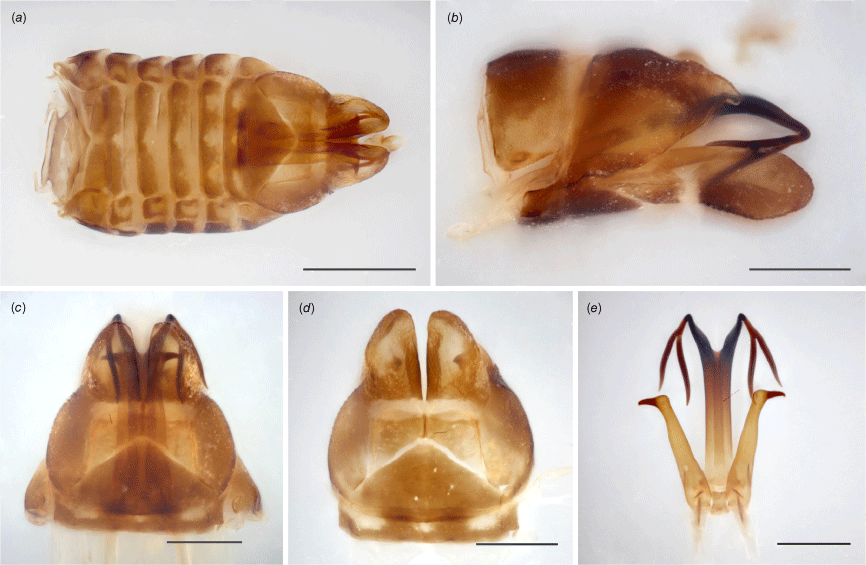
Tan to brown, mottled yellow (preserved specimens); head, pronotum and scutellum predominantly light to dark brown; forewings blotted brown and yellow with few dark brown spots, entirely pigmented and opaque, membrane densely and coarsely punctured throughout; legs stripped yellow and brown. Head distinctly laminate, crown projected well beyond eyes, anterior margin converging to a point at coronal suture, where crown is longest; in anteroventral view, vertex broad, anterior margin broadly arched, ecdysial line nearly semicircular, frontoclypeus robust and strongly convex, posterior margin somewhat acute. Pronotum flat in dorsal view, margins subparallel. Forewings arched dorsally, regularly curved toward acute apex; venation reticulate on clavus, showing additional transverse veins between CuP, Pcu and A1. Hindwings absent. Male pygofer with subgenital plate scoop-like, slender in ventral view, strongly constricted at middle, expanded into round apical lobe in lateral view; lateral plate somewhat triangular, with small dorsal finger-like projection, slightly curved downwards in lateral view, plate partially enclosed dorsally, pair of tooth-like tubercles, each tooth located on internal side of plate, visible in ventral view, curved towards each other; parameres elongate, shank cylindrical, slightly tapering toward apical hook, abruptly bent and spine-like at the tip; aedeagus tubular, shaft slender, projected upward, apically bifid, each segment further divided into two thin branches, the one closer to aedeagus slightly longer.
Specimens currently identified as Austrolopa brunensis represent a wide geographic range, encompassing nearly all states in Australia (Fletcher 2012). Considerable variation in colour was reported from various localities in New South Wales and Western Australia (Fletcher 2012; Moir et al. 2003). These observations indicate the need for careful revision of Austrolopa in Australia to determine whether this genus includes new species with fewer or specific host plant associations. In the interim, we restrict our concept of the species to include representatives from Tasmania that closely match Evans’ original description (Evans 1937). Male genitalic features appear to be extremely useful for species-level identification. Austrolopa tasmaniensis Evans, 1947 (p. 140) is kept in synonymy until the primary type can be verified.
Evans (1937) used the word ‘type’ in the singular to refer to the male name-bearing specimen of Austrolopa brunensis. Illustrations do not feature the actual specimen, only structures of the male genitalia (paramere, aedeagus and subgenital plate; Plate XV, fig. 16a–c). The type locality, repository and collector name are mentioned in the original description (‘Australian Museum, Sydney’; ‘Bruni Island’ [sic]; ‘Coll. A.M. Lea’). The type is currently missing, however, two female specimens matching Evans’ (1937) description were located at the Australian Museum. Collection labels list the same provenance of the type (‘Bruni [sic] I. \ Tas: Lea’) and a species identification label features the author’s handwriting. A card containing a male genitalic dissection is attached to a third female specimen (K594499) identified by Evans as Austrolopa brunensis. The genitalic structures are very similar to those in the original description, and the preservation technique is identical to other dissections appended to specimens that Evans identified and described. The only remnant of the primary type is possibly the male genitalia and this was likely incorrectly appended by Evans – or subsequently by someone else – to a conspecific specimen from a different collecting series. The identification of Austrolopa brunensis should currently be based on the original description, with attention to the female specimens with closely matching labels.
AUSTRALIA: Tasmania: Bruny Island, ‘Bruni [sic] I. \ Tas: Lea’; ‘Austrolopa \ brunensis Ev \ J. W. Evans det.’ (two female specimens on the same pin, K290272); Lake St Clair National Park (one female, K594423); Arthur Road? [label illegible] (one female, K594499; specimen intact, male genitalia on the same pin); Risdon, low shrubs (one male, K594479; one female, K594422); Hobart (one female, K594482) . All specimens at the AM.
Austrolopa species in lateral view. (a) Austrolopa brunensis Evans 1937, female (K290272); (b) Austrolopa brunensis, male (K594479); (c) Austrolopa kingensis Evans, 1939, lectotype male (K594426); (d) Austrolopa kingensis, female (incorrectly marked as a holotype, K290273); (e) Austrolopa botanica sp. nov., holotype female (ANIC 20-006240); (f) Austrolopa rotunda sp. nov., holotype female (ANIC 20-011403); (g) Austrolopa sublima sp. nov., holotype male (ANIC 20-006241); (h) Austrolopa victoriensis Evans, 1939.
Austrolopa species in dorsal view. (a) Austrolopa brunensis Evans 1937, female (K290272); (b) Austrolopa kingensis Evans, 1939, lectotype male (K594426); (c) Austrolopa botanica sp. nov., holotype female (ANIC 20-006240); (d) Austrolopa rotunda sp. nov., holotype female (ANIC 20-011403); (e) Austrolopa sublima sp. nov., holotype male (ANIC 20-006241); (f) Austrolopa victoriensis Evans, 1939, holotype female.

Austrolopa species, view of face from anteroventral view. (a) Austrolopa brunensis Evans 1937, female (K290272); (b) Austrolopa kingensis Evans, 1939, lectotype male (K594426); (c) Austrolopa botanica sp. nov., holotype female (ANIC 20-006240); (d) Austrolopa rotunda sp. nov., holotype female (ANIC 20-011403); (e) Austrolopa sublima sp. nov., holotype male (ANIC 20-006241); (f) Austrolopa victoriensis Evans, 1939, holotype female.
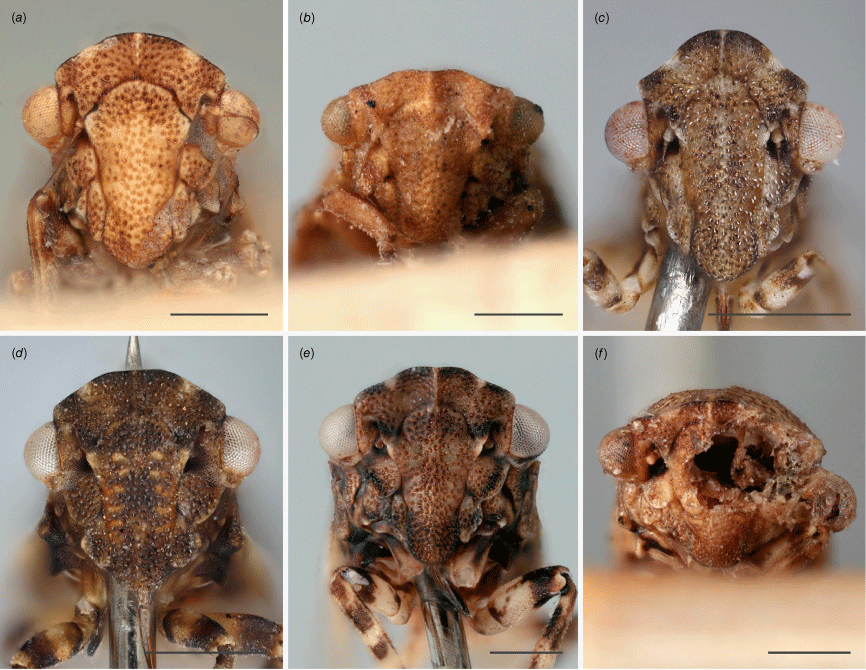
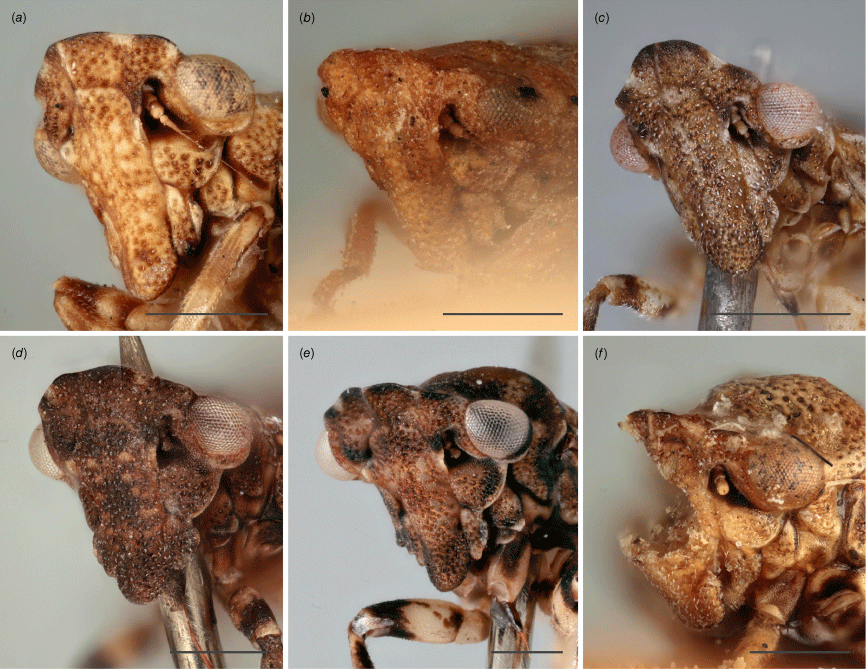
Austrolopa species, view of face from laterofrontal view. (a) Austrolopa brunensis Evans 1937, female (K290272)' (b) Austrolopa kingensis Evans, 1939, lectotype male (K594426); (c) Austrolopa botanica sp. nov., holotype female (ANIC 20-006240); (d) Austrolopa rotunda sp. nov., holotype female (ANIC 20-011403); (e) Austrolopa sublima sp. nov., holotype male (ANIC 20-006241); (f) Austrolopa victoriensis Evans, 1939, holotype female.
Austrolopa kingensis Evans, 1937, sp. reval.
(Fig. 2c, d, 3b, 4b, 5b.)
Austrolopa kingensis Evans, 1937, p. 48.
Type locality: King Island, Tasmania, Australia.
Male lectotype/female (mm). Body length: 4.4/4.6; forewing length: 3.19/3.41; head width: 1.34/1.37; crown width: 0.99/1.45; crown length: 0.41/0.46; face length: 0.96/0.98 [estimated due to placement of card].
Golden yellow to tan (preserved specimens); head, pronotum and scutellum predominantly yellow with discreet brown stripes or spots; forewings with pale yellow patches and dark brown spots, entirely pigmented and opaque, membrane densely and coarsely punctured throughout; legs mostly yellow. Head moderately laminate, crown projected well beyond eyes, anterior margin converging to a point at coronal suture, where crown is longest; in anteroventral view, vertex narrow, anterior margin angled, nearly flat in middle, ecdysial line forming a wide semi-ellipsis, frontoclypeus robust and distinctly depressed at sides, posterior margin truncate. Pronotum flat in dorsal view, margins subparallel. Forewings feebly arched dorsally, regularly curved toward acute apex; venation weakly reticulate on clavus. Hindwings absent.
The species is named after the type locality.
Lectotype designation for Austrolopa kingensis Evans, 1937, p. 48.
Evans (1937) used the word ‘type’ in the singular to refer to the male name-bearing specimen of Austrolopa kingensis. Illustrations from the original description include the dorsal habitus of an adult specimen, a hind leg (presumably), a forewing and the male aedeagus (Plate XV, fig. 15a–d). The type locality and repository are mentioned (‘Australian Museum, Sydney’ and ‘King Island’) but no other details are provided (e.g. date or collector). Four specimens matching Evans’ (1937) description that we consider conspecific were in the Australian Museum. Collection labels list the same provenance (‘King I. \ Tas: Lea’) and a species identification label features the author’s handwriting. The specimens are mounted on two cards, one containing one male and one female, and the other containing two females that are marked as types. The type labels do not specifically state the nominal taxon associated with the designation. An older handwritten label on copy paper reads ‘type’ and a second label reads ‘holotype’ in typesetting. These seem to have been appended to the specimens at different points in time and not necessarily by the author. Although the word ‘type’ used to refer to a single specimen implies a holotype designation, consistent curatorial issues recently observed in the Evans collection have undermined our confidence that the type series was accurately depicted in this instance. Among these four eligible name-bearing types, there is a male that closely matches the description of Austrolopa kingensis. The aedeagus, a small tubular structure lacking any projections is partially visible in this specimen, presumably allowing an illustration to have been produced. This is potentially a strong distinguishing feature of this species but needs to be confirmed when additional specimens are available for dissection. The lectotype herein designated (illustrated in Fig. 2c, 3c, 4c and 5c) is glued to the left side of the card adjacent to a female that is not considered part of the type series. This is also the case with the other two female specimens that are incorrectly marked as types; one of these females is also illustrated (Fig. 2d).
AUSTRALIA: Tasmania: King Island, ‘King I. \ Tas: Lea’; ‘Austrolopa kingensis \ EV \ J. W. Evans det.’; ‘Australian Museum \ K594426’; ‘ASCT HE019289’ (two specimens on the same pin: lectotype male, here designated, on left side of card; female on right side of card, not considered part of the type series; K594426); two females on the same card with same collection data as lectotype, incorrectly marked as types (additional labels: ‘[handwritten] Type’, ‘[red label] HOLOTYPE’; K290273). All specimens are in the AM.
Austrolopa botanica Evangelista, sp. nov.
ZooBank: urn:lsid:zoobank.org:act:754F83A5-098E-441C-87E8-8287Aj3A96D75
Type locality: Canberra, Australian Capital Territory, Australia.
Female holotype (mm). Body length: 3.63; forewing length: 2.64; head width: 1.26; crown width: 0.90; crown length: 0.49; face length: 1.26.
Light brown to greyish-yellow (preserved specimen); head, pronotum and scutellum predominantly brown, crown anterior margin with two yellow patches located near each ocelli, scutellum with two yellow spots on sides; forewings with dark brown patches more concentrated basally, veins light brown, membrane greyish-yellow, partially translucent, densely and coarsely punctured throughout except for small area near apex; legs stripped brown and yellow. Head strongly laminate, crown broad, projected well beyond eyes, anterior margin converging into acute angle at coronal suture, where crown is longest; in anteroventral view, vertex broad, ecdysial line strongly pointed upwards, frontoclypeus robust, strongly convex, posterior margin somewhat acute. Pronotum flat in dorsal view, margins subparallel. Forewings nearly straight dorsally, curved slightly near tip toward acute apex; venation distinctly reticulate on clavus. Hindwings absent. Male genitalia unknown.
DNA sequences are available for holotype that is included in the phylogenetic analysis presented in this study (voucher MEM698; NCBI BioSample Accession: SAMN37932475).
Austrolopa rotunda Evangelista, sp. nov.
ZooBank: urn:lsid:zoobank.org:act:1D0368EF-34AE-4AA1-81C5-6214CA6ADFA0
Type locality: Namadgi National Park, Australian Capital Territory, Australia.
Female holotype (mm). Body length: 4.18; forewing length: 3.3; head width: 1.43; crown width: 1.04; crown length: 0.44; face length: 1.34.
Dark brown with slight red tinge (preserved specimen); crown and scutellum darker, nearly black, with pale yellow spots, vertex spotted yellow, especially on frontoclypeus; pronotum brown; forewings mostly dark brown spotted greyish-yellow, densely and coarsely punctured throughout; legs stripped yellow and brown, more predominantly brown. Head strongly laminate, crown broad, projected well beyond eyes, anterior margin converging into acute angle at coronal suture, where crown is longest; in anteroventral view, vertex broad, ecdysial line semicircular, frontoclypeus robust, somewhat flat, slightly tapering toward posterior margin, somewhat truncate. Pronotum flat in dorsal view, margins subparallel. Forewings arched dorsally, slightly angled near tip, apex acute, discreetly pointed upward; venation distinctly reticulate on clavus. Hindwings absent. Male genitalia unknown.
Austrolopa sublima Evangelista, sp. nov.
ZooBank: urn:lsid:zoobank.org:act:B7FA5BD0-904F-4DFB-B58B-FA9313FD9F8F
Type locality: Kosciuszko National Park, New South Wales, Australia.
Male holotype (mm). Body length: 5.06; forewing length: 4.29; head width: 1.73; crown width: 1.21; crown length: 0.49; face length: 1.43.
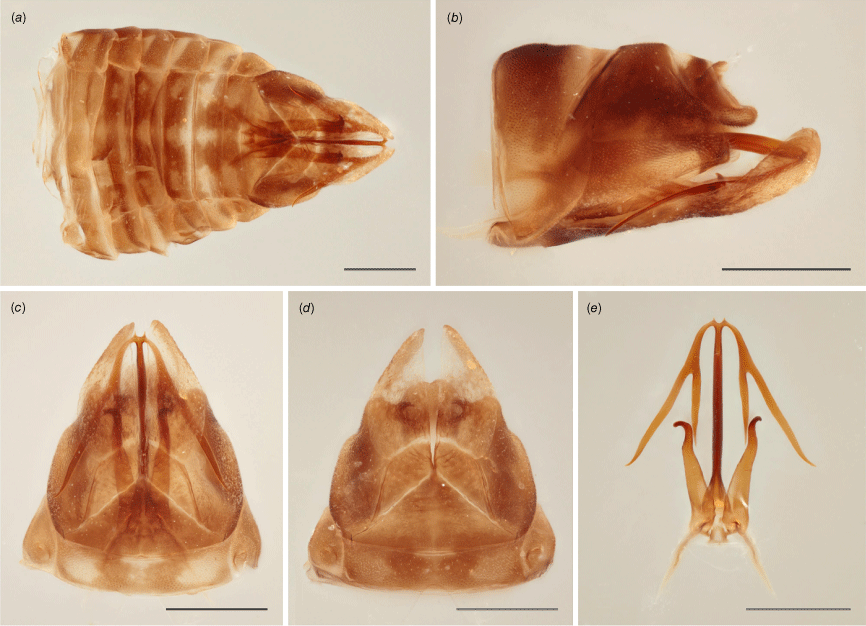
Brown with tinge of yellow; crown with three longitudinal yellow lines, one along coronal suture and the other two on each side adjacent to ocelli; pronotum and scutellum with slightly darker areas and pale yellow spots; in anteroventral view, vertex darker anteriorly with two yellow spots, irregular transverse dark band near ecdysial line and at apex of frontoclypeus; forewings mottled brown and greyish-yellow, densely and coarsely punctate throughout; legs stripped yellow and brown. Head strongly laminate, crown broad, projected well beyond eyes, anterior margin converging into round point at coronal suture, where crown is longest; in anteroventral view, vertex broad, ecdysial line semi-circular, frontoclypeus robust, sightly depressed at sides and tapering toward posterior margin, feebly round. Pronotum humped in dorsal view, anterior margin round. Forewings nearly straight dorsally, gradually curved toward round apex; venation weakly reticulate on clavus. Hindwings present (see description of genus). Male pygofer with subgenital plate scoop-like, slender in ventral view, widest medially, tapering toward apex; lateral plate somewhat triangular with no visible projection in lateral view, plate partially enclosed dorsally forming a pair of tooth-like tubercles, each located on internal side of plate, visible in ventral view, curved towards each other; parameres elongate, shank cylindrical, slightly tapering toward apical hook, curved and round at the tip; aedeagus tubular, shaft slender, projected upward, apically bifid, each segment further divided into two branches, nearly as thick as shaft, the one closer to aedeagus distinctly shorter.
The holotype is a large specimen with functional hindwings and distinctive male genitalia (Fig. 7a–e). DNA sequences are available for the holotype that is included in the phylogenetic analysis presented in this study (voucher MEM695, NCBI BioSample Accession: SAMN37932477).
The name is derived from the Latin ‘sublimis’, in the sense of lofty and high, referring to the type locality close to the highest elevation in Australia.
Austrolopa victoriensis Evans, 1939
Austrolopa victoriensis Evans, 1939, p. 44.
Type locality: Warburton, Victoria, Australia.
Body length: 4.24; forewing length: 3.43; head width: 1.51; crown width: 1.05; crown length: 0.3; face length: 2.01 [estimated due to placement of card].
Golden yellow to light brown (preserved specimen); head, pronotum and scutellum predominantly brown, crown slightly darker along coronal suture, scutellum nearly black on edges, forming a large semicircle in middle; forewings with dark brown patches, part of venation darker, membrane amber hyaline, partially translucent, densely and coarsely punctured along coastal margin and clavus, mid longitudinal section and apex membranous; legs presumably yellow. Head weakly laminate, crown evenly long throughout, projected for very short distance beyond eyes, anterior margin broadly arched; in anteroventral view, vertex narrow, approximately half as long in middle, ecdysial line semi-circular, frontoclypeus robust, strongly convex, posterior margin truncate. Pronotum distinctly humped in dorsal view, anterior margin curved. Forewings nearly straight dorsally, curved abruptly toward round apex; venation weakly reticulate on clavus. Hindwings present. Male genitalia unknown.
Evans (1939) diagnosed Austrolopa victoriensis based on the shape of the crown, narrow and evenly long throughout. Although the species is currently known only from the female holotype, the appearance is distinctive and there is a unique combination of features such as a humped pronotum in profile, partially membranous forewings and the presence of hindwings. Venation of hindwings could not be examined because the holotype is attached to a card and covered in glue. The head of the holotype is damaged (Fig. 4f, 5f) but images of a previously acquired intact specimen (Fig. 2h, 3f) illustrate the diagnostic features presented here.
Discussion
The unique appearance of Ulopinae leahoppers underlies the increasing phylogenetic relevance, as these are key to understanding the paraphyly of leafhoppers with respect to treehoppers. Clarifying the phylogenetic placement in relation to other subfamilies will illuminate the evolution of interesting morphological and behavioural traits in this major evolutionary transition. Our phylogenetic matrix includes the largest sampling of ulopine leafhoppers to date, with representatives of four of the five currently recognised tribes. At subfamily level, the resulting topologies provide strong support for the monophyly of Ulopinae and a monophyletic Ulopini + Cephalelini; however, these two tribes are not monophyletic with respect to each other. This study also demonstrates that the current diversity of Australian Ulopinae is underestimated, and cryptic species of Austrolopa can be distinguished based on morphological characters and genetic divergence.
Position of Ulopinae with respect to other membracoid taxa
Contextualising a greater diversity of Ulopinae also provides a structure for additional insight concerning the position within Cicadomorpha. Although multiple studies have posited that treehoppers are derived from within Cicadellidae, no classification changes have been accepted due to the lack of clarity regarding the number of major lineages of Cicadellidae and which of those are closest to treehoppers. Recent phylogenomic studies (Dietrich et al. 2017; Skinner et al. 2019; Hu et al. 2023) that included fewer Ulopinae but a broader sampling of cicadellid subfamilies have largely agreed that Ulopinae is closely related to treehoppers. However, conflict persists regarding the placement of Eurymelinae (sensu lato) and Megophthalminae. For example, Hu et al. (2023) recovered a clade consisting of Megophthalminae and Ulopinae as sister to the treehoppers and suggested that Eurymelinae was not monophyletic, whereas Eurymelinae is recovered monophyletic in Dietrich et al. (2017). Most parameter regimes in our study find Ulopinae sister to a clade containing Eurymelinae, Megophthalminae and treehoppers. Ulopinae is recovered as a sister to treehoppers when ribosomal genes are excluded (Analyses 6 and 7), however these deeper nodes have received low branch support. A c odon model framework within the treehoppers recovers Melizoderidae inside Membracidae with low support (Analysis 7), as Hu et al. (2023) observed from the phylogenomic analysis of transcriptome data.
Evolution of Australian Ulopinae
Morphological evidence provides additional support for the monophyly of Ulopinae in agreement with the present analyses. The mesanepisternum with a horizontal keel and distinct cleft separating the maxillary plate and gena are distinctive features not otherwise present in Cicadellidae. Diagnostic characters previously attributed to Ulopini by Emeljanov (1996) are also present in Cephalelini, including the high incidence of hindwing absence and the convex, ovoid forewing shape. Although a comprehensive treatment is still lacking, these shared features provide support for a monophyletic Cephalelini + Ulopini, pending a future phylogenetic reassessment to ascertain the placement of the constituent genera. The ulopine tribes Mesargini, Coloborrhini and Monteithiini are smaller groups that are relatively well characterised based on aspects of the head, wing and male genitalia (Evans 1968; Emeljanov 1996). However, unusual new taxa featuring characters of both Mesargini and Coloborrhini await tribal assignment and may necessitate further clarification of these tribes (Dai et al. 2012).
We convincingly recover an Australian clade of Ulopinae and amonophyletic Austrolopa. The gCF_N of 2 for the position of Taslopa sister to Austrolopa + Paracephaleus, compared to the low sCF (31) for Taslopa as sister to Paracephaleus are both low enough to be essentially indistinguishable from random. Therefore, with the current sampling the Australian endemic Ulopinae is best considered a polyphyletic group. This could be resolved with additional taxa and by incorporating more nuclear protein-coding genes into future analyses.
Paracephaleus may be non-monophyletic with respect to other genera but additional genera of Cephalelini and Ulopini should be studied before proposing any changes. The terminal representing Paracephaleus dobsonensis has low gene coverage and missing data can induce a bias in our results. Microlopa does not have a strongly projected head but otherwise displays morphological congruencies with Cephalelini. As the name suggests, slender Cephalelini have remarkably elongated heads that combined with the cryptic colours renders these outstanding mimics of grass seeds. These insects have strong host plant associations, feeding almost exclusively on members of the Restionaceae. South African representatives of Cephalelini were included in Dietrich et al. (2017) and Cao et al. (2022) but ribosomal and mitochondrial genes could not be mined from those terminals. Our study suggests that Cephalelini may not be monophyletic, Microlopa may be misplaced and Paracephaleus as currently delimited may include more than one genus-level lineage.
Taxonomy of Australian Ulopinae
Austrolopa can be easily recognised based on a combination of features of the head, forewing and male genitalia. A compelling putative synapomorphy for Austrolopa is the elongate, spatulate frontoclypeus that entirely conceals the anteclypeus in frontal view, a unique feature in Ulopinae. Austrolopa was previously known from two species: A. brunensis, originally described from Bruny Island in Tasmania but reportedly distributed across Australia and A. victoriensis from the state of Victoria that is known only from the female holotype. Geographic distribution and host plant associations are reported from focal surveys and institutional databases, and most records are derived from Moir et al. (2003) and Fletcher (2012). These sources list a wide range of host associations in Austrolopa in Western Australia and New South Wales, and noted substantial morphological variation in geographic range and population level differences in colour associated with certain hosts.
Hindwings are absent in all known Austrolopa species except A. victoriensis and A. sublima sp. nov., both known only from the holotype specimens. Forewings are used primarily to protect the body, therefore species with short hind legs and no hindwings have reduced dispersal capacity. Evans (1939) posited that differences in the head and thorax structure between A. brunensis and A. victoriensis could be due to muscle reduction because of the diminished ability of A. brunensis to jump and fly. Based on the genomic and morphological diversity of species treated herein, we demonstrate that populations previously ascribed to A. brunensis represent new species. Our findings strongly suggest that species of Austrolopa have narrow ranges and are perhaps further restricted to certain elevations or host plants. For example, the three new species of Austrolopa are described from adjacent mountain ranges in a small area of South-eastern New South Wales. Austrolopa brunensis and A. kingensis were described from small Tasmanian islands. Our study provides evidence that A. brunensis is an assemblage of different species instead of representing one widely distributed polyphagous leafhopper. Host records are reported at the genus level until the species-level diversity in Austrolopa is further clarified.
Our findings illuminate the evolution of worldwide Ulopinae and establish a foundation for understanding the phylogenetic relationships of the Australian groups. These also help illustrate that members of the Australian Ulopinae are far more diverse than currently circumscribed, with non-monophyletic tribes and genera needing reappraisal and potential for a deep restructuring of the classification. The lack of an adequate reference set with which to compare and validate new input data from the Australian fauna is a challenge that this study begins to address. Phylogenomic data generated help establish a foundation to galvanise Australian leafhopper systematics under a modern revisionary framework.
Supplementary material
Supplementary Figures and Tables are included with this submission and are available online.
Data availability
The data that support this study are available in the article and accompanying online supplementary material. Genomic assemblies were submitted to NCBI as SRA files and will be shared upon acceptance of the manuscript. Matrices used in the phylogenetic analyses will be publicly available through Dryad. The examined material is deposited in the Australian Museum, Australian National Insect Collection, Queensland Museum, South Australian Museum, Victoria Museum and Western Australian Museum.
Acknowledgements
The authors thank the following museum staff for facilitating access to relevant holdings, especially primary types: Derek Smith and Russel Cox (Australian Museum); Ben Parslow (South Australian Museum); Genefor Walker-Smith and Catriona McPhee (Victoria Museum); Susan Wright, Karin Koch and Geoff Thompson (Queensland Museum). We are indebted to our colleagues at the Australian Biological Resources Study, National Parks Australia and CSIRO who helped manage the contract and funds (Helen Cross, Jo Harding, Erika Cleaver and Suzanne Blankley).
References
Allio R, Schomaker-Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F (2020) MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Molecular Ecology Resources 20(4), 892-905.
| Crossref | Google Scholar | PubMed |
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19(5), 455-477.
| Crossref | Google Scholar | PubMed |
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-20.
| Crossref | Google Scholar | PubMed |
Borowiec ML (2016) AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ 4, e1660.
| Crossref | Google Scholar | PubMed |
Bushnell B, Rood J, Singer E (2017) BBMerge – accurate paired shotgun read merging via overlap. PLoS One 12(10), e0185056.
| Crossref | Google Scholar | PubMed |
Cai L, Zhang H, Davis CC (2022) PhyloHerb: a high-throughput phylogenomic pipeline for processing genome skimming data. Applications in Plant Sciences 10(3), e11475.
| Crossref | Google Scholar | PubMed |
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421.
| Crossref | Google Scholar | PubMed |
Cao Y, Dietrich CH, Zahniser JN, Dmitriev DA (2022) Dense sampling of taxa and characters improves phylogenetic resolution among deltocephaline leafhoppers (Hemiptera: Cicadellidae: Deltocephalinae). Systematic Entomology 47(3), 430-444.
| Crossref | Google Scholar |
Cryan JR, Urban JM (2012) Higher-level phylogeny of the insect order Hemiptera: is Auchenorrhyncha really paraphyletic? Systematic Entomology 37(1), 7-21.
| Crossref | Google Scholar |
Dai W, Viraktamath CA, Zhang Y (2012) Ulopsina, a remarkable new ulopine leafhopper genus from China. Journal of Insect Science 12, 70.
| Crossref | Google Scholar | PubMed |
Dietrich CH (2005) Keys to the families of Cicadomorpha and subfamilies and tribes of Cicadellidae (Hemiptera: Auchenorrhyncha). Florida Entomologist 88(4), 502-517.
| Crossref | Google Scholar |
Dietrich CH, Rakitov RA, Holmes JL, Black WC (2001) Phylogeny of the major lineages of Membracoidea (Insecta: Hemiptera: Cicadomorpha) based on 28S rDNA sequences. Molecular Phylogenetics and Evolution 18(2), 293-305.
| Crossref | Google Scholar | PubMed |
Dietrich CH, Allen JM, Lemmon AR, Lemmon EM, Takiya DM, Evangelista O, Walden KKO, Grady PGS, Johnson KP (2017) Anchored hybrid enrichment-based phylogenomics of leafhoppers and treehoppers (Hemiptera: Cicadomorpha: Membracoidea). Insect Systematics and Diversity 1(1), 57-72.
| Crossref | Google Scholar |
Dietrich CH, Dmitriev DA, Takiya DM, Thomas MJ, Webb MD, Zahniser JN, Zhang Y (2022) Morphology-based phylogenetic analysis of Membracoidea (Hemiptera: Cicadomorpha) with placement of fossil taxa and description of a new subfamily. Insect Systematics and Diversity 6(5), 1-30 7.
| Crossref | Google Scholar |
Dmitriev DA (2010) Homologies of the head of Membracoidea based on nymphal morphology with notes on other groups of Auchenorrhyncha (Hemiptera). European Journal of Entomology 107, 597-613.
| Crossref | Google Scholar |
Emeljanov AF (1996) To the knowledge of leafhoppers of the family Ulopidae (Homoptera, Cicadina). Entomologicheskoe Obozrenie 75(2), 278-293.
| Google Scholar |
Encinas-Viso F, Bovill J, Albrecht DE, Florez‐Fernandez J, Lessard B, Lumbers J, Rodriguez J, Schmidt‐Lebuhn A, Zwick A, Milla L (2022) Pollen DNA metabarcoding reveals cryptic diversity and high spatial turnover in alpine plant–pollinator networks. Molecular Ecology 32, 6377-6393.
| Crossref | Google Scholar |
Evangelista O, Florez-V C, Sakakibara AM (2014) The identity of the treehopper genus Dysyncritus Fowler, with descriptions of new related taxa (Hemiptera: Membracidae: Heteronotinae). Zootaxa 3847(4), 495-532.
| Google Scholar |
Evans JW (1937) Australian Leafhoppers (Jassoidea: Homoptera) Part 4. — (Ledridae, Ulopidae and Euscelidae, Paradorydiini). Papers and Proceedings of the Royal Society of Tasmania 1936, 37-50.
| Google Scholar |
Evans JW (1939) Some new Australian Leafhoppers (Homoptera: Jassoidea). Transactions of the Royal Society of South Australia 63(1), 44-50.
| Google Scholar |
Evans JW (1947) Some new Ulopinae (Homoptera, Jassidae). Annals and Magazine of Natural History 14(11), 140-150.
| Google Scholar |
Evans JW (1951) Some notes on the classification of the leafhoppers (Jassoidea, Homoptera) with special reference to the Nearctic fauna. Commentationes Biologicae Societas Scientiarum Fennica 12, 1-11.
| Google Scholar |
Evans JW (1966) The leafhoppers and froghoppers of Australia and New Zealand (Homoptera: Cicadelloidea and Cercopoidea). Australian Museum Memoir 12, 1-347.
| Crossref | Google Scholar |
Evans JW (1968) Some relict New Guinea leaf hoppers and their significance in relation to the comparative morphology of the head and thorax of the Homoptera Auchenorrhyncha. Pacific Insects 10, 215-229.
| Google Scholar |
Evans JW (1977) The leafhoppers and froghoppers of Australia and New Zealand (Homoptera: Cicadelloidea and Cercopoidea). Part 2. Records of the Australian Museum 31(3), 83-129.
| Crossref | Google Scholar |
Fletcher MJ (2009) Identification keys and checklists for the leafhoppers, planthoppers and their relatives occurring in Australia and neighbouring areas (Hemiptera: Auchenorrhyncha). (New South Wales Government, Department of Primary Industries, Biosecurity Collections) Available at https://idtools.dpi.nsw.gov.au/keys/auch/index.html
Fletcher MJ (2012) Genus Austrolopa Evans, 1937. (Australian Faunal Directory. Australian Biological Resources Study: Canberra, ACT, Australia) Available at https://biodiversity.org.au/afd/taxa/Austrolopa
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29, 644-652.
| Crossref | Google Scholar | PubMed |
Hamilton KGA (1983) Classification, morphology and phylogeny of the family Cicadellidae (Rhynchota: Homoptera) In ‘Proceedings of the 1st International Workshop on Biotaxonomy, Classification and Biology of Leafhoppers and Planthoppers of Economic Importance’, 4–7 October 1982, London, UK. (Eds WJ Knight, NC Pant, TS Robertson, MR Wilson) pp. 15–37. (Commonwealth Institute of Entomology: London, UK)
Hamilton KGA (1990) Chapter 6. Homoptera. In ‘Insects from the Santana Formation, Lower Cretaceous of Brazil’. (Ed. DA Grimaldi) Bulletin of the American Museum of Natural History, vol. 195, pp. 82–122. (American Museum of Natural History) Available at https://flow.hemiptera-databases.org/flowpdf/349.pdf
Harvey MS (2002) Short-range endemism amongst the Australian fauna: some examples from non-marine environments. Invertebrate Systematics 16(4), 555-570.
| Crossref | Google Scholar |
Hu Y, Dietrich CH, Skinner RK, Zhang Y (2023) Phylogeny of Membracoidea (Hemiptera: Auchenorrhyncha) based on transcriptome data. Systematic Entomology 48(1), 97-110.
| Crossref | Google Scholar |
Johnson KP, Dietrich CH, Friedrich F, Beutel RG, Wipfler B, Peters RS, Allen JM, Petersen M, Donath A, Walden KKO, Kozlov AM, Podsiadlowski L, Mayer C, Meusemann K, Vasilikopoulos A, Waterhouse RM, Cameron SL, Weirauch C, Swanson DR, Percy DM, Hardy NB, Terry I, Liu S, Zhou X, Misof B, Robertson HM, Yoshizawa K (2018) Phylogenomics and the evolution of hemipteroid insects. Proceedings of the National Academy of Sciences of the United States of America 115(50), 12775-12780.
| Crossref | Google Scholar | PubMed |
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587-589.
| Crossref | Google Scholar | PubMed |
Krugner R, Sisterson MS, Backus EA, Burbank LP, Redak RA (2019) Sharpshooters: a review of what moves Xylella fastidiosa. Austral Entomology 58(2), 248-267.
| Crossref | Google Scholar |
Le SQ, Dang CC, Gascuel O (2012) Modeling protein evolution with several amino acid replacement matrices depending on site rates. Molecular Biology and Evolution 29(10), 2921-2936.
| Crossref | Google Scholar |
Leinonen R, Sugawara H, Shumway M, on behalf of the International Nucleotide Sequence Database Collaboration (2011) The sequence read archive. Nucleic acids research 39(suppl_1), D19-D21.
| Crossref | Google Scholar | PubMed |
Marsh JR, Bal P, Fraser H, Greenville A, Latty T, Moir ML, Rumpff L, Umbers K, Woinarski JC (2023) Impacts of the 2019–20 wildfires on Australian invertebrates. Australia’s Megafires: Biodiversity Impacts and Lessons from 2019–2020. Project 8.3.1, final report. (NESP Threatened Species Recovery Hub: Brisbane, Qld, Australia)
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37, 1530-1534.
| Crossref | Google Scholar | PubMed |
Moir ML, Majer JD, Fletcher MJ (2003) New records for Hemiptera species in Western Australia. Records of the Western Australian Museum 21(4), 353-357.
| Crossref | Google Scholar |
Rakitov RA (1996) Post-moulting behaviour associated with Malpighian tubule secretions in leafhoppers and treehoppers (Auchenorrhyncha: Membracoidea). European Journal of Entomology 93, 167-184.
| Google Scholar |
Rakitov RA (1998) On differentiation of cicadellid leg chaetotaxy (Homoptera: Auchenorrhyncha: Membracoidea). Russian Entomological Journal 6, 7-27.
| Google Scholar |
Rakitov RA, Godoy C (2005) New egg-powdering sharpshooters (Hemiptera: Cicadellidae: Proconiini) from Costa Rica. Annals of the Entomological Society of America 98(4), 444-457.
| Crossref | Google Scholar |
Rosauer D, Laffan SW, Crisp MD, Donnellan SC, Cook LG (2009) Phylogenetic endemism: a new approach for identifying geographical concentrations of evolutionary history. Molecular Ecology 18(19), 4061-72.
| Crossref | Google Scholar | PubMed |
Schmidt HA, Strimmer K, Vingron M, Von Haeseler A (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18(3), 502-504.
| Crossref | Google Scholar | PubMed |
Skinner RK, Dietrich CH, Walden KKO, Gordon E, Sweet AD, Podsiadlowski L, Petersen M, Simon C, Takiya DM, Johnson KP (2019) Phylogenomics of Auchenorrhyncha (Insecta: Hemiptera) using transcriptomes: examining controversial relationships via degeneracy coding and interrogation of gene conflict. Systematic Entomology 45(1), 85-113.
| Crossref | Google Scholar |
Wang J-J, Wu Y-F, Yang M-F, Dai R-H (2020) The phylogenetic implications of the mitochondrial genomes of Macropsis notata and Oncopsis nigrofasciata. Frontiers in Genetics 11, 443.
| Crossref | Google Scholar |