‘Where is my family?’ Molecular and morphological data reveal the phylogenetic position and diversity of the enigmatic handsome fungus beetle genus Anamycetaea Strohecker, 1975 (Coleoptera, Coccinelloidea)
Wioletta Tomaszewska A * , Karol Szawaryn
A * , Karol Szawaryn  A and Emmanuel Arriaga-Varela
A and Emmanuel Arriaga-Varela  A
A
A Museum and Institute of Zoology, Polish Academy of Sciences, Wilcza 64, P-00-679 Warsaw, Poland.
Invertebrate Systematics 37(4) 231-253 https://doi.org/10.1071/IS22053
Submitted: 22 October 2022 Accepted: 14 March 2023 Published: 3 April 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution 4.0 International License (CC BY)
Abstract
The genus Anamycetaea Strohecker, 1975, established for Anamycetaea keralae, a single species from India, was originally placed in the diverse endomychid subfamily Mycetaeinae and has subsequently been considered a member of the subfamily Anamorphinae based on closed mesocoxal cavities, a postulated synapomorphy of this group. Recent molecular research resulted in raising Anamorphinae to family level and revealed this group to be distantly related to Endomychidae sensu stricto. However, Anamycetaea has been ‘neglected’ since description. Our detailed study of this genus has been possible due to new material collected from Oriental and Australian regions. Striking overall similarity to the endomychine genus Tharina and a tentorium with anterior arms fused medially (separated in almost all Anamorphidae) have raised our doubts and led to further investigation of the phylogenetic placement of this enigmatic genus within Endomychidae sensu lato (handsome fungus beetles). Phylogenetic analyses of molecular and morphological datasets were conducted under Bayesian (BI), maximum likelihood (ML) and parsimony (MP) frameworks. Our results recovered Anamycetaea as belonging to the family Endomychidae, in the subfamily Endomychinae, distant from Anamorphidae. The close affinity to Stenotarsus and allies was strongly supported in all analyses. Based on material studied, A. keralae is described in detail here and includes description of previously unknown male genitalia. Four new species are also described, extending the ragne of the genus to the Australian region: Anamycetaea borneensis sp. nov. (from Borneo), A. novoguineensis sp. nov. and A. papuensis sp. nov. (from Papua New Guinea) and A. queenslandica sp. nov. (from Australia). Illustrations of morphological details and diagnoses are provided for each species. A key to the species of the genus is also presented.
ZooBank: urn:lsid:zoobank.org:pub:90BAA954-7849-4FA9-997B-061FE7BB5702
Keywords: Anamorphidae, Australian region, Endomychidae, entomology, new species, Oriental region, phylogenetic position, taxonomy.
Introduction
The most effective approach for the classification of organisms is to incorporate a pluralistic source of information and integrate this into a robust hypothesis that reflects the phylogenetic relationships among organisms (Dayrat 2005; Will et al. 2005; Padial et al. 2010). Classification schemes that rely solely on one or only a few characters of the external morphology can lead to artificial grouping due to the presence of homoplastic states (Tarasov and Solodovnikov 2011). In some groups of insects where species are rarely collected and rarely studied in detail, misclassifications are common.
The genus Anamycetaea was described by Strohecker (1975) to accommodate the new species, A. keralae Strohecker, 1975 from India. Strohecker (1975) placed this new genus in the diverse endomychid subfamily Mycetaeinae knowing that this subfamily was a highly heterogenous group, united only by a rather superficial combination of characters such as small size, very rarely longer than 2.5 mm; tarsi 3-segmented and linear or distinctly 4-segmented; and antennae composed of 8–11 antennomeres. According to this definition (Strohecker 1953), Mycetaeinae contained a loosely associated variety of small fungus beetles including taxa currently classified in different families or subfamilies (e.g. Pakaluk et al. 1994; Tomaszewska 2000, 2005; Robertson et al. 2015). Only one specimen of Anamycetaea was available for the description, and mouthparts, internal head structures and genitalia were not studied by Strohecker (1975).
At that time of description, phylogenetic hypotheses of the higher taxa of Endomychidae (handsome fungus beetles) did not exist and the classification was based on overall similarities, therefore the placement of Anamycetaea in the subfamily Mycetaeinae by Strohecker was artificial.
Sasaji (1978) was the first author to discover that the mesocoxal cavities were laterally closed and the anterior arms of the tentorium were separate throughout the length in some genera included in a broadly defined Mycetaeinae of Strohecker (1953) (e.g. Mychothenus Strohecker, 1953, Bystodes Strohecker, 1953, Bystus Guérin-Méneville, 1857 and Dialexia Gorham, 1891). Based on these characters, previously unknown within the family Endomychidae, Sasaji (1978) established the subfamily Anamorphinae (=Mychotheninae) that was subsequently elevated to family status (Sasaji 1987, 1990). The later action, however, did not receive a great deal of attention from researchers (Pakaluk et al. 1994; Lawrence and Newton 1995).
The first modern classification of Endomychidae that was based on a cladistic analysis of morphological characters (Tomaszewska 2000) revealed the subfamily Mycetaeinae that included only two genera, Mycetaea Stephens, 1829 and Agaricophilus Motschulsky, 1838. All genera of the former Mycetaeinae with closed mesocoxal cavities (including Anamycetaea) were classified as members of the subfamily Anamorphinae. Recently, the concept of the family Endomychidae that contains 12 subfamilies (e.g. Tomaszewska 2000, 2005, 2010; Shockley et al. 2009) has been changed and the family is currently restricted to 9 subfamilies, whereas former Anamorphinae, Eupsilobiinae and Mycetaeinae have been elevated to family level in the superfamily Coccinelloidea (Robertson et al. 2015).
After 47 years since the description of the monotypic genus Anamycetaea based on a single specimen of undetermined sex (Strohecker 1975), additional material from Borneo, Australia and Papua New Guinea have allowed us to study the morphological features including mouthparts, tentorium and genitalia in detail, and present comprehensive descriptions of the genus and species. Four new species are described here from far beyond the original type locality, considerably extending the range of occurrence of this genus and suggesting that more undescribed species from the Oriental and Australian regions may be waiting to be discovered.
Morphology of Anamycetaea species showing striking overall similarity to that of the endomychine genus Tharina Arriaga-Varela, Tomaszewska & Fikáček, 2019 and the tentorium with anterior arms widely fused medially, unlike in Anamorphidae, led us to question the phylogenetic position of this enigmatic genus within Endomychidae sensu lato (handsome fungus beetles). Therefore, the main aim of this study, based on the availability of relatively rich genus material, was to conduct phylogenetic analyses of morphological and molecular data, to clarify the taxonomic placement of Anamycetaea.
Materials and methods
Morphological material – preparation and processing
The specimens studied during this contribution are deposited at the following institutions:
ANIC, Australian National Insect Collection, CSIRO, Canberra, Australia;
IECA, Institute of Entomology, Biology Centre ASCR, České Budějovice, Czech Republic;
IRSNB, Institute Royal des Sciences Naturelles de Belgique, Brussels, Belgium;
MHNG, Museum d’Histoire Naturelle, Geneve, Switzerland;
MIZ, Museum and Institute of Zoology PAS, Warszawa, Poland;
MNHN, Muséum National dʼHistoire Naturelle, Paris;
NMPC, National Museum, Prague, Czech Republic;
NAIC, National Agricultural Insect Collection, Kila Kila, Port Moresby, Papua New Guinea;
QMB, Queensland Museum Brisbane, Australia.
Anamycetaea keralae is redescribed and new species are described and listed in alphabetical order. Label information is transcribed verbatim. Genitalia and other morphological structures were dissected, cleared in 10% KOH solution and rinsed with distilled water, and subsequently transferred to glycerol and examined on slides. After examination, the genitalia were transferred to microvials and pinned beneath the specimen. Measurements were made using an ocular micrometre attached to an Olympus SZX16 dissecting microscope and recorded as follows: total body length, from apical margin of clypeus to apex of elytra; width, across both elytra in the widest part; pronotal length, from the middle of anterior margin to margin of basal foramen; pronotal width, across widest part; and elytral length, along suture including scutellum. Habitus colour photographs were taken with a 4K Ultra-high accuracy microscope VHX-7000 Series by KEYENCE in the Museum and Institute of Zoology PAS in Warsaw and subsequently modified in Adobe Photoshop CS5. The photographs of the genitalia and other disarticulated morphological structures were taken using an Olympus DP23 digital camera attached to an Olympus BX43F compound microscope; final images were produced using Helicon Focus 5.0 × 64 and Adobe Photoshop CS6 software. Scanning electron micrographs were taken using a HITACHI S-3400N microscope under low vacuum conditions in the Electron Microscopy Laboratory at the MIZ PAS. Morphological terminology follows Tomaszewska (2010) and Lawrence et al. (2011) and the description of the aedeagus is based on the resting position in the abdomen.
Taxonomic acts established in this work have been registered in ZooBank (see below) together with the electronic publication LSID.
Molecular analyses
One of the main goals of our study was to resolve the taxonomic position of Anamycetaea within Endomychidae sensu lato of the superfamily Coccinelloidea. Therefore most of the molecular data used herein originated from Robertson et al. (2015), to which new sequences of two new species described here were added: Anamycetaea novoguineensis sp. nov. and A. papuensis sp. nov., from Papua New Guinea. Specimens of newly sequenced species were recently collected, kept in 95% ethanol and stored at −20°C.
Endomychidae belongs to the coccinellid group of Coccinelloidea sensu Robertson et al. (2015) containing the currently recognised families Akalyptoischiidae, Latridiidae, Alexiidae, Anamorphidae, Corylophidae, Endomychidae, Mycetaeidae, Eupsilobiidae and Coccinellidae. Members of Anamorphidae, Endomychidae, Mycetaeidae and Eupsilobiidae (=Endomychidae sensu lato) were considered as the ingroup, and representatives of the remaining five families were treated as outgroups. The tree was rooted using two species of the bothriderid group of Coccinelloidea: Oxylaemus californicus Crotch (Teredidae) and Discoloma sp. (Discolomatidae). The final molecular dataset is composed of 218 species (Akalyptoischiidae 3 spp., Latridiidae 20 spp., Alexiidae 2 spp., Anamorphidae 15 spp. (including two new species of Anamycetaea), Corylophidae 37 spp., Endomychidae 48 spp., Mycetaeidae 1 sp., Eupsilobiidae 1 sp., Coccinellidae 89 spp., Discolomatidae 1 sp. and Teredidae 1 sp. (Supplementary Table S1).
In our dataset, we included the eight molecular markers used by Robertson et al. (2015): nuclear 18S rDNA (18S), 28S rDNA (28S), histone subunit 3 (H3) and carbamoyl-phosphate synthetase (CAD); and mitochondrial 12S rDNA (12S), 16S rDNA (16S), cytochrome-c oxidase subunit I (COI) and cytochrome-c oxidase subunit II (COII). Primers and protocols for the amplification and sequencing of target genes used in that work are outlined in Robertson et al. (2013). The primers used for the amplification of sequences from the two newly added specimens and specifications of the PCR protocols are listed in Supplementary Table S2. Amplification of the CAD gene was attempted without success. Since the long hypervariable section towards the 5′ region of gene 28s as obtained by Robertson et al. (2015) produces multiple gaps and cannot be unambiguously aligned, we removed that part of ~2700 bp. Total length of the sequence by gene included in the final alignment is as follows (bp range in the final alignment is provided in brackets): COI, 1239 bp (1–1239); COII, 630 bp (1240–1869); H3, 372 bp (1870–2196); CAD, 966 bp (2197–3150); 12S, 397 bp (3151–3549); 16S, 539 bp (3550–4088); 18S, 2013 bp (4089–6102); and 28S, 1031 bp (6103–7134).
The trace files of the new sequences obtained from Anamycetaea spp. were inspected, assembled and edited in Geneious (ver. 9.1.8, see https://www.geneious.com/; Kearse et al. 2012). All newly generated sequences were submitted to GenBank under accession numbers OQ428620, OQ428621, OQ433780, OQ433781 and OQ435982–OQ435987. The list of vouchers, associated data and GenBank accession numbers of the sequences used in this study are listed in Supplementary Table S1. Sequences were aligned in Geneious using the ClustalW (ver. 1.8.1, see http://www.clustal.org/clustal2/; Thompson et al. 2003) algorithm with default settings. The alignment of DNA sequences was trivial for COI, COII, H3 and CAD. Alignments for rDNA sequences (12S, 16S, 18S and 28S) were inspected by eye and corrected manually in Geneious. Concatenation of sequences was performed manually in Geneious software. The final concatenated aligned matrix can be found as Supplementary File S1. The concatenated alignments were used to infer the phylogenetic relationships using Bayesian inference (BI) and maximum likelihood (ML). Protein-coding genes were partitioned by codon position and ribosomal genes were analysed unpartitioned. The ML analysis was performed in W-IQ-TREE (ver. 1.5, see http://iqtree.cibiv.univie.ac.at/; Nguyen et al. 2015; Trifinopoulos et al. 2016). An ultrafast bootstrap (UFB) with 1000 repetitions (Minh et al. 2013) was applied to estimate the support for the recovered nodes. The BI analysis was performed using MrBayes (ver. 3.2.6, see https://github.com/NBISweden/MrBayes/; Ronquist et al. 2012) implemented on the CIPRES Science Gateway (ver. 3.3, see http://www.phylo.org/; Miller et al. 2010). Four simultaneous independent MCMC runs with six chains each and 20 million generations were used for the analysis, with a tree sampled every 10 000 generations to calculate posterior probabilities (PP). The convergence of the runs and associated parameters were assessed in TRACER (ver. 1.7, see https://github.com/beast-dev/tracer/releases/tag/v1.7.1; Rambaut et al. 2018). A conservative 25% burn-in fraction was excluded from the data prior to the construction of the final maximum credibility tree.
Morphological dataset and analysis
Being aware of certain contradictions between the results of analyses based solely on molecular data (Robertson et al. 2015) or morphology (Tomaszewska 2000, 2005) in relation to Endomychidae, a cladistic analysis of morphological characters was also conducted. The aim was to find the closest relatives of the genus Anamycetaea and recognise the proper family (and subfamily) placement within handsome fungus beetles and among relatives, therefore the analysis of morphological features was treated as another independent test of the phylogenetic position for this genus.
Taxon sampling and characters used for the analysis were broadly based on Tomaszewska (2000, 2005) with some modifications made in an attempt to provide resolution among Anamycetaea and possibly related taxa. A broad representation of Endomychidae sensu lato (Endomychidae, Anamorphidae, Eupsilobiidae and Mycetaeidae) was sampled, including the type species for genera, subfamilies and families when specimens were available for study, and these taxa were treated as ingroup; members of Coccinellidae and Corylophidae were treated as closely related outgroups. Hobartius eucalypti (Blackburn) (Hobartiidae) was used as a more distant outgroup. In total, our morphological data matrix comprised 42 taxa (34 ingroup taxa and eight outgroups) scored for 48 multistate characters. Unknown character states were treated as missing and coded with ‘?’. The list of morphological characters and the character matrix can be found in Supplementary Table S3 and Supplementary File S2 respectively.
The maximum parsimony (MP) analysis of our dataset was conducted in TNT (ver. 1.5, see https://www.lillo.org.ar/phylogeny/tnt/; Goloboff and Catalano 2016) to find the most parsimonious trees (MPTs), using the New Technology (NT) option and in turn, the Driven Search with Sectorial Search, Ratchet, Drift and Tree fusing options activated with standard settings; and the Traditional Search (TS) option under the following parameters: memory set to hold 1 000 000 trees, tree bisection – reconnection (TBR) branch-swapping algorithm with 1000 replications saving 100 trees per replicate; zero-length branches collapsed after the search. All characters were treated as unordered and analyses were first performed under equal weights.
Subsequently, implied weighting options were used to reduce the effects of homoplasy. Other settings were unchanged. The analysis was repeated with values of the concavity constant K ranging from 3 to 10. Character mapping was done in Winclada (ver. 1.00.08, see www.diversityoflife.org/winclada; Nixon 2002) using unambiguous optimisation.
Results
Phylogenetic analyses of molecular data
The analyses of the molecular dataset under BI (Fig. 1) and ML resulted in similar topologies, differing slightly only in terminal bifurcations among some closely related species (see Supplementary Fig. S1 and Supplementary File S3 for complete BI and ML trees respectively). Species of Anamycetaea are recovered in both trees as a clade related to Stenotarsus and allies within the subfamily Endomychinae of ‘endomychine complex’ within Endomychidae sensu stricto of Robertson et al. (2015). The genus Anamycetaea is confirmed to be unrelated to Anamorphidae by the molecular analyses.
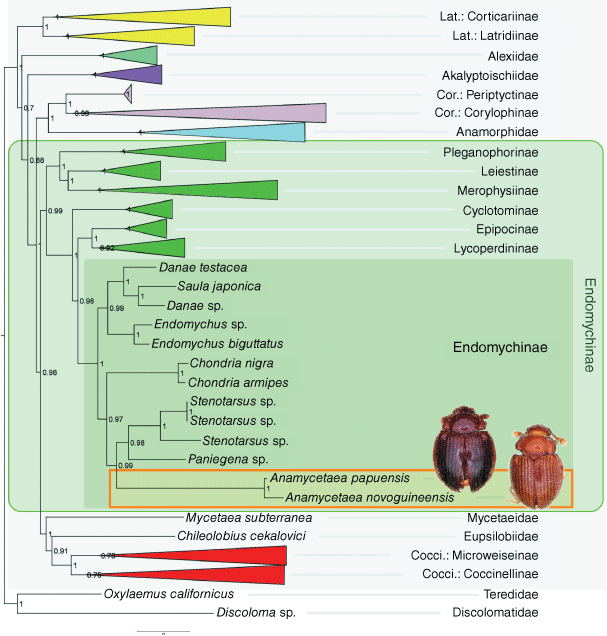
|
As illustrated in Fig. 1, major clades in the tree are recovered with high or maximum support based on the reulsts of the BI analysis. Relationships between families of the coccinellid group of Coccinelloidea agree with those reported by Robertson et al. (2015) with the exception of the relative positions of Akalyptoischiidae and Alexiidae. Species of Anamycetaea are recovered as a clade and the genus forms a sister group with Stenotarsus + Paniegena with high support (PP = 0.99). This group is sister to Chondria within the subfamily Endomychinae (as defined by Robertson et al. 2015). In molecular analyses, Anamorphidae forms the sister group to Corylophidae and together, these are the sister group to the remaining Endomychidae sensu lato and Coccinellidae.
Phylogenetic analysis of the morphological dataset
The analysis of morphological characters conducted under different search strategies resulted in very similar resolutions.
The maximum parsimony analysis under both Traditional Search (MP TS) and New Technology Search (MP NT) resulted in two most parsimonious trees (MPT) each. Strict consensus calculated from two trees for each option (TS and NT) showed the same resolution with length (L) of 135 steps, consistency index (CI) = 42 and retention index (RI) = 77. The consensus tree (Supplementary Fig. S2) shows Endomychidae sensu lato as a monophyletic group with the exception of Eupsilobiidae that is recovered as sister group to Coccinellidae. The major clades Corylophidae, Coccinellidae + Eupsilobiidae, and the remaining Endomychidae sensu lato are recovered as an unresolved trichotomy.
MP analysis under implied weighting (IW) with concavity constant K set as K = 3, 5, 10 resulted in a single tree (for each K parameter) of the same resolution and the parameters L = 133 steps, CI = 43 and RI = 78, in both the TS and NT search options. The tree obtained under IW is very similar to the strict consensus tree from the TS and NT options but is fully resolved showing Corylophidae as sister group to (Coccinellidae + Eupsilobiidae) + Endomychidae sensu lato (except for Eupsilobiidae)). We chose the resulting single tree from the MP TS analysis under IW with K = 3 as our preferred tree, showing the best-resolved topology (Fig. 2) that mostly agrees with the results of Tomaszewska (2005) and at least partly agrees with those of Robertson et al. (2015).
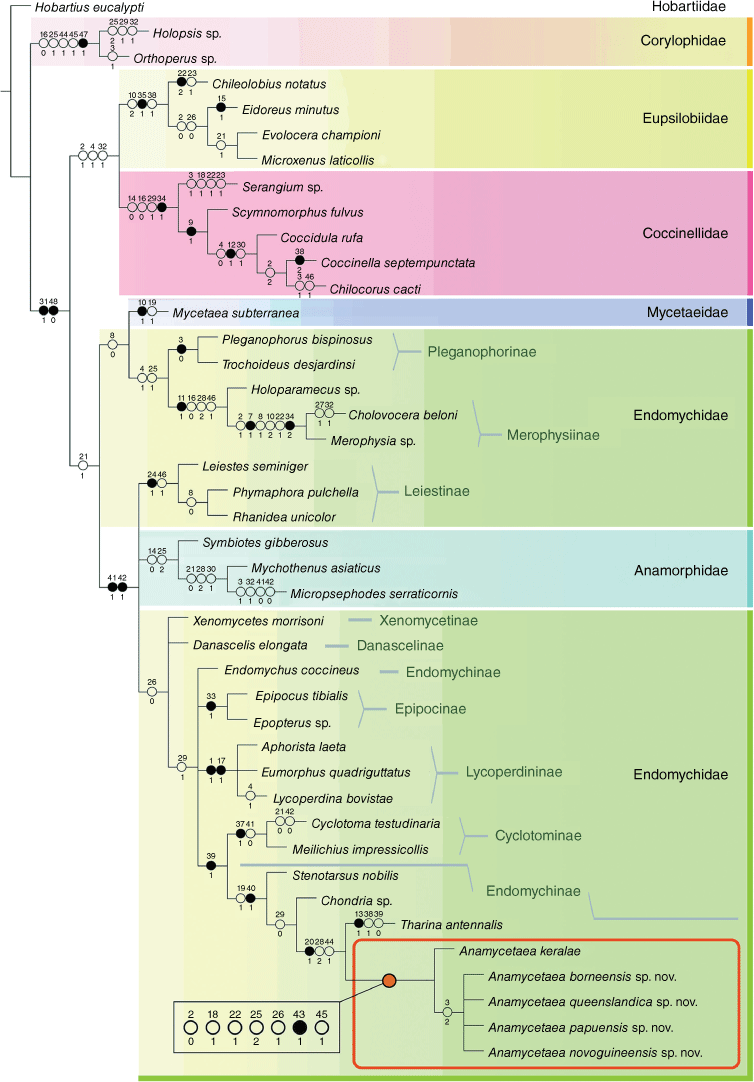
|
Unsurprisingly, the results of our morphology-based analysis contradict the current molecular-based classification of the family Endomychidae (Robertson et al. 2015) to some extent (see Discussion). We recovered Endomychidae as a monophyletic group including Mycetaeidae and Anamorphidae, whereas Mycetaeidae is recovered as sister group to Eupsilobiidae + Coccinellidae, and Anamorphidae as sister to Corylophidae in Robertson et al. (2015). Leiestinae and Anamorphidae are recovered in a large clade comprising Xenomycetinae, Danascelinae and ‘Higher Endomychidae’ of Tomaszewska (2005) (=endomychine complex sensu Robertson et al. 2015), whereas Leiestinae is a sister group to Pleganophorinae + Merophysiinae in Robertson et al. (2015). Endomychus coccineus forms a separate line, outside of a clade with former Stenotarsinae, making the currently defined subfamily Endomychinae (Robertson et al. 2015) polyphyletic. The subfamily Cyclotominae is recovered as sister group to Endomychinae (except Endomychus), whereas Cyclotominae is sister group to all remaining taxa of the endomychine complex in Robertson et al. (2015).
The results of our study emphasised taxonomic and classification problems of the handsome fungus beetles related to the morphological plasticity that may lead to misleading interpretations about affinities and the development of incorrect classification systems. Being aware of limitations of analyses based on morphological characters only, we will address problems listed above (including monophyly of the subfamily Endomychinae) in further research with an integrative approach (E. Arriaga-Varela, W. Tomaszewska and K. Szawaryn, in prep.). We followed the current molecular-based classification and treated a clade containing former Stenotarsinae genera that included Anamycetaea as the subfamily Endomychinae for the purposes of this study
Phylogenetic placement and taxonomy of Anamycetaea
The ML, BI and MP analyses showed Anamycetaea to be monophyletic, and provided unequivocal support for the placement of this genus within the subfamily Endomychinae, in a clade with Stenotarsus, Paniegena, Tharina and Chondria. The molecular analyses recovered the relationships within this clade as Chondria + (Anamycetaea + (Paniegena + Stenotarsus)) (Paniegena included only in molecular analysis); whereas morphological analysis recovered these relationships as Stenotarsus + (Chondria + (Anamycetaea + Tharina)) (Tharina included only in morphological analysis).
Anamycetaea is highly supported as a monophyletic group (BS = 100; PP = 1.00) and is morphologically well defined by one synapomorphy: prosternum strongly carinate (43:1); and six homoplastic character states: head with short antennal grooves (2:0); pronotal base distinctly, comparatively widely bordered (18:1); prothoracic ventral antennal grooves present on prosternum (22:1); mesocoxal cavity widely closed outwardly (25:2); mesotrochantin concealed (26:1); and maxillary palpi 3-segmented (45:1). The sister relationship with Tharina in morphological analysis is strongly supported by a synapomorphy: pronotal anterior margin bordered with crenulate bordering line (20:1); and two homoplastic character states: tarsal formula 3-3-3 (28:2); and labial palpi 2-segmented (44:1).
Taxonomy
Order COLEOPTERA Linnaeus, 1758
Suborder POLYPHAGA Emery, 1886
Superfamily COCCINELLOIDEA Latreille, 1807
Family ENDOMYCHIDAE Leach, 1815
Subfamily ENDOMYCHINAE Leach, 1815
Genus Anamycetaea Strohecker, 1975
Redescription
Body
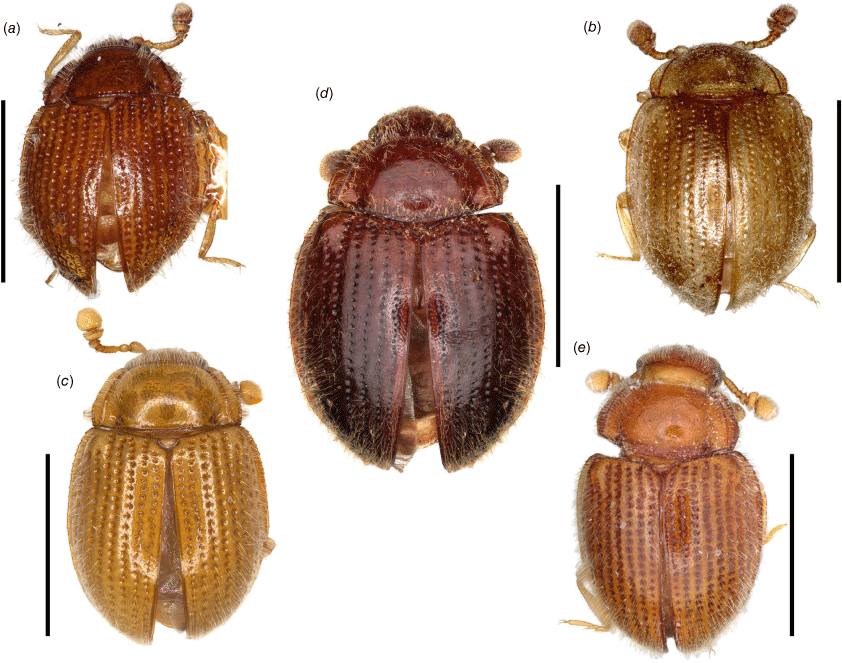
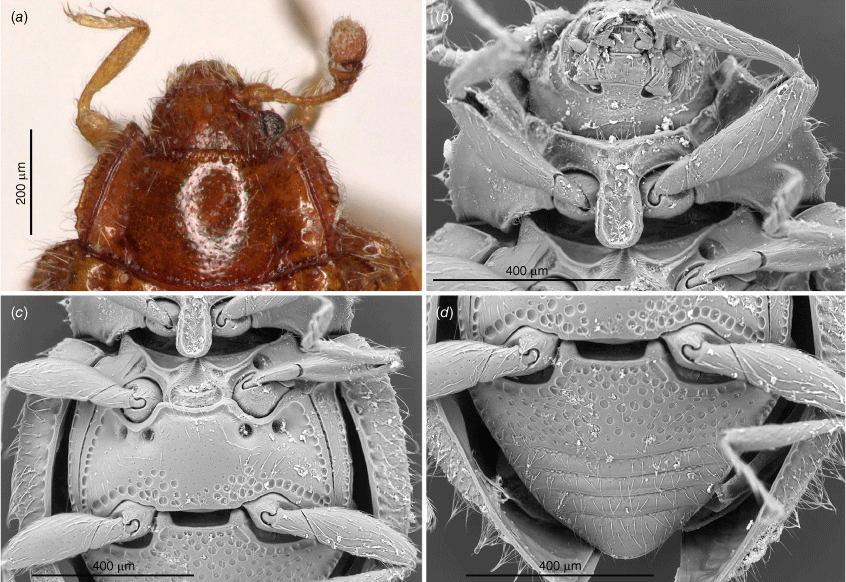
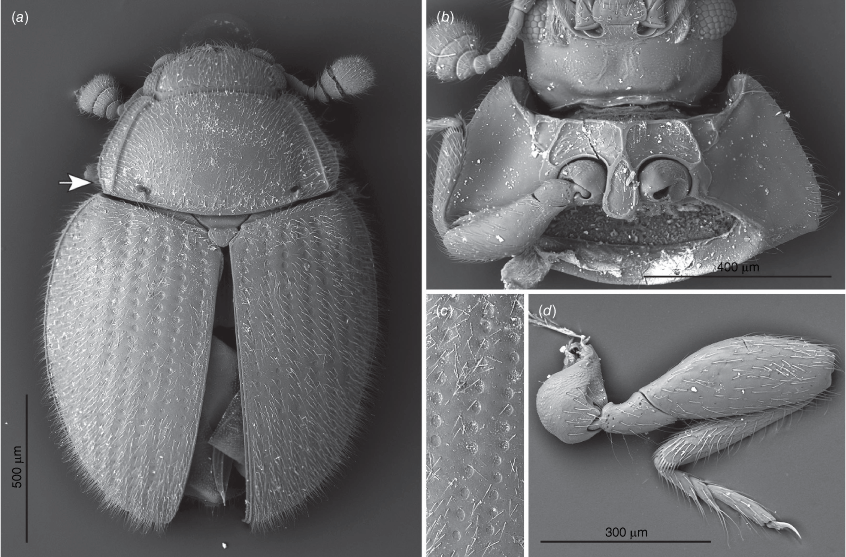
Length: 1.4–2.4 mm, shape elongate–oval (Fig. 3), moderately to strongly convex; shiny, covered with moderately dense, long, suberect or erect, pale setae; elytra covered with regular rows of large punctures. Colour uniformly pale yellowish-brown to chestnut brown.
Head
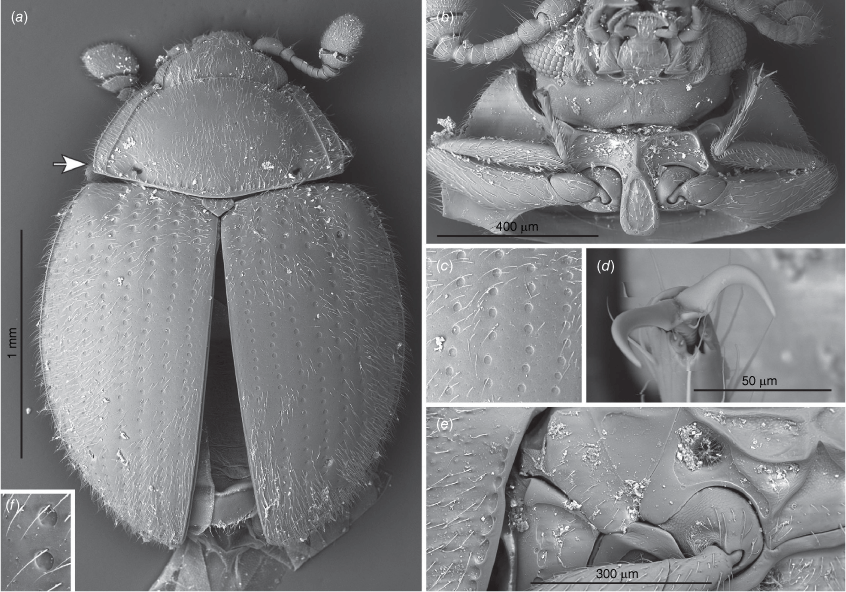
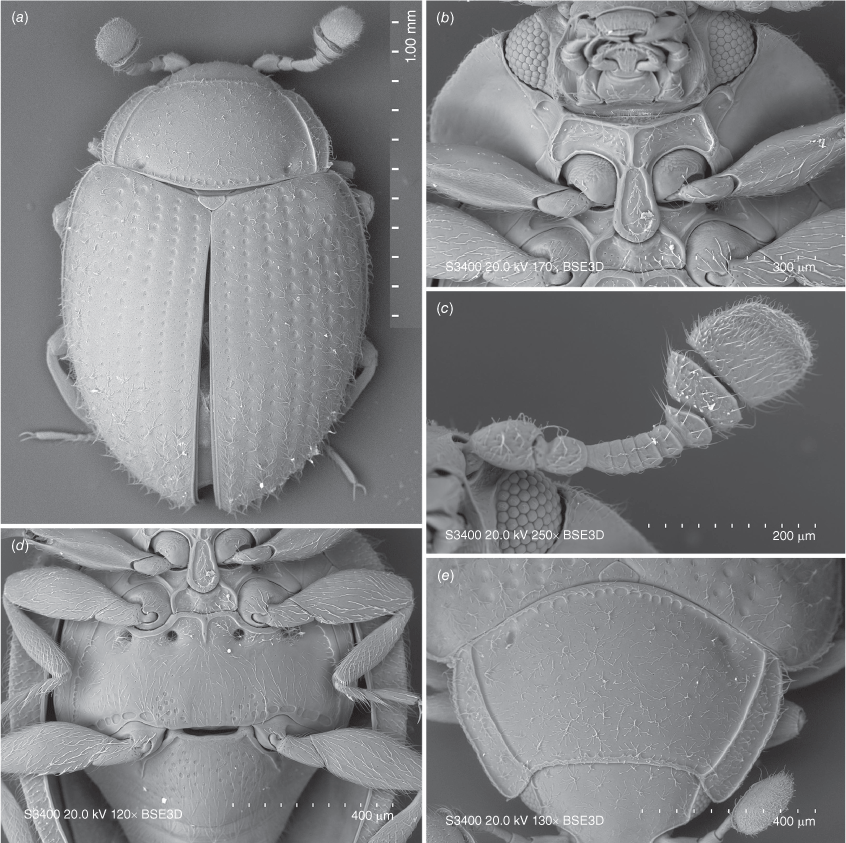
Transverse; eyes large, oval, moderately prominent, coarsely faceted; interfacetal setae absent (Fig. 6b, 9b, 10f). Antennal sockets visible from above (Fig. 5a). Antenna slightly shorter than head and prothorax together, composed of 10 (Fig. 6c, 7a, 9a, 10b) or 11 antennomeres (Fig. 5a) with large, wide, compact club composed of three antennomeres; terminal antennomere largest, oval, penultimate antennomere transverse and as wide as terminal one, antepenultimate antennomere shorter and narrower than penultimate antennomere (sometimes club may appear 2-segmented). Ventral antennal grooves present, reaching no further than hind margin of eyes. Mandible (Fig. 11d) with large, somewhat chisel-shaped apical tooth and small subapical tooth; mola large, transversely ridged; submola large, membranous. Maxilla with 3-segmented palp (Fig. 10c); palpomeres 1 and 3 distinctly elongate, palpomere 2 very short, transverse; maxillary galea and lacinia covered apically with stiff, broad setae; terminal palpomere obliquely truncate at apex. Labium (Fig. 5b, 10c) with palpi very widely separated basally; terminal palpomere subquadrate, obliquely truncate at apex; mentum widest near base, flat. Tentorium (Fig. 11b) with anterior arms broadly fused medially and widely divergent anteriorly; posterior arms widely divergent; corpotentorial brigde absent.
|
Prothorax
Pronotum (Fig. 5a, 6e, 7a, 9a, 10d) transverse, widest near base, ~2 times as wide as long, almost as wide basally as base of elytra; hardly to very distinctly crenulate laterally, widely bordered with carinae, area between lateral edge and carina weakly deflexed; margined anteriorly and basally with crenulate line; basal sulcus hardly marked to absent with lateral pits present; anterior angles weakly to distinctly produced, blunt to acute; posterior angles right-angled to obtuse; lateral margins weakly to distinctly rounded. Pronotal disc convex. Prosternum (Fig. 5b, 6b, 7b, 9b, 10f) strongly carinate with weak to distinct cavities antero-laterally that may serve as antennal grooves; prosternal process comparatively broad and long, margined by carina forming more or less distinct triangular or oval shape, widening towards apex, reaching far beyond front coxae and resting in fossa on mesoventral process; widely separating procoxae.
Pterothorax
Scutellar shield small, transverse, somewhat pentagonal (Fig. 5a, 6a, 7a, 9a, 10a). Mesoventrite (Fig 5c, 6d) strongly carinate with a pair of large pits antero-laterally; intercoxal process irregularly pentagonal, widely separating mesocoxae, strongly narrowing anteriorly and provided with deep fossa for apex of prosternal process. Mesocoxal cavities widely closed outwardly; trochantin concealed (Fig. 9e). Elytron (Fig. 6a, 7a, 9a, 10a) elongate–oval, convex with regular rows of large punctures and minute, irregularly distributed, setose punctae; lateral margins narrow, visible from above at least along basal half-length of each elytron; epipleuron comparatively narrow, incomplete apically. Metaventrite (Fig. 5c, 6d, 10e) with anterior margin distinctly bordered, and provided with two pairs of large, postcoxal pits; discrimen absent; submarginal rows of large foveolate impressions at lateral and posterior margins present, sometimes punctures are incomplete appearing as a crenulate line. Metacoxae (Fig. 5d, 10g) transverse–oval, widely separated. Hind wings well developed; anal lobe absent, single anal vein strongly reduced, medial fleck undivided.
Legs
Trochanterofemoral attachment oblique; femora subcylindrical with weak cavity along inner margin to receive tibia; tibia gradually widening towards tarsus, surrounded by short, stout spines. Tarsal formula 3-3-3 in both sexes (Fig. 7d); tarsomeres simple with tarsomere 2 weakly lobed ventrally; tarsal claws simple, at most with weak basal angulation; empodium distinct, unisetose.
Abdomen (Fig. 5d, i, 8i, 10g, 11i)
With five freely articulated ventrites; ventrite 1 as long as ventrites 2–5 together. Abdominal ventrite 1 with a row of foveolate punctures below each metacoxa and sometimes punctures are incomplete forming a crenulate submarginal line.
|
Male terminalia and genitalia
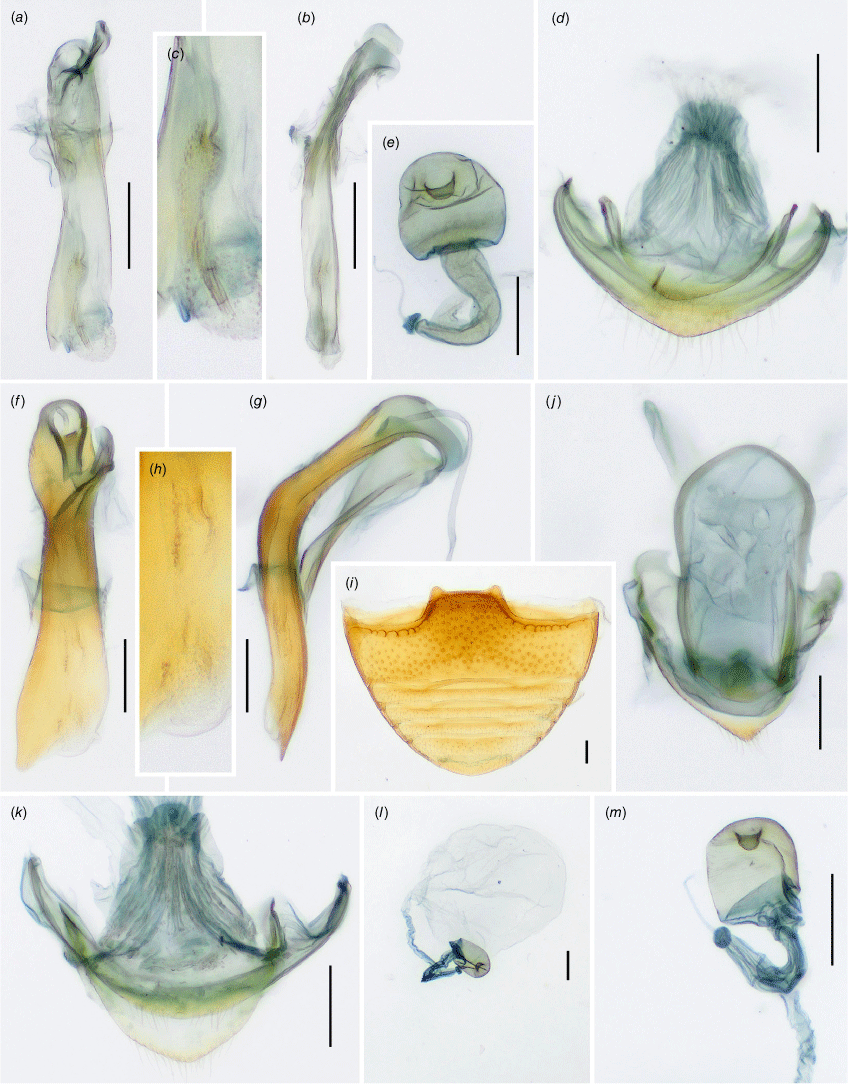
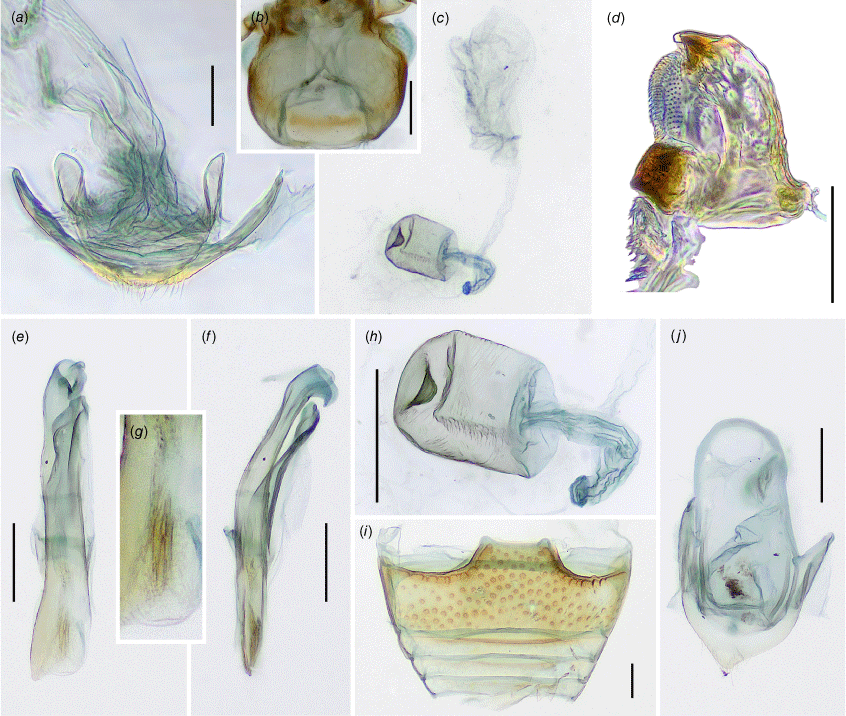
Sternite 9 of male genital segment (Fig. 5e, j, 8j, 11j) weakly developed, membranous with a pair of sclerotised apophyses fused roundly at apex and with a pair of short, additional apophyses basally. Aedeagus (Fig. 5a–c, f–h, 8a–c, f–h, 11e–g) well sclerotised, comparatively long, curved; resting on the side when retracted. Tegmen in form of short ring in mid-length of median lobe (=penis), tegminal strut long, weakly sclerotised, flattened.
Female genitalia
Ovipositor (Fig. 8d, k, 11a) weakly sclerotised without coxites; bursa copulatrix short, with apical outlet of sperm duct; sperm duct attached to broad connection between spermatheca and accessory gland. Spermatheca (Fig. 8e, m, 11h) short–oval, weakly sclerotised, with small sclerotised pocket at apex; accessory gland (Fig. 8l, 11c) large, rounded and membranous.
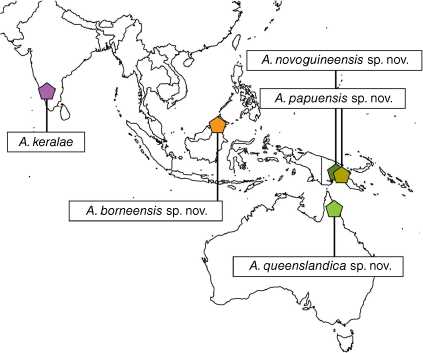
Distribution
Australia (Queensland), India (Kerala), Malaysia (Borneo), Papua New Guinea (Fig. 4).
Species descriptions
Anamycetaea keralae Strohecker, 1975
Type material examined
Holotype. Male, ‘INDIA, Kerala, Cardamon H, 25.xi.72, Valara Fall 450–500 m, Besuchet, Löbl, Mussard/Anamycetaea keralae, Holotype, Strohecker 1974’ (MHNG).
Diagnosis
This species can be easily distinguished from congeners by the antennae having 11 antennomeres instead of 10 (Fig. 5a), and by large foveolate punctures on the metaventrite near internal border of metacoxa and on abdominal ventrite 1 (Fig. 5c, d). In Anamycetaea borneensis sp. nov. similar patches of foveolate punctures are present, however, A. keralae can be easily distinguished from that species by the antennae composed of 11 antennomeres, the larger foveolate punctures on the metaventrite and abodminal ventrite 1 (Fig. 5c, d), and by lacking the longitudinal median carina in the anterior border of the metaventrite and a pair of oblique carinae in the anterior margin of abdominal ventrite 1, below each metacoxa (Fig. 5d, 6d).
Redescription
Body (Fig. 3a)
1.4 mm long, 1.38 times as long as wide. Colouration reddish-brown.
Head
Antenna with 11 antennomeres; antennomeres 3 and 11 slightly longer than wide. Distance between labial palps at insertion approximately as wide as the length of palpomere 1 along external margin (Fig. 5b).
Prothorax
Pronotum (Fig. 5a) 2.1 times as wide as long, widest near basal fifth, 1.7 times wider at widest part than on front angles; lateral margins continuously curved, coarsely crenulate throughout; area between lateral edge and carina weakly broadening from anterior angle along 2/5 length, subsequently strongly narrowing posteriorly, ~0.5 times as wide near posterior angle as at anterior. Prosternum (Fig. 5b) with area anterior to procoxae surrounded by carinae ~6 times as wide as long; with short, weakly developed longitudinal carina in front of prosternal intercoxal process (Fig. 5b); prosternal process ~1.8 times longer than wide.
Pterothorax
Elytra 1.00 mm long, as long as wide, 3.3 times as long as pronotum and 1.35 times as wide as pronotum. Third elytral stria with ~13 punctures in anterior half. Metaventrite (Fig. 5c) with anterior margin regularly bordered, with patches of large foveolate punctures near internal border of metacoxae.
Abdomen
Ventrite 1 with anterior margin regularly bordered (Fig. 5d, 6d); with surface covered with large foveolate punctures.
Male terminalia and genitalia
Sternite 9 of male genital segment (Fig. 6e) weakly developed, membranous; tergite 10 rounded apically. Penis (Fig. 6a–c) in inner view with large, lateral, subtriangular membranous projection, with numerous distinct v-shaped sclerites in median part; in lateral view sinuate, broad in basal part and narrowing toward apex, apex pointed, slightly curved outwards.
Female genitalia
Unknown.
Remarks
As pointed out by Strohecker (1975) in the original description, the holotype is a teratological individual as the left eye is not developed.
Distribution
India: Kerala (Fig. 4).
Anamycetaea borneensis, sp. nov.
Type material examined
Holotype. Male, MALAYSIA, Borneo: ‘Sabah: Kibongol V., 7 km N Tambunan, 700 m, 20.V.1987, Burckhardt, Löbl’ (MHNG).
|
Diagnosis
This species can be easily distinguished from all congeners in having the anterior margin of the metaventrite provided with a longitudinal, pointed median carina reaching anterior fourth length of metaventrite (Fig. 7d), the anterior margin of abdominal ventrite 1 with oblique carina below each metacoxa reaching approximately half length of ventrite (Fig. 6i), the posterior margin of metaventrite with patches of foveolate punctures near internal margins of metacoxae (Fig. 7d) and strongly transverse antennomeres 3–7 (Fig. 7c). Anamycetaea keralae has similar patches of foveolate punctures near the posterior margin of the metaventrite (Fig. 5c), however in A. borneensis sp. nov. these punctures are distinctly smaller than in A. keralae (Fig. 5c, 7d). Moreover A. borneensis can be easily distinguished from that species in having antenna composed of 10 antennomeres (Fig. 7c), and the longitudinal carinae on metaventrite and abdominal ventrite 1 (as described above) (Fig. 6i).
Description
1.56 mm long, 1.6 times as long as wide. Colouration pale brown.
Head
Antenna (Fig. 7c) with 10 antennomeres; antennomere 3–9 shorter than wide; 10 approximately as long as wide. Distance between labial palpi at insertion ~2/3 the length of palpomere 1 at external margin (Fig. 7b).
Prothorax
Pronotum (Fig. 7e) 1.9 times as wide as long, widest around basal 2/5, 1.55 times wider at widest part than on front angles; lateral margins curved at basal half, converging anteriorly; area between lateral edge and carina widest near anterior angles and subsequently scarcely narrowing posteriorly, more distinctly along basal fifth, ~0.65 times as wide at posterior angle as at maximum width. Prosternum (Fig. 7b) with area anterior to procoxae surrounded by carinae ~5 times wider than long; with long, distinct longitudinal carina in front of prosternal process; triangular area of prosternal process ~1.7 times longer than wide.
Pterothorax
Elytra (Fig. 7a) 1.05 mm long, approximately as long as wide, 2.9 times as long as pronotum and 1.4 times as wide as pronotum. Third elytral stria with ~11–12 punctures in anterior half, distance between punctures ~1.5–2.0 puncture diameters; foveolate punctures possessing additional point (Fig. 7a). Metaventrite (Fig. 7d) with anterior margin provided with median, pointed longitudinal carina reaching anterior fourth length of metaventrite and posterior margin with patches of large foveolate punctures near internal margins of metacoxae.
Abdomen
Abdominal ventrite 1 with anterior margin provided with oblique carina near internal margin of each metacoxa reaching approximately half length of ventrite (Fig. 6i); foveolate punctures moderately large, widely distributed.
Male terminalia and genitalia
Sternite 9 of male genital segment poorly developed (Fig. 6j), membranous; tergite 10 narrowly rounded at apex. Penis in inner view (Fig. 6f) with sides sub-parallel for most of the length and strongly narrowing apically, endophallus (Fig. 6h) with distinct sclerite with a hook-shaped apex; in lateral view (Fig. 6g) continuously rounded, broad in basal part and narrowing toward apex, apex pointed.
Female genitalia
Unknown.
Etymology
The name of this new species refers to Borneo island, the type locality.
Distribution
Malaysia (Borneo) (Fig. 4).
Anamycetaea novoguineensis, sp. nov.
Type material examined
Holotype. Male: ‘PAPUA NEW GUINEA, Mt Wilhelm transect: 200 m (–5.739897, 145.3297). FIT understorey. 28-30.x.2012, FIT-MW200-K-2/8-d04 (P0789). Coll. by Dilu, Ray, Novotny, Leponce’ (MIZ).
Paratypes. ‘PAPUA NEW GUINEA, Mt Wilhelm transect: 200 m (–5.739897, 145.3297). FIT understorey. 9-11.xi.2012, FIT-MW200-P-8/8-d16 (P0835, vial 14281). Coll. by Dilu, Ray, Novotny, Leponce’ (1 female: MIZ); same data except: ‘30.x-1.xi.2012, FIT-MW200-P-3/8-d06 (P0830 vial 14283)’ (1 male: MIZ); same data except: ‘7-11.xi.2012, FIT-MW200-O-7/8-d14 (P0826 vial 7460)’ (1 female: MIZ); same data except: ‘28-30.x.2012, FIT-MW200-M-2/8-d04 (P0805 vial 7486)’ (1 female: MIZ); same data except: ‘30.x-1.xi.2012, FIT-MW200-P-3/8-d06 (P0846 vial 14338)’ (1 female: MIZ); same data except: ‘3-5.xi.2012, FIT-MW200-N-5/8-d10 (P0816 vial 06577)’ (1 female: MIZ); same data except: ‘7-9.xi.2012, FIT-MW200-Q-7/8-d14 (P0842 vial 7490)’ (1 female: MIZ); same data except: ‘28-30.x.2012, FIT-MW200-M-2/8-d04 (P0837 vial 7477’ (1 female: MIZ); ‘PAPUA NEW GUINEA, Mt Wilhelm transect: 200 m (–5.741031, 145.3294). FIT understorey. 2-4.xi.2012, FIT-MW200-B-5/8-d09 (P0720, vial 7171). Coll. by Dilu, Ray, Novotny, Leponce.’ (1 male: MIZ); ‘PAPUA NEW GUINEA, Wanang transect: 175 m (–5.22767, 145.0797). FIT understorey. 24-26.xi.2012, FIT-WAN-P-4/8-d08(P0671 vial 17649. Coll. by Gewa, Damag, Novotny, Leponce’ (1 female: NMPC); same data except: ‘18-20.xi.2012, FIT-WAN-B-1/8-d01 (P0556 vial 22298)’ (1 female: NAIC); same data except: ‘24-26.xi.2012, FIT-WAN-N-4/8-d08 (P0655 vial 2455)’ (1 female: MNHN); same data except: ‘25-26.xi.2012, FIT-WAN-S-4/8-d08 (P0695 vial 2439)’ (1 female: MIZ); same data except: ‘20.xi-2.xii.2012, FIT-WAN-F-7/8-d13 (P0594 vial 17790)’ (1 female: IECA); same data except: ‘20-22.xi.2012, FIT-WAN-D-2/8-d03 (P0573 vial 17617)’ (1x: MIZ); same data except: ‘20-22.xi.2012, FIT-WAN-I-2/8-d03 (P0613 vial 17622)’ (1 female: IRSNB).
Diagnosis
This species is very similar to both Anamycetea queenslandica sp. nov. and A. papuensis sp. nov. in having the antenna composed of 10 antennomeres, the simply bordered anterior margins of the metaventrite and abdominal ventrite 1 without longitudinal carinae and the posterior margin of the metaventrite without patches of foveolate punctures. However, A. novoguineensis can be distinguished from these species by the antennomere 3 ~1.5 times as long as wide (Fig. 8b) (approximately as long as wide in A. queenslandica), the area between lateral pronotal edge and carina narrowed at posterior angle to ~0.3 times the maximum width (Fig. 8a) (scarcely narrowed in A. queenslandica and ~0.65 as wide as maximum width in A. papuensis), the elytra with proportionally larger foveolate punctures, with third stria provided with ~12–13 punctures in anterior half and distance between punctures ~0.5–1.0 puncture diameters (Fig. 8a, c) (~1.5–2.0 diameters in A. queenslandica and A. papuensis).
|
Description
1.4–1.8 mm long, 1.45 times as long as wide. Colouration pale chestnut brown.
Head
Antenna with 10 antennomeres; antennomere 3~1.5 times as long as wide, antennomere 10 approximately as long as wide. Distance between labial palpi at insertion approximately as wide as the length of palpomere 1 at external margin.
Prothorax
Pronotum 2.0 times as wide as long, widest near basal fifth, 1.7 times wider at widest part than on front angles; lateral margins almost straight, converging anteriorly, rounded at basal fifth, weakly crenulate at basal half; area between lateral edge and carina weakly broadening towards basal fifth, subsequently strongly narrowing posteriorly to ~0.3 times the maximum width. Prosternum (Fig. 8b) with area anterior to procoxae surrounded by carinae, almost 5 times as wide as long; with long, distinct longitudinal carina in front of procoxal process; prosternal process ~1.7 times longer than wide.
Pterothorax
Elytra (Fig. 8a) 1.00–1.10 mm long, approximately as long as wide, 2.6–2.7 times as long as pronotum and 1.3 times as wide as pronotum. Third elytral stria with ~12–13 punctures in anterior half, distance between punctures ~0.5–1.0 puncture diameters (Fig. 8c); foveolate punctures smooth inside, without additional point (Fig. 8c). Metaventrite with anterior margin regularly bordered and posterior margin without foveolate punctures near metacoxae.
Abdomen
Abdominal ventrite 1 with anterior margin regularly bordered; foveolate punctures moderately large.
Male terminalia and genitalia
Sternite 9 of male genital segment weakly developed, membranous; tergite 10 slightly acuminate apically. Penis (Fig. 9a–c) in inner view in basal half with sides sub-parallel, slightly widening apically, with membranous gonopore at apex, with distinct paired endosclerite in apical part; in lateral view comparatively narrow, weakly curved, almost sub-parallel, with blunt apex.
Female terminalia and genitalia
Ovipositor (Fig. 9d) with bursa copulatrix short; proctiger subacuminate apically. Spermatheca (Fig. 9e) short–oval, approximately as long as wide, comparatively well sclerotised; accessory gland approximately as large as spermatheca, membranous, rounded.
Etymology
The name of this new species refers to the island, New Guinea, the type locality.
Habitat
This species is known from mixed alluvium forests on the lower part (175–200 m ASL) of Mount Wilhelm, central Papua New Guinea. All known specimens were collected using flight intercept traps in the forest understorey.
Distribution
Papua New Guinea (Fig. 4).
Anamycetaea papuensis, sp. nov.
Type material examined
Holotype. Male: ‘PAPUA NEW GUINEA, Mt Wilhelm transect: 1200 m (–5.720874, 145.2695). FIT understorey. 8.x-11.xi.2012, FIT-MW1200-E-8/8-d15 (P1527, vial 16879). Coll. by Philip, Alois, Novotny, Leponce’ (MIZ).
Paratypes. ‘PAPUA NEW GUINEA, Mt Wilhelm transect: 1200 m (–5.720874, 145.2695). FIT understorey. 31.x-2.xi.2012, FIT-MW1200-J-4/8-d07 (P1563, vial 17310). Coll. by Philip, Alois, Novotny, Leponce’ (1 male: MIZ); same data except: ‘6-8.xi.2012, FIT-MW1200-D-7/8-d13 (P1518 vial 17054)’ (1: MIZ); same data except: ‘26-28.x.2012, FIT-MW1200-K-1/8-d02 (P1568, vial 17122)’ (1 female: MIZ); same data except: ‘30.x-1.xi.2012, FIT-MW1200-L-3/8-d06 (P01578 vial 17625)’ (1 female: IRNSB); same data except: ‘29-31.x.2012, FIT-MW1200-I-3/8-d05 (P1554 vial 17160)’ (1 female: MIZ): same data except: ‘28-30.x.2012, FIT-MW1200-K-2/8-d04 (P1569 vial 17302)’ (1 female: MIZ); same data except: ‘30.x-01.xi.2012, FIT-MW1200-O-3/8-d06 (P1602 vial 18868)’ (1 female: NAIC); ‘PAPUA NEW GUINEA, Mt Wilhelm transect: 1200 m (–5.721022, 145.2703). FIT understorey. 2-4.xi.2012, FIT-MW1200-B-5/8-d09 (P1500 vial 14070). Coll. by Dilu, Ray, Novotny, Leponce’ (1 female: MIZ); ‘PAPUA NEW GUINEA, Mt Wilhelm transect: 700 m (–5.731961, 145.2522). FIT understorey. 28-30.x.2012, FIT-MW700-M-2/8-d04 (P1195). Coll. by Philip, Alois, Novotny, Leponce’ (1 female: MNHN); ‘PAPUA NEW GUINEA, Mt Wilhelm transect: 700 m (–5.732514, 145.2568). FIT understorey. 8-10.xi.2012, FIT-MW700-C-8/8-d15 (P1121 vial 7340). Coll. by Keltim, Uma, Novotny, Leponce’ (1 female: IRSNB).
Diagnosis
This species is very similar to Anamycetea queenslandica sp. nov. and A. novoguineensis sp. nov. in having the antennae composed of 10 antennomeres, the anterior margins of the metaventrite and abdominal ventrite 1 simply bordered without longitudinal carinae and the posterior margin of the metaventrite without patches of foveolate punctures. However, A. papuensis can be distinguished from these species by the larger body size (Fig. 3d), the longer antennae with antennomere 3 ~1.4 times as long as wide (Fig. 10a, b) (approximately as long as wide in A. queenslandica), the area between lateral pronotal edge and carina ~0.65 as wide at posterior angle as at maximum width (Fig. 10a) (strongly narrowed to ~0.3 times the maximum width in A. novoguineensis; Fig. 8a and scarcely narrowed in A. queenslandica Fig. 11d), the elytra with smaller foveolate punctures and third elytral stria with ~14–15 punctures in anterior half, with distance between punctures ~1.5–2.0 puncture diameters (Fig. 10a) (~0.5–1.0 diameters in A. novoguineensis).
Description
1.9–2.4 mm long, 1.50 times as long as wide. Colouration dark chestnut brown.
Head
Antenna with 10 antennomeres; antennomere 3 ~1.4 times as long as wide, antennomere 10 slightly longer than wide. Distance between labial palpi at insertion approximately as wide as the length of palpomere 1 at external margin (Fig. 10b).
Prothorax
Pronotum (Fig. 10a) 2.3 times as wide as long, widest near basal fifth, 1.6 times wider at widest part than on front angles; lateral margins almost straight, converging anteriorly, weakly crenulate at basal half; area between lateral edge and carina of the same width from anterior angle to basal fifth, subsequently distinctly narrowing posteriorly to ~0.65 times as wide near posterior angle as at anterior. Prosternum (Fig. 10b) with area anterior to procoxae surrounded by carinae, ~5 times as wide as long; with long, distinct longitudinal carina in front of procoxal process; prosternal process ~1.6 times longer than wide.
Pterothorax
Elytra (Fig. 10a) 1.50–1.60 mm long, approximately as long as wide, 2.9–3.0 times as long as pronotum and 1.32–1.37 times as wide as pronotum. Third elytral stria with ~14–15 punctures in anterior half, distance between punctures ~1.5–2.0 puncture diameters; the foveolate punctures smooth inside, without additional point (Fig. 10c, f). Metaventrite with anterior margin regularly bordered and posterior margin without patches of foveolate punctures near metacoxae.
Abdomen (Fig. 9i)
Abdominal ventrite 1 with anterior margin regularly bordered; foveolate punctures moderately large.
Male terminalia and genitalia
Sternite 9 of male genital segment (Fig. 9j) comparatively well developed, sclerotised at least medially, somewhat angular posteriorly; tergite 10 acuminate apically. Penis (Fig. 9f–h) in inner view distinctly widening apically, with small membranous gonopore at apex, with a few indistinct endosclerites; in lateral view strongly curved, almost sub-parallel, with pointed apex.
Female terminalia and genitalia
Ovipositor (Fig. 9k) with bursa copulatrix short; proctiger subacuminate at apex. Spermatheca (Fig. 9m) regularly short–oval, ~1.3 times longer than wide, weakly sclerotised; accessory gland (Fig. 9l) very large, much larger than spermatheca, membranous, rounded.
Etymology
The name of this new species refers to the country of origin, Papua New Guinea.
Habitat
This species is known from mixed evergreen forests on the low slopes of Mount Wilhelm (700–1200 m ASL) in central Papua New Guinea. All known specimens were collected using flight intercept traps in the forest understorey.
Distribution
Papua New Guinea (Fig. 4).
Anamycetaea queenslandica, sp. nov.
Type material examined
Holotype. Male, AUSTRALIA: ‘Daintree NE QLD, Thompson Creek, 16.06.31S 145.26.25E, 140 m, Log Knock dn, 08.12.98, Coll. Simon Grove’ (ANIC).
Paratypes. AUSTRALIA, ‘12.44S 143.14E QLD, 3 km ENE Mt Tozer, 28 June–4 July 1986, J.C. Cardale, ex pantraps’ (1 female: ANIC); ‘Lockerbie, Cape York, N. QLD, 6–10.VI.1969, G.B. Monteith/U.Q.I.C., Loan 979’ (1 male: QMB; 1 female, totally dissected MIZ).
Diagnosis
This species is very similar to Anamycetea papuensis sp. nov. and A. novoguineensis sp. nov. in having the antenna composed of 10 antennomeres, the simply bordered anterior margins of the metaventrite, abdominal ventrite 1 without longitudinal carinae and the posterior margin of the metaventrite without patches of foveolate punctures. However, A. queenslandica can be distinguished from these species by the shorter antennae with antennomere 3 approximately as long as wide (Fig. 11b), the pronotum with area between lateral edge and carina nearly of the same width throughout the length (Fig. 11a, d) and the foveolate punctures on the elytra possessing an additional point (Fig. 11h).
Description
1.58 mm long, 1.5 times as long as wide. Colouration pale to dark reddish-brown.
Head
Antenna (Fig. 11b) with 10 antennomeres; antennomere 3 subquadrate, 10 wider than long. Distance between labial palpi at insertion slightly shorter than length of palpomere 1 at external margin (Fig. 11c).
Prothorax
Pronotum (Fig. 11d) 1.85 times as wide as long, widest near basal 2/5, 1.6 times wider at widest part than on front angles; lateral margins almost continuously curved, weakly crenulate throughout; area between lateral edge and carina approximately the same width throughout the length. Prosternum (Fig. 11f) with area anterior to procoxae surrouned by carinae, ~4 times wider than long; with long, distinct longitudinal carina in front of procoxal process; subtriangular area of prosternal process ~2.3 times longer than wide.
Pterothorax
Elytra (Fig. 11a) 1.05 mm long, 1.0 times as long as wide, 2.6 times as long as pronotum and 1.4 times as wide as pronotum. Third elytral stria with ~10–11 punctures in anterior half, distance between punctures ~1.5–2.0 puncture diameters; foveolate punctures possessing additional point (Fig. 11h). Metaventrite (Fig. 11e) with anterior margin without longitudinal carina and posterior margin without patches of foveolate punctures.
Abdomen
Abdominal ventrite 1 (Fig. 11g, 12i) with anterior margin regularly bordered; foveolate punctures large and widely scattered.
Male terminalia and genitalia
Sternite 9 of male genital segment (Fig. 12j) weakly sclerotised at apical margin, rounded at both sides; tergite 10 acuminate medially. Penis (Fig. 12e–g) in inner view with sides sub-parallel along basal 2/3, slightly widening apically, with small membranous gonopore at apex, with distinct endosclerite in apical part; in lateral view weakly curved, narrower along basal half, with blunt apex.
Female terminalia and genitalia
Ovipositor (Fig. 12a) with bursa copulatrix elongate; proctiger rounded at apex. Spermatheca (Fig. 12h) regularly short–oval, scarcely longer than wide, weakly sclerotised; accessory gland larger than spermatheca, membranous, oval (Fig. 12c).
Etymology
The name of this new species refers to the type locality, Queensland in Australia.
Habitat
Specimens of this species were collected on logs using pan traps.
Distribution
Australia: Queensland (Fig. 4).
Key to the species of Anamycetaea|
1. Antenna with 10 antennomeres (Fig. 7c, 11b); lateral margins of pronotum weakly crenulate with at most small, blunt teeth (Fig. 7a, 8a, 10a, 11d); prosternum with long, well developed median carina in front of prosternal process (Fig. 7b, 8b, 10b, 11f); abdominal ventrite 1 with comparatively smaller foveolate punctures (Fig. 6i, 7d, 9i, 11g, 12i) |
|
2. Metaventrite regularly bordered anteriorly, without longitudinal carina (Fig. 11e); abdominal ventrite 1 with anterior margin regularly bordered (Fig. 9i, 11g); metaventrite without patches of foveolate punctures near internal margins of metacoxae (Fig. 11e) |
|
3. Antennomere 3 at least 1.3 times as long as wide (Fig. 8b); area between lateral pronotal edge and carina narrowed towards posterior angle to at least 0.65 times the max. width (Fig. 8a, 10a); large foveolate punctures on elytra without additional point (Fig. 8c, 10c, f); (Papua New Guinea) |
|
4. Body length 1.4–1.8 mm (Fig. 3e, 8a); elytra with larger foveolate puctures, third elytral stria with ~12–13 punctures in anterior half, with distance between punctures ~0.5–1.0 puncture diameters (Fig. 8c); area between lateral edge and carina strongly narrowed at posterior angle to ~0.3 times the maximum width (Fig. 8a); aedeagus as in Fig. 9a–c |
Discussion
Anamycetaea was neglected for decades since the original description by Strohecker (1975). This monotypic genus based on a single specimen was a mystery and the holotype was not studied in detail until the present project. The classification was therefore based on a limited set of morphological characters.
The placement of this genus in the subfamily Mycetaeinae by Strohecker (1975) was mainly based on overall similarities to other genera included in Mycetaeinae, according to Strohecker’s (1953) concept of this subfamily. At that time, Mycetaeinae was formed by a heterogeneous assemblage of beetles with linear tarsi that included members of currently recognised endomychid subfamilies (e.g. Xenomycetinae, Leiestinae; Strohecker 1962; Pakaluk et al. 1994; Tomaszewska 2000, 2005) or even independent families (e.g. Mycetaeidae and Anamorphidae; Robertson et al. 2015).
The placement of Anamorphinae in the Cerylonid Series of Crowson (1955) (currently superfamily Coccinelloidea) has been a subject of discussion over the time. Sasaji’s (1978) discovery of laterally closed mesocoxal cavities in some genera of Mycetaeinae and the tentorium with anterior arms widely separated, characters not previously known within the family, brought further clarity to the Mycetaeinae problem. Sasaji (1978) established the subfamily Mychotheninae based on unique characters discovered and subsequently elevated this to family status (Sasaji 1987, 1990). In subsequent research this group was regarded as a subfamily within Endomychidae (under the name Anamorphinae due to a nomenclatorial priority) and the family status was not widely accepted (Pakaluk et al. 1994; Lawrence and Newton 1995). Also, the first phylogenetic study on the family Endomychidae by Tomaszewska (2000) and the followed-up study (Tomaszewska 2005) supported the monophyly of Anamorphinae within Endomychidae. The character postulated as synapomorphy for this subfamily was the mesocoxal cavities that were widely closed outwardly by sterna (Tomaszewska 2000) and all former Mycetaeinae genera having closed mesocoxal cavities were subsequently classified in Anamorphinae (Shockley et al. 2009).
However, the hypotheses of the relationships of the handsome fungus beetles based on morphological evidence (Tomaszewska 2000, 2005) versus those based on molecular sequences (Robertson et al. 2015) are contradictory in terms of boundaries of Endomychidae as a monophyletic taxon and the sister relationships. Tomaszewska (2000, 2005) recovered a monophyletic Endomychidae containing 12 subfamilies and placement as sister group to Coccinellidae and Coccinellidae + Corylophidae respectively. This scheme was rejected by Robertson et al. (2015) based on results of molecular analysis The later study confirmed Anamorphidae as an independent lineage as hypothesised by Sasaji (1978) and related to Corylophidae. All earlier molecular analyses that included anamorphines recovered these outside the core Endomychidae (e.g. Hunt et al. (2007) recovered Anamorphinae as sister to Alexiidae and Robertson et al. (2008, 2013) recovered Anamorphinae as sister to Corylophidae, whereas in Bocak et al. (2014) Anamorphinae is nested within Corylophidae).
Despite some morphological features that separate Anamorphinae from core Endomychidae, as outlined in Robertson et al. (2015), none of these seem to be an unambiguous synapomorphy of this group, i.e. adults with anterior arms of tentorium separate (fused in core Endomychidae, except one species of Merophysiinae), mesocoxal cavities widely closed by the meso- and metaventrite (open to mesepimeron in core endomychids, except Merophysiinae and Pleganophorinae, where these are narrowly closed), pretarsal claws modified (not in all Anamorphidae) and penis broad, stout, weakly curved with endophallic sclerites (penis variable in core Endomychidae). By contrast, there are numerous characters that unite Anamorphidae with the remaining handsome fungus beetles, including the presence of frontoclypeal suture on the head, pronotum with basal or lateral sulci or modifications, round procoxa, the presence of postcoxal pits on meso- and metaventrite and five pairs of abdominal spiracles. At the same time, these morphological characters placed Anamorphidae distantly from Corylophidae in our tree resulting from the morphological data analysis (Fig. 2). We support the monophyly of Anamorphidae (within Endomychidae) by the tentorium with anterior arms separate and mesocoxal cavity widely closed outwardly. Both characters were revealed as homoplastic.
The incongruence between the morphological and molecular analyses renders currently accepted groupings that are based on Robertson et al. (2015) difficult to diagnose and in which morphological homologies are not clear. The relationships among members of Endomychidae sensu lato including Mycetaeidae, Eupsilobiidae, Anamorphidae and also Coccinellidae remain to be fully clarified. Similarly, the systematic position of the endomychid subfamilies Danascelinae and Xenomycetinae remains to be resolved since these were not represented in the dataset of the analysis of Robertson et al. (2015). These problems will be addressed in the ongoing research of Arriaga-Varela, Tomaszewska & Szawaryn (in prep.) in which a full-evidence approach is being implemented.
Regardless of whether Anamorphidae is a monophyletic group at family level outside core Endomychidae (as revealed in our molecular analysis, Fig. 1) or a subfamily within Endomychidae (as shown in the tree resulting from our morphological analysis, Fig. 2), the genus Anamycetaea with five currently known species is recovered as unrelated to that group. The analyses of both molecular and morphological datasets revealed the genus as a distinct clade within the endomychine complex of Endomychidae sensu stricto. The phylogenetic placement as a part of the Stenotarsus clade in the subfamily Endomychinae is strongly supported in all our analyses.
The subfamily Endomychinae is strongly supported in our Bayesian (PP = 1.0) and ML analyses (BS = 99), although this was recovered as a monophyletic group without Endomychus in the morphological analysis. Based on morphology, there is one synapomorphy, ovipositor with deeply divided basal part of coxites (40:1) and one homoplastic character, pronotum with wide lateral margins (19:1) that support this group.
The clade Chondria + (Tharina + Anamycetaea) is united by a homoplastic feature, penultimate mesotarsomere slightly reduced and not enclosed within lobe of antepenultimate tarsomere (29:0). Some similarities observed between Anamycetaea with Tharina appeared as supporting characters resulting from our analysis, including pronotal anterior margin bordered with crenulate bordering line (20:1), tarsi 3-segmented (28:2) and 2-segmented labial palpi (44:1).
Our morphological analysis is simultaneously a confirmation of the placement of Tharina within Endomychinae. Arriaga-Varela et al. (2019) placed this unusual genus from the Neotropics in Endomychinae based on the general appearance suggesting a possible relationship with pantropical Stenotarsus or the Oriental and Australasian Chondria. Arriaga-Varela et al. (2019) pointed out that the ‘superficial similarities’ of Tharina with representatives of Stenotarsus and Chondria, such as transverse pronotum with wide, raised lateral margins, a pair of arcuate foveae at pronotal base, the general shape of the meso- and metaventrite, and the small body size and narrow tarsi with weakly lobed second tarsomere, must be tested.
Representatives of Tharina and Anamycetaea represent the smallest members of the subfamily Endomychinae (1.50–2.30 mm), and the three segmented tarsi in some species of Tharina and all species of Anamycetaea are unique within Endomychinae and in the endomychine complex sensu Robertson et al. (2015) (=‘higher Endomychidae’ sensu Tomaszewska 2005). Additionally, both genera show similarities in the mouth parts such as the two segmented labial palpi, and the stiff, broad setae in the maxillary galea and lacinia. One of the most intriguing issues to be addressed in further phylogenetic research is the biogeography of the Endomychinae, and relationships of such distantly distributed taxa as Tharina and Anamycetaea.
Anamycetaea has been restricted to Kerala, India but is now known from a much broader range, reaching through Malaysia (Borneo) to Papua New Guinea and Australia (Fig. 4). This is a small, homogenous group within Endomychinae, differing from all other genera of this subfamily in having a strongly carinate prosternum. Species of Anamycetaea are very similar to each other however, having distinct external distinguishing characters and the study of the male genitalia dispels doubts about the correct identification.
Collecting data and known ranges of the Anamycetaea species show a general allopatric distribution pattern. Even the two species from Papua New Guinea, A. novoguineensis and A. papuensis, described from Mount Wilhelm (Madang Province) were collected at different elevations, 175–200 and 700–1200 m ASL respectively, areas of distribution did not overlap and the species inhabited distinct microhabitats (mixed alluvium forest and mixed evergreen forest respectively). Similar species distribution patterns on Mount Wilhelm occur in other beetle groups such as megasternine hydrophilids (Szczepański et al. 2018).
Owing to the small amount of material available for study, the larvae of Anamycetaea and biological characteristics of these beetles remain a mystery.
Supplementary material
The tables, figures and files listed below are available as part of the supplementary material. Supplementary Table S1 lists specimens for molecular analyses with GenBank accession numbers. Supplementary Table S2 lists primers used for sequences newly obtained in this study. Supplementary File S1 provides a concatenate-aligned molecular matrix. Supplementary Table S3 lists morphological characters (modified after Tomaszewska 2000, 2005). Supplementary File S2 provides a morphological data matrix used for cladistic analysis in TNT format. Supplementary Fig. S1 shows a Bayesian inference analysis of the molecular dataset, full tree. Supplementary File S3 provides a Bayesian inference analysis of the molecular dataset, machine-readable full tree (NEXUS file). Supplementary Fig. S2 shows a maximum-likelihood analysis of the molecular dataset, full tree. Supplementary File S4 provides a maximum-likelihood analysis of the molecular dataset, machine-readable full tree (NEXUS file). Supplementary Fig. S3 shows a maximum-parsimony analysis of the morphological matrix – strict consensus of two most parsimonious trees obtained under equal weights. Supplementary File S5 provides a maximum-parsimony analysis of the morphological matrix – strict consensus of two most parsimonious trees obtained under equal weights, machine-readable tree. Supplementary material is available online.
Data availability
The sequence data that support this study are available at NCBI, with corresponding accessions available in Supplementary Table S1.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
This work was partly supported by the National Science Centre of Poland (Narodowe Centrum Nauki; grant number 2020/36/C/NZ8/00584 to E. Arriaga-Varela)
Acknowledgements
Adam Ślipiński (ANIC), Giulio Cuccodoro (MHNG) and Geoff Monteith (QMB) are acknowledged for the loan of the museum specimens used in this study. We thank Carl Wardaugh (Scion, New Zealand Forest Research Institute, New Zealand), Vojtěch Novotný (Faculty of Science, University of South Bohemia, Czech Republic) and Maurice Leponce (Royal Belgian Institute of Natural Sciences, Operational Directorate Natural Environment, Belgium) for allowing us to study the specimens collected in the framework of the ‘Our Planet Reviewed Papua-New-Guinea 2012–2013’ project. This project was set up by Pro-Natura International, the National Museum of Natural History (MNHN, France) and the Institut de Recher-che pour le Développement (IRD, France) in partnership with the Royal Belgian Institute of Natural Sciences, the New Guinea Binatang Research Center, the University of Papua New Guinea and the Divine Word University of Madang. Core funding was provided by the Prince Albert II of Monaco Foundation, the Stavros Niarchos Foundation, the Total Foundation, the Fondation d’entreprise EDF, the Fonds Pacifique, Spiecapag, Entrepose Contracting, the New-Caledonia Government, the Reef Foundation, FNRS (Belgium) and the Belgian National Lottery. All participants in this collaborative project, as part of the IBISCA experts' network patronned by Prof. R. K. Kitching, are thanked for their contributions. Specimens from this project were exported under the permit #012297 issued by the Department of Environment and Conservation (DEC, Port Moresby). We thank Magdalena Kowalewska-Groszkowska (MIZ) for help with SEM illustrations and Aleksandra Gwiazdowska (MIZ) for help with obtaining molecular sequences used in this study.
References
Arriaga-Varela, E, Tomaszewska, W, and Fikáček, M (2019). A new genus of Endomychinae (Coleoptera: Endomychidae) from the Neotropics with unusual mouthparts. Neotropical Entomology 48, 290–301.| A new genus of Endomychinae (Coleoptera: Endomychidae) from the Neotropics with unusual mouthparts.Crossref | GoogleScholarGoogle Scholar |
Bocak, L, Barton, C, Crampton-Platt, A, Chesters, D, Ahrens, D, and Vogler, AP (2014). Building the Coleoptera tree-of-life for >8000 species: composition of public DNA data and fit with Linnaean classification. Systematic Entomology 39, 97–110.
| Building the Coleoptera tree-of-life for >8000 species: composition of public DNA data and fit with Linnaean classification.Crossref | GoogleScholarGoogle Scholar |
Crowson RA (1955) ‘The Natural Classification of the Families of Coleoptera.’ (Nathaniel Lloyd: London, UK)
Dayrat, B (2005). Towards integrative taxonomy. Biological Journal of the Linnean Society 85, 407–415.
| Towards integrative taxonomy.Crossref | GoogleScholarGoogle Scholar |
Goloboff, PA, and Catalano, SA (2016). TNT version 1.5, including a full implementation of phylogenetic morphometrics. Cladistics 32, 221–238.
| TNT version 1.5, including a full implementation of phylogenetic morphometrics.Crossref | GoogleScholarGoogle Scholar |
Hunt, T, Bergsten, J, Levkanicova, Z, Papadopoulou, A, John, OS, Wild, R, Hammond, PM, Ahrens, D, Balke, M, Caterino, MS, Go’mez-Zurita, J, Ribera, I, Barraclough, TG, Bocakova, M, Bocak, L, and Vogler, AP (2007). A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318, 1913–1916.
| A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation.Crossref | GoogleScholarGoogle Scholar |
Kearse, M, Moir, R, Wilson, A, Stones-Havas, S, Cheung, M, Sturrock, S, Buxton, S, Cooper, A, Markowitz, S, Duran, C, Thierer, T, Ashton, B, Meintjes, P, and Drummond, A (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649.
| Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Crossref | GoogleScholarGoogle Scholar |
Lawrence JF, Newton AF (1995) Families and subfamilies of Coleoptera (with selected genera, notes, references and data on family-group names). In ‘Biology, phylogeny and classification of Coleoptera. Papers celebrating the 80th Birthday of Roy A. Crowson. Vol. 2’. (Eds J Pakaluk, SA Ślipiński) pp. 779–1006. (Muzeum i Instytut Zoologii PAN: Warsaw, Poland)
Lawrence, JF, Ślipiński, A, Seago, AE, Thayer, MK, Newton, AF, and Marvaldi, AE (2011). Phylogeny of the Coleoptera based on morphological characters of adults and larvae. Annales Zoologici 61, 1–217.
| Phylogeny of the Coleoptera based on morphological characters of adults and larvae.Crossref | GoogleScholarGoogle Scholar |
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In ‘2010 Gateway Computing Environments Workshop (GCE)’, 14 November 2010, New Orleans, LA, USA. INSPEC Accession Number 11705685. (IEEE)
| Crossref |
Minh, BQ, Nguyen, MAT, and von Haeseler, A (2013). Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30, 1188–1195.
| Ultrafast approximation for phylogenetic bootstrap.Crossref | GoogleScholarGoogle Scholar |
Nguyen, LT, Schmidt, HA, Von Haeseler, A, and Minh, BQ (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32, 268–274.
| IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies.Crossref | GoogleScholarGoogle Scholar |
Nixon KC (2002) ‘WinClada.’ (Published by the author: Ithaca, NY, USA)
Padial, JM, Miralles, A, De la Riva, I, and Vences, M (2010). The integrative future of taxonomy. Frontiers in Zoology 7, 16.
| The integrative future of taxonomy.Crossref | GoogleScholarGoogle Scholar |
Pakaluk, J, Ślipiński, SA, and Lawrence, J (1994). Current classification and family-group names in Cucujoidea (Coleoptera). Genus 5, 223–268.
Rambaut, A, Drummond, AJ, Xie, D, Baele, G, and Suchard, MA (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67, 901–904.
| Posterior summarization in Bayesian phylogenetics using Tracer 1.7.Crossref | GoogleScholarGoogle Scholar |
Robertson, JA, Whiting, MF, and McHugh, JV (2008). Searching for natural lineages within the Cerylonid Series (Coleoptera: Cucujoidea). Molecular Phylogenetics and Evolution 46, 193–205.
| Searching for natural lineages within the Cerylonid Series (Coleoptera: Cucujoidea).Crossref | GoogleScholarGoogle Scholar |
Robertson, JA, Ślipiński, A, Hiatt, K, Miller, KB, Whiting, MF, and McHugh, JV (2013). Molecules, morphology and minute hooded beetles: a phylogenetic study with implications for the evolution and classification of Corylophidae (Coleoptera: Cucujoidea). Systematic Entomology 38, 209–232.
| Molecules, morphology and minute hooded beetles: a phylogenetic study with implications for the evolution and classification of Corylophidae (Coleoptera: Cucujoidea).Crossref | GoogleScholarGoogle Scholar |
Robertson, JA, Ślipiński, A, Moulton, M, Shockley, FW, Giorgi, A, Lord, NP, McKenna, DD, Tomaszewska, W, Forrester, J, Miller, KB, Whiting, MF, and McHugh, JV (2015). Phylogeny and classification of Cucujoidea and the recognition of a new superfamily Coccinelloidea (Coleoptera: Cucujiformia). Systematic Entomology 40, 745–778.
| Phylogeny and classification of Cucujoidea and the recognition of a new superfamily Coccinelloidea (Coleoptera: Cucujiformia).Crossref | GoogleScholarGoogle Scholar |
Ronquist, F, Teslenko, M, Van Der Mark, P, Ayres, DL, Darling, A, Höhna, S, Larget, B, Liu, L, Suchard, MA, and Huelsenbeck, JP (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542.
| MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space.Crossref | GoogleScholarGoogle Scholar |
Sasaji, H (1978). Notes on the Japanese Endomychidae, with an establishment of a new subfamily (Coleoptera). The Memoirs of the Faculty of Education, Fukui University Series II (Natural Science) 28, 1–31.
Sasaji, H (1987). On the higher classification of the Endomychidae and their relative families (Coleoptera). Entomological Journal of Fukui 1, 44–51.
Sasaji, H (1990). The family Mychothenidae of Japan (Coleoptera). Esakia (Special Issue) 1, 65–75.
| The family Mychothenidae of Japan (Coleoptera).Crossref | GoogleScholarGoogle Scholar |
Shockley, FW, Tomaszewska, KW, and McHugh, JV (2009). An annotated checklist of the handsome fungus beetles of the world (Coleoptera: Cucujoidea: Endomychidae). Zootaxa 1999, 1–113.
| An annotated checklist of the handsome fungus beetles of the world (Coleoptera: Cucujoidea: Endomychidae).Crossref | GoogleScholarGoogle Scholar |
Strohecker HF (1953) Coleoptera Fam. Endomychidae. In ‘Genera Insectorum’. (Ed. P Wytsman) pp. 1–145. (Desmet-Verneuil: Bruxelles, Belgium)
Strohecker HF (1962) Key to the Genera of the United States (Endomychidae). In ‘The Beetles of the United States (A Manual for Identification). Part V. Suborder Polyphaga (cont.). Series Cucujiformia (cont.). Tenebrionoidea. Cucujoidea’. (Ed. RH Arnett) p. 801. (Catholic University of America Press: Washington, DC, USA)
Strohecker, HF (1975). Several new Endomychidae from India (Coleoptera). Revue Suisse de Zoologie 82, 625–628.
| Several new Endomychidae from India (Coleoptera).Crossref | GoogleScholarGoogle Scholar |
Szczepański, WT, Vondráček, D, Seidel, M, Wardhaugh, C, and Fikáček, M (2018). High diversity of Cetiocyon beetles (Coleoptera: Hydrophilidae) along an elevational gradient on Mt Wilhelm, New Guinea, with new records from the Bird’s Head Peninsula. Arthropod Systematics & Phylogeny 76, 323–347.
Tarasov, SI, and Solodovnikov, AY (2011). Phylogenetic analyses reveal reliable morphological markers to classify mega-diversity in Onthophagini dung beetles (Coleoptera: Scarabaeidae: Scarabaeinae). Cladistics 27, 490–528.
| Phylogenetic analyses reveal reliable morphological markers to classify mega-diversity in Onthophagini dung beetles (Coleoptera: Scarabaeidae: Scarabaeinae).Crossref | GoogleScholarGoogle Scholar |
Thompson JD, Gibson TJ, Higgins DG (2003) Multiple sequence alignment using ClustalW and ClustalX. In ‘Current protocols in Bioinformatics’. pp. 2.3.1–2.3.22. (Wiley)
| Crossref |
Tomaszewska, KW (2000). Morphology, phylogeny and classification of adult Endomychidae (Coleoptera: Cucujoidea). Annales Zoologici 50, 449–558.
Tomaszewska, W (2005). Phylogeny and generic classification of the subfamily Lycoperdininae with re-analysis of the family Endomychidae (Coleoptera: Cucujoidea). Annales Zoologici 55, 1–172.
Tomaszewska W (2010) Endomychidae Leach, 1815. In ‘Handbook of Zoology. Vol. 2, Coleoptera’. (Eds RAB Leschen, RG Beutel, JF Lawrence) pp. 442–454. (Walter de Gruyter GmbH and Co. KG)
Trifinopoulos, J, Nguyen, LT, von Haeseler, A, and Minh, BQ (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44, W232–W235.
| W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis.Crossref | GoogleScholarGoogle Scholar |
Will, KW, Mishler, BD, and Wheeler, QD (2005). The perils of DNA barcoding and the need for integrative taxonomy. Systematic Biology 54, 844–851.
| The perils of DNA barcoding and the need for integrative taxonomy.Crossref | GoogleScholarGoogle Scholar |