Arabidopsis plasma membrane intrinsic protein (AtPIP2;1) is implicated in a salinity conditional influence on seed germination
Phan Thi Thanh Hoai A B C * , Jiaen Qiu A B , Michael Groszmann
A B C * , Jiaen Qiu A B , Michael Groszmann  D , Annamaria De Rosa
D , Annamaria De Rosa  D , Stephen D. Tyerman
D , Stephen D. Tyerman  A B and Caitlin S. Byrt
A B and Caitlin S. Byrt  A B D *
A B D *
A Australian Research Council Centre of Excellence in Plant Energy Biology, Waite Research Precinct, University of Adelaide, Glen Osmond, SA, Australia.
B School of Agriculture, Food and Wine, and Waite Research Institute, Waite Research Precinct, University of Adelaide, Glen Osmond, SA, Australia.
C Faculty of Agriculture and Forestry, Tay Nguyen University, 567 Le Duan, BMT, Dak Lak, Vietnam.
D Australian Research Council Centre of Excellence for Translational Photosynthesis, Division of Plant Sciences, Research School of Biology, The Australian National University, Acton, ACT 2601, Australia.
Handling Editor: Frans Maathuis
Functional Plant Biology 50(8) 633-648 https://doi.org/10.1071/FP22260
Submitted: 9 November 2022 Accepted: 15 May 2023 Published: 6 June 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Dynamic changes in aquaporin gene expression occur during seed germination. One example is the ~30-fold increase in Arabidopsis thaliana PIP2;1 transcripts within 24 h of seed imbibition. To investigate whether AtPIP2;1 can influence seed germination wild-type Columbia-0, single (Atpip2;1) and double (Atpip2;1-Atpip2;2) loss-of-function mutants, along with transgenic 2x35S::AtPIP2;1 over-expressing (OE) lines and null-segregant controls, were examined. The various genotypes were germinated in control and saline (75 mM NaCl treatment) conditions and tested for germination efficiency, imbibed seed maximum cross sectional (MCS) area, imbibed seed mass, and seed Na+ and K+ content. Seed lacking functional AtPIP2;1 and/or AtPIP2;2 proteins or constitutively over-expressing AtPIP2;1, had delayed germination in saline conditions relative to wild-type and null-segregant seed, respectively. Exposure to saline germination conditions resulted in Atpip2;1 mutants having greater imbibed seed mass and less accumulated Na+ than wild-type, whereas lines over-expressing AtPIP2;1 had reduced imbibed seed mass and greater seed K+ content than null-segregant control seed. The results imply a role for AtPIP2;1 in seed germination processes, whether directly through its capacity for water and ion transport or H2O2 signalling, or indirectly through potentially triggering dynamic differential regulation of other aquaporins expressed during germination. Future research will aid in dissecting the aquaporin functions influencing germination and may lead to novel solutions for optimising germination in sub-optimal conditions, such as saline soils.
Keywords: aquaporins, Arabidopsis spp., hydraulic conductivity, ion channel, ion transport, physiology, salinity, seed viability, signalling, sodium transport, sucrose, transporter proteins.
Introduction
Seed germination involves many different cellular activities, such as hydration, mRNA transcription and translation, cell extension, cell cycle activation, mechanisms of reparation and organelle reassembly. These processes are influenced by hormone signalling and environmental factors (Bewley et al. 2013; Rosental et al. 2014). The first stage of germination is imbibition, which triggers the metabolic activity required to generate resources for translating proteins, for cell growth and division, and subsequent radicle protrusion (Rosental et al. 2014). Imbibition involves water absorption into a quiescent, dormant and dry seed. Water absorption initiates energy metabolism, regulation of oxidative status, DNA repair, cell cycle activation, energy reserve mobilisation and modification of hormonal status (Lutts et al. 2016). Water influx during seed imbibition is thought to occur mainly via a small opening called the micropyle (Manz et al. 2005). Previous studies have explored aquaporin roles in water transport in seeds, but it remains unclear what proportion of water movement across membranes in imbibing seeds is via aquaporins relative to other mechanisms (Bewley 1997; Fenner and Thompson 2005; Vander Willigen et al. 2006; Lutts et al. 2016).
Aquaporins belong to an extensive ancient family of major intrinsic proteins (MIPs). They function in facilitating passive movement of water and other solutes across membranes in plants and animals. Plant aquaporins can be subdivided into subfamilies according to their sequence homology (Chaumont et al. 2001; Abascal et al. 2014; Tyerman et al. 2021). Seed embryonic axes are known to be equipped with aquaporins (Obroucheva 2013). Tonoplast intrinsic proteins (TIPs) are thought to provide water inflow to enlarging vacuoles in maturing seeds and during vacuole transformation to protein-storage vacuoles, and water outflow from water displacement by deposited proteins (Maurel et al. 1997; Zhang et al. 2007; Zhou et al. 2007). Plasma membrane intrinsic protein (PIP) isoforms have been implicated in influencing seed germination responses to osmotic stress. For example, silencing of OsPIP1;1 and OsPIP1;3 reduced seed germination and over-expression of OsPIP1;3 promoted germination under water-stress conditions (Liu et al. 2007). The roles of OsPIPs in germination have been associated with their responses to nitric oxide and reactive oxygen species signalling (Guha et al. 2021; Matsunami et al. 2022).
In maturing Arabidopsis thaliana embryos, transcripts of 6 out of 35 aquaporins encoded in the genome are notably abundant and these include AtPIP1;3, AtPIP1;4, AtPIP2;7/2;8, AtTIP3;1 and AtTIP3;2, while the most abundant aquaporin transcripts in dry seed are AtNIP1;2, AtPIP1;2, AtSIP1;1 and AtTIP3;2 (Hoai et al. 2020). In particular, AtTIP3;1 and AtTIP3;2 accumulate during seed maturation, and these two isoforms along with AtTIP4;1 have roles influencing germination (Footitt et al. 2019). AtTIP3 isoforms were shown to act antagonistically to modulate responses of Arabidopsis seed germination to ABA where TIP3;1 and TIP4;1 are upregulated during seed germination under osmotic stress conditions (Footitt et al. 2019). Analysis of Arabidopsis lines lacking or over-expressing AtTIP3 and AtTIP4 isoforms revealed that AtTIPs have roles in sensing of soil temperature and moisture content (Footitt et al. 2019). During germination TIP3;1 transcript abundance decreases and transcripts for PIP2;1, PIP2;7, TIP1;1, TIP1;2 and TIP4;1 become more abundant indicating that this set of aquaporins are candidates for being involved in rapid cellular expansion that occurs during post-germination growth (Hoai et al. 2020). In Zea mays, expression of ZmPIP1s, ZmPIP2s, ZmTIP1-1, ZmTIP2s and ZmTIP3-3 is relatively high in roots during early seed germination around 48–60 h after imbibition in comparison to expression of the ACTIN 1 housekeeping gene, indicating that multiple aquaporins are likely to be involved in early seedling growth (Su et al. 2022).
Individual aquaporin isoforms can have multiple roles (Tyerman et al. 2021). For example, AtPIP2;1 has been reported to be involved in transport of water, H2O2 and CO2 in plant cells (Dynowski et al. 2008; Bienert and Chaumont 2014; Grondin et al. 2015; Wang et al. 2016; Groszmann et al. 2017; Rodrigues et al. 2017); in roots AtPIP2;1 has been reported to be involved in lateral root emergence regulated by auxin (Péret et al. 2012; Verdoucq et al. 2014); and AtPIP2;1 has been proposed as a candidate for having a non-selective cation channel function (Byrt et al. 2017). Regulation of expression of AtPIP2;1 has been observed to be influenced by changes in abscisic acid, ethylene, H2O2 and salicylic acid signalling, and AtPIP2;1 permeability has been reported to be influenced by changes in pH, calcium and phosphorylation (Qing et al. 2016; McGaughey et al. 2018). These observations indicate that the potential roles for AtPIP2;1 in other aspects of plant physiology, such as in seed germination, may be complex. During seed development, dormancy and germination, aquaporins are differentially regulated (Belmonte et al. 2013; Narsai et al. 2017; Footitt et al. 2019; Hoai et al. 2020). When aquaporin water permeability was inhibited using mercury in germinating Arabidopsis seed the water uptake accompanying expansion and growth was restricted (Vander Willigen et al. 2006). Although aquaporins are expected to contribute to regulation of water flow in seeds the complement of different roles that aquaporins play during seed germination is yet to be resolved (Vander Willigen et al. 2006; Gattolin et al. 2011; Hoai et al. 2020).
Aquaporins can help germinating seeds to adjust to unfavourable conditions in the environments in which the seed is germinating, such as osmotic stress caused by salinity (Liu et al. 2013a; Footitt et al. 2019). Soil salinity is one of the most common environmental stressors influencing plant performance, particularly seed development and germination (Ayers and Hayward 1949). Soil salinity can affect seed germination by decreasing water uptake into seed and by causing seed to accumulate NaCl. This decrease in water uptake and accumulation of NaCl can lead to a slower rate of germination, percentage germination and limit early seedling growth. Plants grown in saline conditions experience both osmotic stress and ionic stress (Munns and Tester 2008). Similarly, salinity stress can prevent or delay seed germination either by osmotic effects or ion toxicity or these combined stressors (Nasri et al. 2016; Wang et al. 2016). Previous analysis of the Arabidopsis electronic Fluorescent Pictograph (eFP) Browser data 2.0 (http://bar.utoronto.ca/) revealed that during seed imbibition and germination aquaporins from the PIP2 subfamily are expressed from early imbibition to 48 h of imbibition, particularly AtPIP2;7 and AtPIP2;1 aquaporins (Hoai et al. 2020). These PIPs are candidates for having roles in water and hydrogen peroxide permeation, but also monovalent cation permeation that may depend on post-translational modifications (Byrt et al. 2017; Qiu et al. 2020; Groszmann et al. 2021); noting that the role of PIP2;7 in univalent cation transport remains unclear (Kourghi et al. 2017; Groszmann et al. 2021), and PIPs are also likely to be involved in signalling processes (Dynowski et al. 2008; Hooijmaijers et al. 2012; Bienert and Chaumont 2014; Tian et al. 2016). We hypothesise that the phenotype of plant material varying in AtPIP2;1 expression may differ relative to wild-type (WT) control material during germination. Here, we test whether modifying AtPIP2;1 expression alters germination dynamics in control, which refers to water treatment conditions, and saline conditions (75 mM NaCl treatment).
Materials and methods
Plant material, transformation, and growth
Wild-type (WT) Arabidopsis thaliana L. Columbia-0 (Col-0) seed was sourced from the Arabidopsis Biological Resource Centre (ABRC; OH, USA). T-DNA insertion knockout mutant lines (SALK lines), Atpip2;1-1 (6AAS98 Amaze) and Atpip2;1-2 (SM_3_35928) were originally from the Nottingham Arabidopsis Stock Centre and sourced from ABRC (Da Ines et al. 2010). The pip2;1/pip2;2 double mutant Atpip(2;1*2;2) was generated by crossing Atpip2;1 (SM_3_35928) and Atpip2;2 (SAIL_169A03) single mutants (Ceciliato et al. 2019). The 2x35S::AtPIP2;1 (lines 7, 21 and wild-type null (WTnull)) over-expression seeds were from two independent transgenic lines, referred to as line 7 and line 21, which were from a collection of transgenic lines. Line 7 and 21 were the highest expressing independent lines from single insertion events as determined by segregation counts and qPCR (see Supplementary Fig. S1). The origin of the sources of the Col-0 used to create the sets of mutant and transgenic lines were from different laboratories. The origin of the Col-0 was considered as a factor that could potentially influence the data from phenotypic assays; hence, WTnull was included as a control for the transgenic lines to account for potential differences in Col-0 sources and to account for the potential influence of tissue culture on the phenotype for subsequent generations of seed.
AtPIP2;1 coding sequence (CDS) was commercially synthesised (Genscript) as gateway-enabled entry vector with incorporated dicot optimal Kozak translation start site sequence (AGAACCATGGAA). The AtPIP2;1 CDS cassette was transferred/cloned into the pMDC83 plant expression vector of the pMDC Gateway-compatible Agrobacterium sp. binary vector system (Curtis and Grossniklaus 2003) using Gateway LR Clonase II enzyme mix (Invitrogen, Carlsbad, CA, USA). The final transformation cassette consisted of RB-2x35S::AtPIP2;1:nosT-Hygr-LB. All E.coli cloning steps used One Shot OmniMAX 2 T1R Chemically Competent Escherichia coli cells (Invitrogen). The final expression plasmid was sanger sequenced to confirm accuracy of clone using Wizard Plus SV Minipreps DNA Purification Systems (Promega, Alexandria, NSW, Australia), BigDye sequencing chemistry (Thermo Fisher Scientific, Waltham, MA, USA), and ZR DNA Sequencing Clean-Up Kit (Zymo Research). Final expression vector was transformed via electroporation into Agrobacterium tumefaciens strain LBA4044 and PCR genotyped with primers flanking the AtPIP2;1 CDS located upstream in the promoter (2x35S Fwd: 5′ TCATTTGGAGAGGACCTCGA) and downstream in the terminator (NOS-term: 5′ GCAAGACCGGCAACAGGATT).
The RB-2x35S::AtPIP2;1:nosT-Hygr-LB cassette was transformed into Arabidopsis using floral dip method (Clough and Bent 1998). Primary seedling transformants (T1 generation) were selected using RAPID technique (Harrison et al. 2006) and 10 healthy robust seedlings were transferred to soil. Once established, tissues were sampled and the presence of transgene confirmed via PCR genotyping on gDNA using above ‘2x35S Fwd’ and ‘NOS-term’ primer set.
T2 progeny from 10 independently chosen T1 lines were selected on hygromycin (50 mg L−1) MS-based agar medium for 18 days after germination (DAG) at 16 h light, 22°C and light intensities between 100 and 120 μmol m2 s−1. An indication of number T-DNA insertion loci for each line, was determined through segregation counts of hygromycin resistant vs susceptible using >100 seedlings. The expectation being a 3:1 ratio of resistant:susceptible if there is single insertion site, 15:1 (96%) for two insertion sites and essentially 100% for >2 insertion sites. Only transgenic lines containing a single insertion locus were considered for subsequent analysis in this study. To quantify transgene expression levels, three biological replicates per independent transgenic line, each consisting of aerial tissue from eight 18-DAG hygromycin resistant T2 seedlings, were harvested and snap frozen in liquid nitrogen. Tissue was ground using a QIAGEN TissueLyser II and RNA extracted using ISOLATE II RNA Plant Kit (Meridian Bioscience, OH, USA). RNA was quality checked using NanoDrop spectrophotometer (Thermo Fisher Scientific) and diluted to 200 ng μL−1. 1 μg RNA was aliquoted and DNaseI treated as per manufacturer’s instructions (Cat# 18068015; Thermo Fisher Scientific). cDNA was generated using the sensiFAST cDNA Synthesis Kit (Meridian Bioscience) as per manufacturer’s instructions and diluted 1:10. Real time reactions were set up using SensiFAST SYBR Lo-ROX Kit (Meridian Bioscience) chemistry as per manufacturer’s instructions using 1 μL of cDNA (~5 ng). qPCR reactions were performed in a 384-well plate format on a ViiA 7 Real-Time system (Applied Biosystems, Scoresby, Vic., Australia) using cycle format: 95°C 2 min (×1); 95°C 5 s, 60°C 10 s, 72°C 15 s (×40); finished with a melt curve between 95°C and 60°C. QuantStudio Real-Time PCR Software (Applied Biosystems) was used to capture and analyse data. Two primer sets with unique downstream primers were used to enable differential detection between transgenic versus native AtPIP2;1 transcript abundance. For 2x35s:AtPIP2;1:NosT, primers were specific to the 3′ end of AtPIP2;1 CDS (AtPIP2;1 RT Fwd: 5′ ACCAATTCGTTCTGAGAGCTTCA) and within the 3′ transcribed region of NOS-terminator (NosT RT Rev: 5′ GAAATTCGAGCTCCACCGC). For native AtPIP2;1, primers were located in 3′ end of AtPIP2;1 CDS (AtPIP2;1 RT Fwd: 5′ ACCAATTCGTTCTGAGAGCTTCA) and within the AtPIP2;1 3′ UTR (AtPIP2;1 UTR RT Rev: 5′ GATTTGTACAGAGAGATCACAACTTCATT). YLS8 (At5g08290) was used to normalise expression (Czechowski et al. 2005) (YLS8 Fwd: 5′ TTACTGTTTCGGTTGTTCTCCATTT and YLS8 Rev: 5′ CACTGAATCATGTTCGAAGCAAGT).
All experiments were performed on stable T3 transgenic plants or confirmed homozygous mutant lines. All mutant and transgenic lines and WT (Col-0) plants were grown simultaneously in controlled conditions at the Plant Research Centre, Waite Campus, University of Adelaide, to generate comparable seed for subsequent experiments. Seeds of all genotypes were sterilised with chlorine gas and sown on half strength Murashige and Skoog, pH 7.0 medium with 1% phytagel (½ MS). Seeds were stratified in the dark at 4°C for 72 h to let seeds synchronise before germinating. Imbibed seeds were incubated in long-day growth chambers with temperature 19–22°C, continuous light intensity of 90–130 μmol m−2 s−1 and 16 h light/8 h dark for 14 days. At 2 weeks old, seedlings were transferred to pots (1 per pot, 4–5 pots per genotype). Pots were 7.5 cm × 6.0 cm and 6.0 cm in height, containing 170 g autoclaved mix of coco peat 6:4 and ‘Debco seed and cutting premium germinating mix’ at 1:1 ratio. Prior to seedling transplant, 15 pots were placed on a tray and soaked with 2 L of Imidacloprid 0.1 g L−1 (Confidor 200 SC Insecticide, Bayer CropScience Pty Ltd, Hawthorn, Vic., Australia) and Bacillus thuringiensis 5 mL L−1 (BTI Larvicide 1200 ITU mg−1, Amgrow Pty Ltd). Plants were watered every 2–3 days with reverse osmosis water. Young seedlings were covered with a plastic transparent lid for the first 4 weeks to maintain a high humidity for optimal growth. All plants/pots were randomly distributed within trays and their position changed every week to reduce positional biases within the growth chamber (potential for uneven light distribution and air flow variation). When plants bolted and started flowering, each plant was covered by a plastic transparent floral sleeve 20 cm × 50 cm. After siliques turned dark brown plants were placed in a drying area for 2 weeks at 25°C for desiccation. Seed was collected and stored in enclosed 2 mL centrifuge tubes in the dark, at 22°C ready for experiments completed within 3 months from harvesting.
Seed germination assays
Seeds were sterilised with chlorine gas for 2 h and kept in enclosed 2 mL tubes at 22°C. Approximately 150−200 seeds per line were sown on petri dishes (12 × 12 cm) containing ½ MS media, or ½ MS with an additional 75 mM NaCl. Na+ and K+ concentration in ½ MS is 0.104 mM and 10.03 mM, respectively (Murashige and Skoog 1962). A concentration of 75 mM NaCl was chosen because previous studies indicated that this concentration is sufficiently stressful so as to influence Arabidopsis germination and seed ion content, relative to standard non-saline conditions, but is not so severe as to completely prevent germination (Mcdowell et al. 2013; Nasri et al. 2016; Ravindran et al. 2020).
Seed of four individual plants from each genotype was sown onto four plates (representative of four replications). Plates were sealed with micropore tape and placed in a sealed transparent box to maintain optimal moisture conditions. Stratification and growth chamber conditions were as described above.
Seeds were visualised with a binocular microscope at 24 h post-stratification and were classified as ‘germinated’ when their endosperm ruptured (germination sensu stricto). Subsequent to a 24-h time point, seeds were further monitored for germination ‘success’ at 12-h intervals for up to 4 days, then at 24-h intervals for 5, 6 and 9 days after stratification or until no change in scores were observed.
Successful seed germination was assigned when seed clearly showed any of following plant organs: cotyledons and/or hypocotyl and/or radicle. Seeds lacking cotyledons and/or hypocotyl and/or radicle were regarded ‘ungerminated’.
Seed germination data are represented as percentages of germinated seeds (% germination = (germinated seeds × 100)/total sown seeds) as a function of incubation time post-stratification (Bewley et al. 2013).
Time to reach 50% germination (T50) was calculated according to the formula of Coolbear et al. (1984):
where N is final number of germinated seed and ni, nj are total number of germinated seeds by adjacent counts at times ti and tj, respectively, where ni < N/2 < nj.
Monitoring changes in seed mass during imbibition
Seed mass was assayed by weighing 15 mg (using electronic semi-micro balance with 0.00001 g resolution EX125D-OHAUS, Gerard Scientific) and placing seed on pre-prepared 25-mm diameter plastic mesh with 75 μm opening pores, 93 μm thickness (Sefar mesh 03-75/34 ×136; Sefar Pty Ltd Filtration and Metal Mesh; http://www.sefa.com.au) placed on two layers of 11 × 11 cm blue germination paper (Blue Blotter Paper; Anchor Paper Company, http://www.seedpaper.com) in a 12 × 12 cm petri dish. 25 mL MilliQ (MQ) water or 75 mM NaCl solution was poured in Petri dishes and seeds spread into a thin layer on mesh. Time was recorded upon addition of solutions to ensure consistent seed exposure time.
At 30 min, 1 h, 4 h, 8 h and 30 h after sowing, plastic mesh containing seeds were collected and placed on thick layers of tissue paper to absorb all excess water on seed. A very thin and flat spatula was used to collect all seeds from plastic mesh and mass was recorded by as above. Seeds were placed back on plastic mesh and filter paper for subsequent exposure times.
Seed mass data are reported here as fresh seed weight and percentage weight increase over time.
A fresh seed mass was calculated from an average of 15 mg seed. Number of seed was counted by using ImageJ on captured images (Nikon SMZ-800) at 20× magnification.
Percentage mass increase over time was calculated as:
Sodium and potassium content in seeds
Seeds for each genotype were weighed (~15 mg), distributed on plastic mesh (75 μm pores) and placed on filter papers in petri dishes as described above. Twenty five millilitre of MQ water or 75 mM NaCl were supplied to respective Petri dishes for three different imbibing experiments: (1) seeds exposed to MQ water for 50 h (water); (2) seeds exposed to MQ water from 0 to 30 h and then transferred to 75 mM NaCl solution (following Mcdowell et al. 2013; Ravindran et al. 2020) from 31 to 50 h (water-salt); and (3) seeds exposed to 75 mM NaCl for 50 h (salt). Selection of the 30-h timepoint for the applied treatment that involved moving seed from water to 75 mM NaCl was on the basis that Atpip2;1 expression would be expected to double in WT Col-0 seed between 24 h and 48 h after imbibition, based on previous observations by Klepikova et al. (2016), available via http://bar.utoronto.ca/eplant/. Plates with imbibing seed were sealed by micropore tape and placed in long-day chambers under the conditions described in the previous section.
After imbibing treatments, seeds were harvested and washed in 5 mL iso-osmotic D-mannitol solution (i.e. iso-osmotic with 75 mM NaCl; 140 mOsmol kg−1 measured using a Fiske 210 Micro-Sample Osmometer, Fiske Advanced Instruments, USA). Seeds were then placed on 25 mm diameter nitrocellulose membrane filters with 0.45 μm pores (MF-Millipore Tm, Merck Millipore Ltd, Tullagreen, Carrigtwohill, Co. Cork, Ireland) and put in a Millipore Manifold vacuum (XX2702550 Merck Millipore 1225 Sampling Vacuum Manifold, USA) to control the rate that solution passed through the filter. The washing process happened within 1 min per sample to prevent excessive ion loss and ensure uniformity of the wash process between samples. Seeds were oven dried at 65°C for 72 h before dry weight was recorded. Dried seeds were digested in 1% (v/v) nitric acid at 80°C for 4 h. Na+ and K+ contents in samples from seeds were measured using a Sherwood 420 flame photometer (Sherwood, Cambridge, UK) following manufacturer’s instructions.
Statistical analyses
For seed mass and seed ion content, the analysis of genotype collections were split into two analyses: (1) genotype collection 1 consisted of WT Col-0, Atpip2;1-1 and Atpip2;1-2 mutants, and double Atpip2;1*2;2 mutant; and (2) genotype collection 2 consisted of 2x35S::AtPIP2;1 over-expression (OE) line 7, 2x35S::AtPIP2;1 OE line 21 and WTnull, a null segregant line of 2x35S::AtPIP2;1 was used as a control because it had been through same tissue culture process as the two OE lines. There were three experimental replications for each experiment and similar patterns were observed for each replication. To analyse the data for seed germination, a factorial repeated measures two-way ANOVA and Tukey’s HSD pair-wise test was used otherwise factorial repeated measures two-way ANOVA Fisher LSD multiple comparison test was used unless specified otherwise, where P < 0.05 was considered as significant. Data were analysed using GraphPad Prism (ver. 9, San Diego, CA, USA). Data on graphs are presented as mean ± standard error of mean (s.e.m).
Results
Percentage seed germination of Arabidopsis lines differing in abundance of PIP2;1 and PIP2;2
Seed of Arabidopsis Col-0 (WT), Atpip2;1-1 and Atpip2;1-2 single KO mutants, Atpip2;1*Atpip2;2 double mutant, and two independent 2x35S::AtPIP2;1 over-expression (OE) lines, along with its null-segregant control (WTnull), were propagated in consistent conditions for use in experiments to assess whether germination of seed from these genotypes differed between control and saline treatments. Seed germination was recorded over 9 days (216 h) of post-stratification. Col-0, Atpip2;1-1, Atpip2;1-2, Atpip2;1*pip2;2, 2x35S::AtPIP2;1 OE lines and WTnull seeds were germinated on ½ MS media (control treatment) or ½ MS + 75 mM NaCl treatment (salt treatments). At 24 h in control treatment conditions, Col-0 had a percentage of germinated seeds (29.5%) that was greater than the percentage of germinated seed from Atpip2;1-1 and Atpip2;1*2;2, which were in the range of 15.1–17.8 %, (Fig. 1a, b, e, f). At 48 h, the percentage germination of WTnull in ½ MS + 75 mM NaCl was ~98%, which was higher than that of the two 2x35S::AtPIP2;1 OE lines (Fig. 1b, g, h).
Cumulative germination success and time to 50% germination (T50) of Arabidopsis lines differing in PIP abundance in the presence or absence of 75 mM NaCl. Data is reported as percentage of seed germinated under control treatment (½ MS) or saline treatment (½ MS + 75 mM NaCl). All seed with endosperm rupture were recorded over time (0–216 h) upon exposure to 0 mM (a, b) or 75 mM (c, d) added NaCl treatments. Statistical analysis of percent germination data at individual time points after stratification: 24 h (e, f), 48 h (g, h) and 72 h (i, j) and time to reach 50% germination T50 in h (k, l). Data was divided into two groups for analysis: (1) genotype collection 1 consisting of Col-0 ( ), Atpip2;1-1 (
), Atpip2;1-1 ( ), Atpip2;1-2 (
), Atpip2;1-2 ( ), Atpip2;1*2;2 (
), Atpip2;1*2;2 ( ); and (2) genotype collection 2 consists of 2x35S::AtPIP2;1 OE 7 (
); and (2) genotype collection 2 consists of 2x35S::AtPIP2;1 OE 7 ( ), 2x35S::AtPIP2;1 OE 21 (
), 2x35S::AtPIP2;1 OE 21 ( ) and WTnull (
) and WTnull ( ). T-tests indicated that there was no significant difference between WTnull and Col-0. Each panel includes percent seed germination in ½ MS or ½ MS + 75 mM NaCl. Data are means (±s.e.m.) from n = 4–10 plates, with 150–200 seeds per genotype per plate. This experiment was replicated three times, data for experiment replications are included in Figs S2a and S2b. The trends in all three experiment replications were similar; for example, the mean values for Col-0 percentage of germination at 24 h of imbibition in ½ MS and in ½ MS + 75 mM NaCl were 29.51% and 1.72 %, respectively, in replication 1; in replication 2, the values were 28.63% and 1.825 %, respectively; and in replication 3, the values were 25.83% and 2.25%, respectively. Significant difference determined by a factorial repeated measures two-way ANOVA and Tukey’s HSD pair-wise tests at P < 0.05. Different lowercase letters denote significant differences between genotypes within each germination treatment; different capital letters denote significant differences between ½ MS and ½ MS + 75 mM NaCl for each genotype. Absence of error bars indicates s.e. is smaller than symbol.
). T-tests indicated that there was no significant difference between WTnull and Col-0. Each panel includes percent seed germination in ½ MS or ½ MS + 75 mM NaCl. Data are means (±s.e.m.) from n = 4–10 plates, with 150–200 seeds per genotype per plate. This experiment was replicated three times, data for experiment replications are included in Figs S2a and S2b. The trends in all three experiment replications were similar; for example, the mean values for Col-0 percentage of germination at 24 h of imbibition in ½ MS and in ½ MS + 75 mM NaCl were 29.51% and 1.72 %, respectively, in replication 1; in replication 2, the values were 28.63% and 1.825 %, respectively; and in replication 3, the values were 25.83% and 2.25%, respectively. Significant difference determined by a factorial repeated measures two-way ANOVA and Tukey’s HSD pair-wise tests at P < 0.05. Different lowercase letters denote significant differences between genotypes within each germination treatment; different capital letters denote significant differences between ½ MS and ½ MS + 75 mM NaCl for each genotype. Absence of error bars indicates s.e. is smaller than symbol.
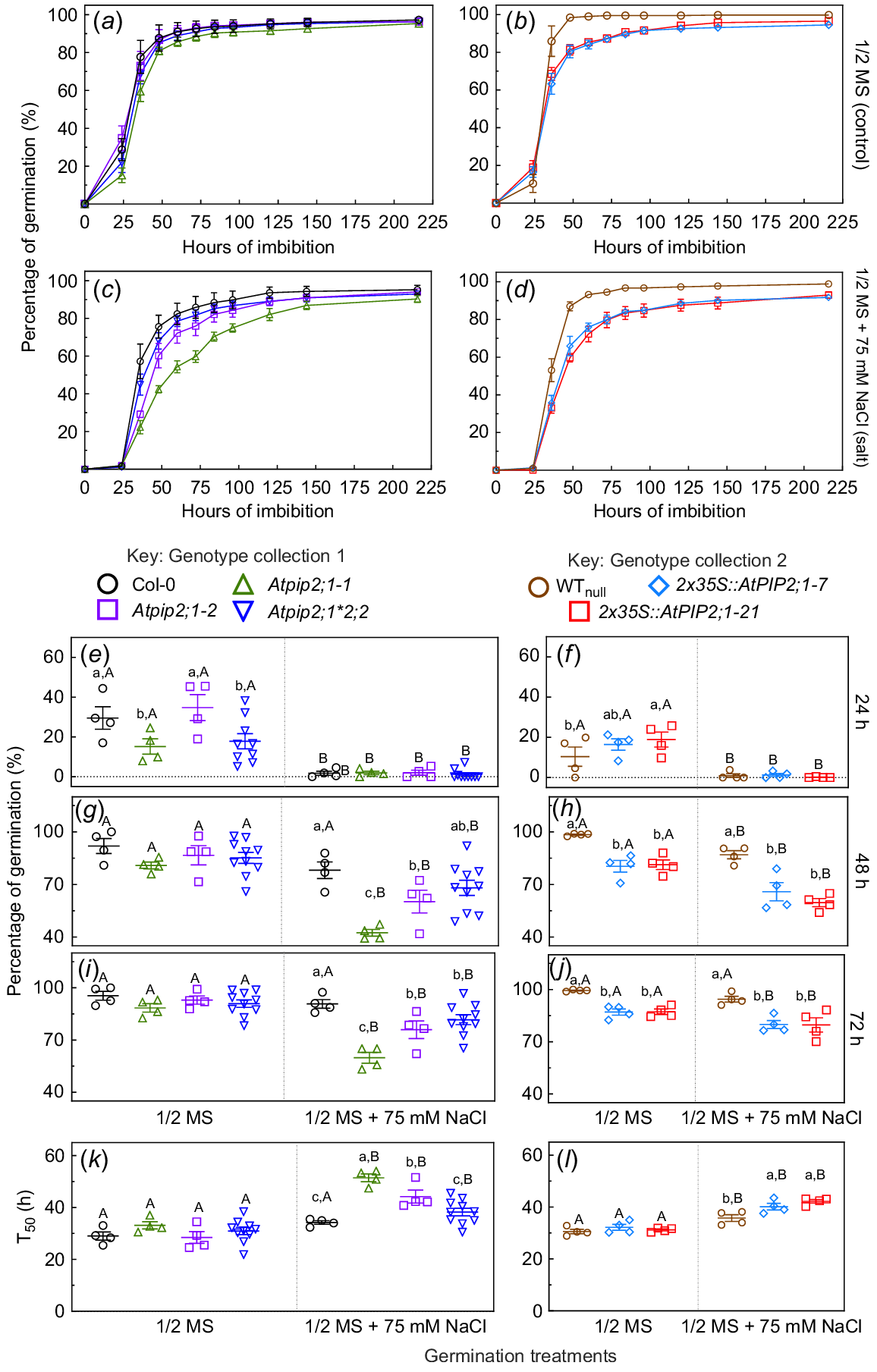
In saline treatment conditions, the seed germination was delayed relative to the control treatment conditions, with the 75 mM NaCl treatment resulting in only ~1% germination at 24 h (Fig. 1c, d) contrasting to control conditions where at 24 h germination ranged between 15% and 35% for all genotypes (Fig. 1a, b). Atpip2;1-1 consistently had the lowest percentage of germination in salt treatment from 48 h to 5 days after stratification (Fig. 1c, d). The percentage of germination of all genotypes after 9 days of exposure to ½ MS media was over 96.7% (s.e. = 0.42%), whereas for seed subjected to the 75 mM NaCl treatment, the total average germination was reduced to 93.8% (s.e. = 0.61%).
No differences in percentage of germination were observed in genotype collection 1, which included Col-0 (WT), Atpip2;1-1, Atpip2;1-2, and Atpip2;1*pip2;2, when seed was germinated in control condition across all time points. Whereas in saline conditions, a reduction in germination compared to Col-0 (WT) seeds was observed at all measured time points for Atpip2;1-1, and at 72 h for Atpip2;1-2 (Fig. 1e, g, i). The Atpip2;1*pip2;2 line did not have significant decreases or increases in germination compared to Col-0 WT seeds.
In genotype collection 2, which included two 2x35S::AtPIP2;1 OE lines and WTnull, the 2x35S::AtPIP2;1 OE seeds (lines 7 and 21) had a significant reduction in percentage of germination relative to WTnull seed in both control and saline treatments at all measured time points (Fig. 1f, h, j).
In control conditions, there were no significant differences between genotypes in both genotype collections for the time when 50% of seed had germinated (T50), which was at 29–33 h post-stratification. However, in salt treatment conditions, Atpip2;1-1 and Atpip2;1-2 had longer T50 compared to WT (51, 44 and 34 h, respectively), OE lines 2x35S::AtPIP2;1 lines 7 and 21 also had longer T50 compared to WTnull (at 40, 42 and 35 h, respectively). (Fig. 1k, l; statistics tests result outcomes are shown in Table S9a, b, c).
Changes in seed mass during imbibition in control and saline conditions
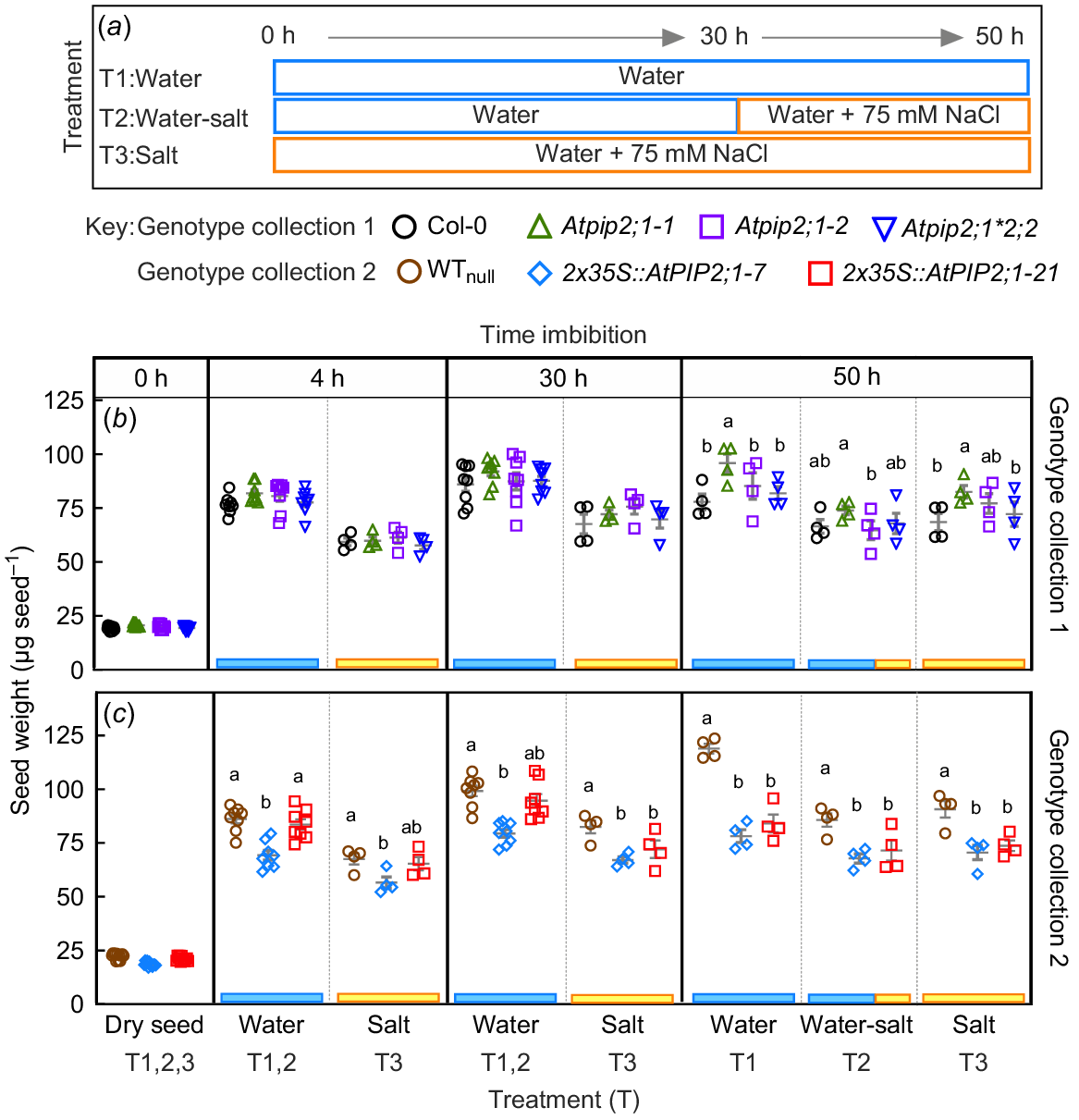
Seed mass was determined by weighing the seed and maximum cross sectional (MCS) area was determined using measurements from high resolution scaled images of seed. MSC was used as a surrogate for seed volume assuming isodiametric swelling. Changes in MCS during imbibition are indicative of seeds accumulating water and swelling. We observed increases in seed mass and MCS area over time for all tested genotypes in control treatment (½ MS) and saline treatment (½ MS + 75 mM NaCl) (Figs S3, S4, S5 and S6). The initial seed mass of the various genotypes fell within 17–20 μg (P > 0.05). However, the rate of seed/seedlings mass increase differed among genotypes during imbibition and germination over 50 h. The relationship between seed weight and MSC was fitted with linear regression lines (Y = B0 + B1 × X, where Y is, seed weight (μg/seed), X is seed MCS area (μg2/seed), B0 is intercept of dry seed weight, B1 is slope of the lines), as expected the relationship was positive (Fig. S6). For values from seed measurements taken from zero time point (Fig. S6a) a single linear line of MCS versus time adequately fit all data in each case (Prism F-test) and at later time points (Fig. S6b) single linear line of MCS versus time adequately fits all data of control treatment, or salt treatment. However, different linear lines were required to fit data for measurements taken following the water to 75 mM salt transfer treatments.
The Atpip2;1-1 had a seedling mass greater than that of Col-0 control and Atpip2;1*2;2 at 50 h of imbibition in control conditions and conditions with 75 mM NaCl (Fig. 2b). In control treatment (water) and water-salt transfer treatments, differences were also observed between Atpip2;1-1 and Atpip2;1-2 lines, with Atpip2;1-1 having a higher seedling mass in both treatments after 50 h of imbibition (Fig. 2b). The weight of WTnull increased faster than weight of 2x35S::AtPIP2;1 OE lines in all imbibition treatments (Fig. 2c).
Changes in seed mass over time during 50 h imbibition in water and saline treatments. Data was divided into two groups for analysis: (1) genotype collection 1 consisting of Col-0 ( ), mutant Atpip2;1-1 (
), mutant Atpip2;1-1 ( ), Atpip2;1-2 (
), Atpip2;1-2 ( ), Atpip2;1*2;2 (
), Atpip2;1*2;2 ( ); and (2) genotype collection 2 consists of over-express lines 2x35S::AtPIP2;1 OE 7 (
); and (2) genotype collection 2 consists of over-express lines 2x35S::AtPIP2;1 OE 7 ( ), 2x35S::AtPIP2;1 OE 21 (
), 2x35S::AtPIP2;1 OE 21 ( ) and their null WTnull (
) and their null WTnull ( ). (a) Summary of the experiment set up consisting of three treatments: T1, 50 h imbibition in water only (water); T2, 30 h imbibition in water followed by 20 h in water containing 75 mM NaCl solution (50 h total, water-salt); and T3, 50 h imbibition in 75 mM NaCl solution (salt). (b, c) Seed mass increases in two genotype collections over 50 h of imbibition. Blue and yellow bars are to indicate the relevant treatment shown in part (a). Dry seed mass data is the average of 600–900 seeds used per genotype on each experimental replicate (plates), n = 4 plates per condition. Data are means and s.e.m. The experiment was repeated times times and showed the same pattern. Significant difference determined by two-way ANOVA and Fisher’s LSD pair-wise tests, P < 0.05. Different letters indicate a statistical difference between genotypes, absence of letters indicates no significant difference.
). (a) Summary of the experiment set up consisting of three treatments: T1, 50 h imbibition in water only (water); T2, 30 h imbibition in water followed by 20 h in water containing 75 mM NaCl solution (50 h total, water-salt); and T3, 50 h imbibition in 75 mM NaCl solution (salt). (b, c) Seed mass increases in two genotype collections over 50 h of imbibition. Blue and yellow bars are to indicate the relevant treatment shown in part (a). Dry seed mass data is the average of 600–900 seeds used per genotype on each experimental replicate (plates), n = 4 plates per condition. Data are means and s.e.m. The experiment was repeated times times and showed the same pattern. Significant difference determined by two-way ANOVA and Fisher’s LSD pair-wise tests, P < 0.05. Different letters indicate a statistical difference between genotypes, absence of letters indicates no significant difference.
Seed MCS area increased upon imbibition in control (½ MS) in all lines tested, with the largest increase observed in genotype collection 1 occurring within 30 min of imbibition, increasing from 11.5–12 to 14–15.5 mm2 (Figs S4 and S5). The rate of MCS area increase slowed down after 30 min through to 30 h of imbibition, with seeds reaching MCS area of ~18 mm2 (Fig. S4a).
Seeds of genotype collection 2 had statistically different dry seed sizes (ranging from 11 to 12.5 mm2), with 2x35S::AtPIP2;1 OE seeds having a smaller seed size than WTnull (Fig. S4b). In control treatment conditions, 2x35S::AtPIP2;1 OE lines had statistically significant reduction in water accumulation rates, relative to WTnull (Fig. S5b). At 30 h imbibition, seed MCS area of Atpip2;1*2;2 was significantly smaller than that of Atpip2;1-2 (Fig. S5). The influence of saline treatments was more evident for seed mass than for seed MCS area (Figs S5c, d and S6). This may be because MCS area was measured from 2D images and thus did not represent seed swelling in all directions.
Sodium and potassium content in seeds with altered PIP2;1 and PIP2;2 abundance
Seeds of AtPIP2;1 loss-of-function mutants, over-expression lines, and their respective controls, were germinated in three different treatments: water (T1); water-salt transfer (T2); and salt (T3) (Fig. 3). As expected, seeds exposed to saline solutions (T2 and T3) had higher Na+ and lower K+ content (per dry weight), compared to seeds germinated in water treatment (T1) (Fig. 3b–e).
(a) Seed Na+ and K+ content for seven genotypes including Col-0 ( ), loss-of-function mutants Atpip2;1-1 (
), loss-of-function mutants Atpip2;1-1 ( ), Atpip2;1-2 (
), Atpip2;1-2 ( ), Atpip2;1*2;2 (
), Atpip2;1*2;2 ( ), and AtPIP2;1 over-expression lines 2x35S::AtPIP2;1 OE 7 (
), and AtPIP2;1 over-expression lines 2x35S::AtPIP2;1 OE 7 ( ), 2x35S::AtPIP2;1 OE 21 (
), 2x35S::AtPIP2;1 OE 21 ( ) and their null WTnull (
) and their null WTnull ( ) exposed to three treatments: (1) water for 50 h (water); (2) 75 mM NaCl solution for 50 h (salt); or (3) 30 h in water then transferred to a solution containing 75 mM NaCl from 30 to 50 h (water-salt). Na+ content (b, c) and K+ content (d, e) and K+/Na+ ratio in seed (f, g) following three treatments (a). At some time points, error bars are so tight as to be obscured by the symbols. Significant difference determined by a factorial repeated measures two-way ANOVA and Tukey’s HSD pair-wise tests at P < 0.05. Different lowercase letters denote significant differences between genotypes within each germination treatment. Absence of error bars indicates s.e. is smaller than symbol. Ion concentration unit, mg per 1 kg dry weight (mg kg−1 DW).
) exposed to three treatments: (1) water for 50 h (water); (2) 75 mM NaCl solution for 50 h (salt); or (3) 30 h in water then transferred to a solution containing 75 mM NaCl from 30 to 50 h (water-salt). Na+ content (b, c) and K+ content (d, e) and K+/Na+ ratio in seed (f, g) following three treatments (a). At some time points, error bars are so tight as to be obscured by the symbols. Significant difference determined by a factorial repeated measures two-way ANOVA and Tukey’s HSD pair-wise tests at P < 0.05. Different lowercase letters denote significant differences between genotypes within each germination treatment. Absence of error bars indicates s.e. is smaller than symbol. Ion concentration unit, mg per 1 kg dry weight (mg kg−1 DW).
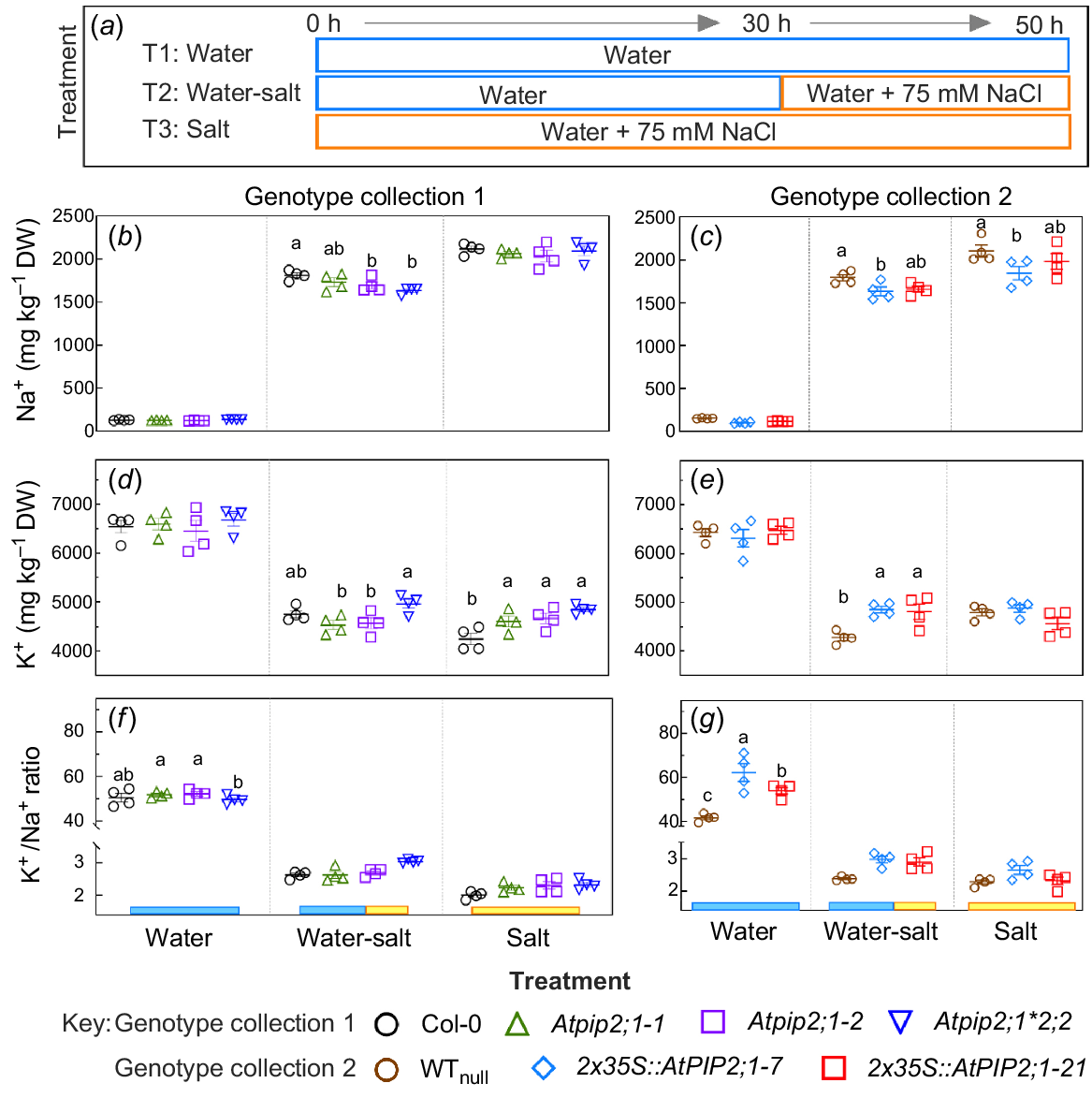
There were no differences between genotypes in relation to Na+ or K+ content of seeds when germinated in water (T1) treatment (Fig. 3b–e), except Atpip2;1*pip2;2 seed did have a lower K+/Na+ ratio than seed for two Atpip2;1 lines. Differences in Na+ and K+ content were observed between genotypes for seeds germinated in water-salt transfer (T2) and salt (T3) treatments, and for dry seed prior to imbibition (Figs 3b–e and S7). Specifically, Na+ content in Col-0 seeds was higher than that of Atpip2;1-2 and Atpip2;1*2;2 seed in water-salt transfer (T2) treatments, but there were no differences in Na+ content between these genotypes when germinated in salt (T3) treatment (Fig. 3b). WTnull seeds had a higher Na+ content than 2x35S::AtPIP2;1-7 OE seeds when germinated in water-salt (T2) and salt (T3) treatments, and also in dry seed prior to imbibition (Figs 3c and S7).
K+ content in seeds of Atpip2;1*Atpip2;2 was higher than that of Atpip2;1-1 and Atpip2;1-2 lines when germinated on water-salt (T2) treatment, but each mutant had similar K+ content to Col-0 control, whereas when germinated solely on salt (T3), Atpip2;1 and Atpip2;1*Atpip2;2 seeds had a higher K+ content than Col-0 control seeds (Fig. 3d).
Both 2x35S::AtPIP2;1 OE lines had a higher K+ content compared to WTnull when germinated on water-salt treatment (T2), and there was no difference in K+ content in these genotypes when germinated in salt treatment conditions (T3, Fig. 3e).
There were no differences in K+/Na+ ratio in seed from salt (T3) treatment and water-salt treatment (T2) (ratio was 2–3), but the K+/Na+ ratio was higher in Atpip2;1-1 and Atpip2;1-2 lines (51.8 and 52.2, respectively) relative to Atpip2;1*2;2 (49.6) in water treatment (Fig. 3f) and WTnull had the lowest K+/Na+ ratio (41.8) followed by 2x35S::AtPIP2;1 OE line 21 (54.1), and 2x35S::AtPIP2;1 OE line 7 had the largest K+/Na+ ratio (62.3) (Fig. 3f, g). In summary of key observations: in the control treatment conditions the trends in germination and ion accumulation were similar in pip2;1 and pip2;1*pip2;2 lines relative to WT (Col-0) and similar in 2x35S::AtPIP2;1 OE lines relative to nulls; but seed mass was higher in the pip2;1 mutant lines than WT and seed mass was lower in 2x35S::AtPIP2;1 OE line than WTnull (Fig. 4). The Atpip2;1*Atpip2;2 double mutant and 2x35S::AtPIP2;1 OE lines had higher K+/Na+ ratios relative to their respective controls.
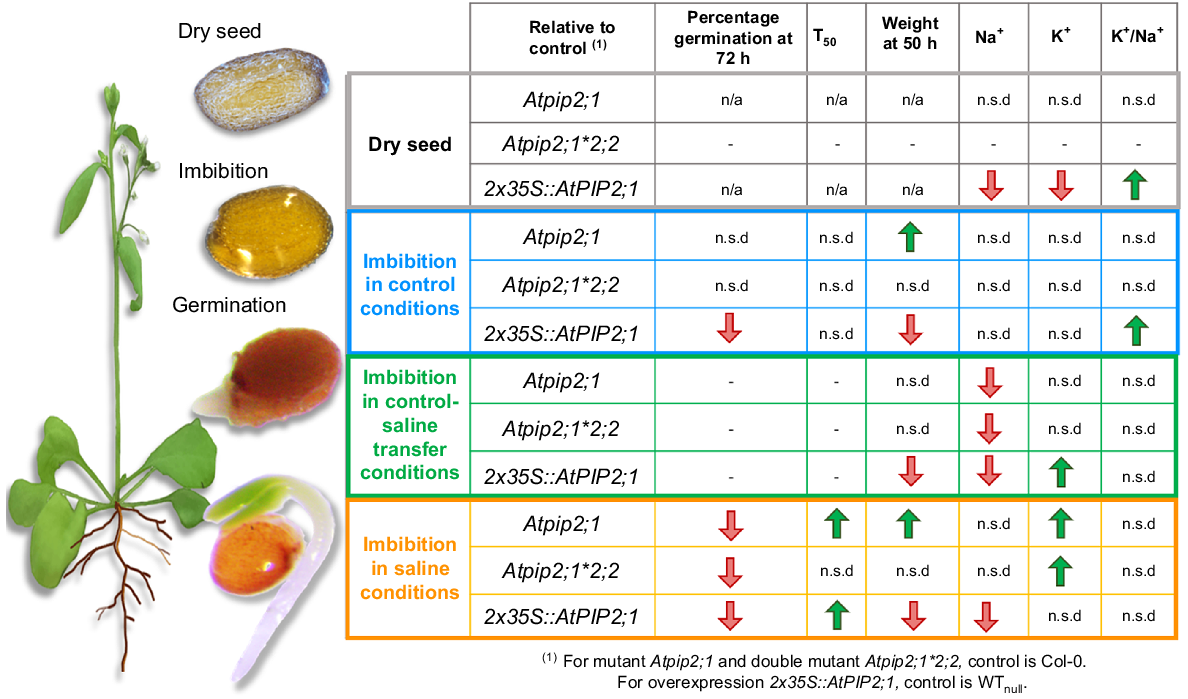
Summary of comparisons for lines lacking PIP2;1, lacking both PIP2;1 and PIP2;2 and lines over-expressing PIP2;1 relative to controls. Data trends for ion content for dry seed and physiological parameters measured during imbibition including ion content, seed percentage of germination, time to reach 50% germination (T50) and seedling weight are represented by either red downward arrows or green upward arrows for mutant relative to Col-0 and transgenic lines relative to WTnull, or no significant difference (n.s.d) if no differences were observed. n/a, not applicable.
Discussion
Atpip2;1 loss-of-function mutants and AtPIP2;1 over-expression line seed both exhibited delayed germination relative to controls when imbibed in saline conditions
In Arabidopsis and in cereals, transcript abundance of various genes encoding for AtPIP1s, AtPIP2s and AtTIPs increase rapidly in seed during imbibition and germination indicating that there are potential roles for multiple aquaporin isoforms from different subfamilies in germination (Matsunami et al. 2022). In germinating Arabidopsis Columbia-0 seed, the abundance of AtPIP2;1, AtPIP2;2 and AtPIP2;7 was previously observed to increase after 3 days soaking in water and in germinated seeds the transcript level for these genes were notably very high in roots (Fig. S8) (Klepikova et al. 2016). Analysis of previously reported transcript data revealed increased AtPIP2;1 expression particularly between 12 and 48 h after start of imbibition, during the time period when germination occurs, which is typically around 24 h after the start of imbibition (Hoai et al. 2020; Fig. S8). Here, we observed that disruption of WT AtPIP2;1 abundance by either loss or increased transcript abundance influenced the seed germination, seed weight and seed Na+ and K+ content when seed was imbibed in a saline treatment (Fig. 4). Both the Atpip2;1*Atpip2;2 double mutant and the 2x35S::AtPIP2;1 OE lines had higher K+/Na+ ratios relative to their respective controls indicating that disruption of AtPIP2;1/AtPIP2;2 function influenced ion homeostasis in Arabidopsis seed.
At 50 h after stratification, the seed from mutant and over-expression lines did not differ from controls in percentage germination when imbibed in water, but when imbibed in saline treatment conditions the percentage germination of seed from mutant and over-expression lines was delayed relative to WT and null controls, respectively, indicating that a decrease or increase of AtPIP2;1 was associated with reduced percentage of germination (Fig. 1, Table S10). This reveals that AtPIP2;1 could potentially have a role in enabling seed to achieve optimal germination success in saline conditions.
Mutant or transgenic line seed with reduced or increased AtPIP2;1 expression tended to differ in mass gain during imbibition relative to their respective control lines (Fig. 2). The loss or over-expression of AtPIP2;1 also resulted in reduced seed Na+ content and greater seed K+ content in mutant and transgenic lines relative to controls (Fig. 3). Hypothetically, greater abundance of AtPIP2;1 in seed might be expected to be associated with more rapid water uptake during imbibition and loss of AtPIP2;1 might be expected to limit water uptake during imbibition, although there are other aquaporins that may also contribute to water uptake, such as AtPIP2;7 and there are likely to be other types of molecular mechanisms that influence water uptake during imbibition. In germinating WT Col-0 seed, transcripts of AtPIP2;1 and AtPIP2;7, followed by AtPIP2;2 were observed to be most abundant relative to the other AtPIP2s (Groszmann et al. 2021). However, relative abundance of transcripts coding for each of 35 Arabidopsis aquaporins in mutant and transgenic lines is yet to be determined, and may not necessarily correspond to the protein abundance for each of the respective aquaporins (Liu 2015), revealing that it may be necessary to test the abundance of the complement of aquaporins in the various genotypes tested to determine the extent to which disruption of AtPIP2;1 function indirectly influences germination due to differential regulation of other aquaporins. In the present study the lines over-expressing AtPIP2;1 had reduced mass and loss-of-function mutants generally had greater mass gain during imbibition than their respective controls (Fig. 2). This indicates that the putative role for AtPIP2;1 in seed germination may not just be related to influencing water uptake or permeability during germination. AtPIP2;1 is permeable to water, CO2, H2O2 and ions (Tyerman et al. 2021), and altered regulation of AtPIP2;1 could influence permeability to a range of different substrates in different ways (Qiu et al. 2020). AtPIP2;1 function is also known to be influenced by differences in hormones and signalling molecules (Daszkowska-Golec 2011; Wojtyla et al. 2016). Moreover, regulation of AtPIP2;1 is known to influence the regulation of other aquaporins (Liu 2015). Therefore, there may be a range of potential factors that could explain why germination of seed with altered AtPIP2;1 abundance is delayed in saline treatment. Suggestions for these hypothetical factors have been categorised as follows:
Transport of either Na+ or K+ via AtPIP2;1 could be important in seed germination in conditions of salt stress, and is likely to be influenced by AtPIP2;1 phosphorylation status (Qiu et al. 2020).
The contribution of AtPIP2;1 to signalling processes, such as distribution of H2O2 signals, could be important in seed germination (Groszmann et al. 2021).
Disruption of AtPIP2;1 could cause differential regulation of other aquaporins which may influence seed germination (Da Ines 2008).
There are limitations to the options for interpretation of the data sets related to the influence of the saline treatment in delaying germination of each of the genotypes. This is because germination delay could confound assessment of comparisons between the genotypes at a given specific time point when comparing the influence of the saline relative to control treatments. There are also a range of other factors that influence the development of plants in saline relative to control conditions. For example, previous studies reported that salt treatment (100 mM NaCl) caused delay in the emergence of lateral roots in Arabidopsis seeds because of possible induced thickening of the Casparian strip (Duan et al. 2013).
Implications for seed germination associated with AtPIP2;1 water and ion permeability
Hypothetically changing the abundance of AtPIP2;1 in seed could influence either seed swelling, and/or radicle cell expansion during seed imbibition and/or germination. Previous studies have demonstrated using either heterologous expression systems or whole Arabidopsis plants that AtPIP2;1 influences water flux; therefore, it is feasible that AtPIP2;1 may also influence water flux in germinating seeds. For example, when AtPIP2;1 was expressed in Xenopus laevis oocytes those oocytes swelled more rapidly than control oocytes when transferred to a hypo-osmotic solution due to AtPIP2;1 facilitating water influx into oocytes (Mahdieh et al. 2008; Byrt et al. 2017; Prado et al. 2019; Qiu et al. 2020; Tyerman et al. 2021). Differences in relative water flux in Atpip2;1 and Atpip2;2 loss-of-function mutant plant rosettes relative to WT controls has also been reported using deuterium as a tracer (Da Ines et al. 2010; Wang et al. 2016). A study by Da Ines et al. (2010) of Arabidopsis rosette water flux revealed that loss of AtPIP2;1 and AtPIP2;2 in different tissues and cells contributed in an integrated fashion to reduced water flux in intact plants. Here, seed swelling of lines with varying AtPIP2;1 abundance, or varying AtPIP2;1 and AtPIP2;2 abundance was investigated by measuring changes in seed area over time when seed was imbibing in control or in saline treatments (Figs S4 and S5). The seed area of Atpip2;1*pip2;2 was similar to that of WT and Atpip2;1 during the first 4 h of imbibition, but at 8 and 30 h imbibition, the relative increase in area of Atpip2;1*pip2;2 was less than that of WT and Atpip2;1 single mutant lines (Fig. S4). The influence that loss of these different PIP2s may have had could potentially have differed in different cell types within the swelling seed during the period of imbibition and germination. Hypothetically it is possible that sub-optimally regulated swelling between different cell types caused by disrupted AtPIP2;1 abundance could have limited the germination efficiency of mutant lines (Fig. 1).
It is expected that as seeds imbibe both the seed weight and the seed MCS would increase. As expected, we observed that the relationship between seed weight and seed MCS area was positive at the zero time point (dry seed) and at timepoints of 4 and 30 h into imbibition, under control treatment and saline treatment treatments (Fig. S6). However, there were significant differences in linear regression lines between 0 mM and 75 mM for P2 but not for P1. This indicates that either seed weight at the same area is lower in 75 mM NaCl, or at the same seed weight the seed area is larger in 75 mM NaCl. Further investigation of seed density in conditions of varying salinity would be required to further investigate how the relationship between seed weight and MSC area.
Previous studies have revealed that salt stress delays seed germination. For example, Bafoil et al. (2019) revealed that seed in 50 mM NaCl had delayed testa rupture relative to seed in conditions without NaCl treatment. As expected, we observed that there was a delay in time to reach 50% germination associated with 75 mM NaCl treatment in both single mutant pip2;1 and over-expression lines, but there was a recovery of percentage germination in double mutant Atpip2;1*2;2 relative to WT Col-0. It is possible that loss of both AtPIP2;1 and AtPIP2;2 in the double mutant leads to different regulation of other mechanisms to compensate, and that this influenced the trends in percentage germination.
AtPIP2;1 involvement in seed signalling could have influenced seed germination
Hydrogen peroxide has multiple signalling roles in seeds including influencing seed germination and responses to stress such as salt stress, and AtPIP2;1 is permeable to H2O2 (Dynowski et al. 2008; Hooijmaijers et al. 2012; Wojtyla et al. 2016; Groszmann et al. 2021). Hypothetically, AtPIP2;1 H2O2 permeability could influence how seed germination responds to stress and this could have been a factor influencing differences in germination in mutant and transgenic seed in saline treatments (Fig. 1). Seed dormancy and germination are known to be regulated by complex signalling processes that involve a range of phytohormones and signals triggered by phytohormone changes may influence AtPIP2;1 functions in seed. Germination is influenced by the abundance of ABA, while gibberellins (GA) and ethylene can restrict the effect of ABA on dormancy and instead GA and ethylene promote germination, meanwhile nitric oxide (NO) is proposed to be implicated in cross talk between ABA and ethylene in seed where NO treatments also tend to promote germination (Arc et al. 2013; Li et al. 2016; Tuan et al. 2021), and potentially some aquaporins may be permeable to NO (Wang and Tajkhorshid 2010). Ethylene treatment was reported to influence C-terminal phosphorylation of AtPIP2;1 and C-terminal phosphorylation of AtPIP2;1 can influence the proportion of AtPIP2;1 in cell plasma membranes and influence AtPIP2;1 water and ion transport (Qing et al. 2016; Qiu et al. 2020). Where there is potentially greater ethylene accumulation under salt stress, this could influence abundance and function of AtPIP2;1 (Wilson et al. 2014).
Auxin is a plant growth hormone that acts as an adaptable trigger for controlling many developmental processes (Vanneste and Friml 2009). Auxin plays a critical role in Arabidopsis seed dormancy that is independent relative to ABA but requires ABA signalling pathways (Liu et al. 2013b). Previously Péret et al. (2012) reported that lateral root emergence is delayed or slowed when AtPIP2;1 abundance is either reduced or increased, in Arabidopsis loss-of-function or over-expression lines, respectively and that auxin signalling influences AtPIP2;1 contribution to hydraulic conductivity to regulate lateral root emergence. Auxin signalling also influences directionality in relation to root growth (Vanneste and Friml 2009; Hu et al. 2021). The analysis of AtPIP2;1 roles in germination here reveals a potential trend for influencing radicle emergence, which is reminiscent of observations for roles of AtPIP2;1 in influencing lateral root emergence where AtPIP2;1 loss-of-function and over-expression lines both have delayed root emergence. To further study whether AtPIP2;1 has a role in radicle emergence, it would be ideal to capture high resolution images of radicle emergence in loss-of-function Atpip2;1 lines and AtPIP2;1 over-expression lines relative to controls in non-saline and saline treatments to check whether or not the radicle appearance differs.
Differential regulation of aquaporins during seed germination
Trends in aquaporin gene transcript abundance differ during different stages of development and phases of germination (Hoai et al. 2020). For example, the abundance of transcripts coding for AtNIP1;2, AtPIP1;2, and AtTIP3;1 are particularly high in dry seed, whereas transcripts coding for AtPIP2;1, AtPIP2;7, AtTIP1;1, AtTIP1;2, and AtTIP4;1 increase either at or immediately after germination (Footitt et al. 2019; Hoai et al. 2020; Su et al. 2022). At the stage of 24 h after imbibition, AtPIP1;2 and AtPIP2;7 transcripts were most abundant, and by 48 h AtPIP1;2, AtPIP2;7 and AtPIP2;1 transcript abundance was notably higher than that of other AtPIPs (Groszmann et al. 2021). This indicates that different aquaporins are required during seed development relative to during seed germination, and it is possible that different aquaporins are required during different phases of seed germination. Three phases of germination can be summarised as follows (Bewley 1997; Weitbrecht et al. 2011): Phase I involves seed swelling; volume and shape change and solute leakage (Simon and Harun 1972; Van Steveninck and Ledeboer 1974; Simon and Mills 1983; Crowe and Crowe 1992; Niemann et al. 2013). In seeds such as white spruce (Picea glauca (Moench) Voss), seed K+ loss during imbibition has been reported, indicating potential for there to be a role in membrane proteins permeable to K+ in this process (Reid et al. 1999). Phase II involves rehydration; solute leakage; resumption of respiration and oxygen consumption; activation of amino acid metabolism and Kreb’s cycle; mRNA, DNA and protein synthesis; formation of new mitochondria; substrate and small molecule diffusion; organelle assembly. Phase III involves cell division; testa rupture and radicle emergence. Previous analysis of aquaporin roles in these phases by Vander Willigen et al. (2006) based on mercury inhibition led to a conclusion that aquaporin functions are unlikely to be involved in Phase I, and are more likely to be involve in Phase III, but the mercury is toxic and could influence other aspects of Phase I biology. Potentially, TIP3s might be the only AQPs involved in mediating seed water uptake in Phase I, but if this was the case they would need to be located in the plasma membrane to contribute (Gattolin et al. 2011). Multiple PIP and TIP1 coding transcripts are abundant during germination, but some polypeptides do not become detectable until approximately 60 h post germination, well into Phase III of water transport in seed (Gattolin et al. 2011). In the context of seed mass changes during imbibition and germination, which may be indicative of solute flux changes, differences between loss of function mutant lines and WT were not observed until 50 h after imbibition, where one of two Atpip2;1 loss-of-function mutant lines exhibited greater seed area than WT controls (Fig. 2b). Whereas for AtPIP2;1 over-expression lines differences were observed 4 h after imbibition and transgenic over-expression line seed did not gain weight as quickly as null control line seed (Fig. 2c). It is difficult to determine which phases of seed germination AtPIP2;1 may play a role in reviewing differences in the timing of onset of phenotypic differences for mutant and over-expression line material relative to controls.
There are potentially hundreds of different types of proteins that could possibly interact with AtPIP2;1 (Bellati et al. 2016), and the implications of loss of AtPIP2;1 function or over-expression of AtPIP2;1 could potentially influence the function of many other proteins, or the proteins interacting with PIP2;1 could confer other functions for PIP2;1 such as sensing (Tyerman et al. 2021). For example, 400 potential AtPIP2;1 and AtPIP2;2 interacting proteins were reported by Bellati et al (2016) and their analysis revealed TIP2;2 as a potential interacting partner, revealing that PIP abundance could impact TIP regulation, and TIPs including TIP3;1, TIP3;2 and TIP4;1 were previously reported to significantly influence Arabidopsis seed germination in response to stress (Footitt et al. 2019). In cereals such as barley (Hordeum vulgare) and rice (Oryza sativa), some of the trends in aquaporin regulation in seeds are similar to those in Arabidopsis, for example, there are key TIPs such as HvTIP3;1 and OsTIP3;1 that are abundant in seeds, and OsTIP3;1 like AtTIP3;1 is influenced by ABA signalling (Takahashi et al. 2004; Utsugi et al. 2015; Han et al. 2018). Transcripts of rice PIPs including OsPIP1;1, OsPIP1;2, OsPIP1;3 and OsPIP2;8 were reported to increase in abundance in germinating seed in response to nitric oxide treatments, further implicating PIPs in processes that influence seed germination (Liu et al. 2007). Loss of OsPIP1;1 and OsPIP1;3 function in rice led to reduced seed germination and OsPIP1;3 over-expression promoted seed germination under water-stressed conditions (Liu et al. 2007). Therefore, changes in abundance of PIP2;1 and PIP2;2 may influence regulation of other aquaporins during seed germination, especially in the case of the double Atpip2;1*pip2;2 mutant where there may be differences in the abundance of PIPs relative to the respective abundance of various PIPs in the single mutant lines. The potential for different regulation of other aquaporins in single and double PIP2 mutants studied here could in part explain why there were different trends in the phenotypes of these different mutant lines. For example, germination, mass and ion content data for the double mutant was more similar to WT than data for the single Atpip2;1 mutants (Figs 1, 2 and 3). There is also potential that the original sources of the Col-0 germplasm used to create the lines included in this study could have varied in their inherent MIP regulation. For example, the two Atpip2;1 mutants (1 and 2) came from two independent sources; the Amaze collection and the John Innes Centre collection. The site of insertion for pip2;1-1 is in the second intron whereas the site of insertion for pip2;1-2 is in the first intron. PIP2 genes and pip2 knockout mutants were previously observed to display distinct co-expression and transcriptional profiles, and PIP2 genes are distinctly co-regulated with other MIP genes, and specific PIP2 proteins can be differentially influenced by PIP1 expression at the protein level (Da Ines 2008). This means that there is the potential that MIP genes other than AtPIP2;1, or other mechanisms, could be influencing the differences in phenotypes between the two mutants from different mutant collections and further investigation is required to test this hypothesis.
Narrowing in on AtPIP2;1 roles in seed germination
Our results indicate a role or possibly multiple roles for AtPIP2;1 in seed germination, but the exact nature of this role(s) remains unclear. It could involve membrane transport, signalling or sensing. For example, rose (Rosa spp.) RhPIP2;1 serves as a key player in orchestrating trade-off between growth and stress survival (Zhang et al. 2019). AtPIP2;1 could similarly be involved in sensing or signalling in seeds. However, aquaporins such as AtPIP2;1 could also be involved in water absorption following germination (Matsunami et al. 2022). The next steps towards determining AtPIP2;1 role(s) in seed germination could involve:
Testing whether Atpip2;1 loss-of-function and AtPIP2;1 over-expression lines have delayed germination in non-saline osmotic stress, such as treatments involving germination tests in solutions containing sorbitol, mannitol or polyethylene glycol (PEG) (Liu et al. 2013c). This may help resolve whether AtPIP2;1 role in germination in saline conditions relates to contributing to osmotic adjustment or sensing osmotic stress.
Testing whether there are differences in the flux of Na+ and K+ from radicles of Atpip2;1 loss-of-function lines and AtPIP2;1 over-expression lines relative to controls, by using approaches such as microelectrode ion flux estimation (Shabala et al. 2003).
Testing the extent to which germination of Atpip2;1 loss-of-function and AtPIP2;1 over-expression lines and control lines change if their seed are exposed to hormone treatments during germination to explore the potential interaction between AtPIP2;1 regulation in response to hormones and regulation of germination.
Testing transcript abundance of the complement of Arabidopsis aquaporin encoding genes during seed imbibition and germination in Atpip2;1 loss-of-function and AtPIP2;1 over-expression lines to determine the extent to which the regulation of other aquaporins change when abundance of AtPIP2;1 is disrupted.
Examination the phosphorylation status of AtPIP2;1 given that this can determine the solute permeation status and membrane location (Johanson et al. 2001; Zhang et al. 2019; Qiu et al. 2020).
Resolving roles of aquaporins in seed germination is relevant to industrial applications such as optimising seed priming techniques (do Espirito Santo Pereira et al. 2021; Pachchigar et al. 2022). Approaches to optimise seed priming are relevant to improving crop yields for future food security (Paul et al. 2022).
Data availability
The data that support this study will be shared upon reasonable request to the corresponding author.
Declaration of funding
The work was supported by the Viet Nam International Cooperation Department (ICD), the Australian Research Council (ARC) Centre of Excellence in Plant Energy Biology (CE140100008), the ARC Centre of Excellence for Translational Photosynthesis (CE140100015), ARC DP190102725, and the Grains Research and Development Corporation (GRDC). CSB received support from ARC FT180100476.
Acknowledgements
We thank Wendy Sullivan for her critical role in providing support in maintaining equipment and resources used for this research.
References
Abascal F, Irisarri I, Zardoya R (2014) Diversity and evolution of membrane intrinsic proteins. Biochimica et Biophysica Acta – General subjects 1840(5), 1468-1481.
| Crossref | Google Scholar |
Arc E, Sechet J, Corbineau F, Rajjou L, Marion-Poll A (2013) ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Frontiers in Plant Science 4, 63.
| Crossref | Google Scholar |
Ayers AD, Hayward HE (1949) A method for measuring the effects of soil salinity on seed germination with observations on several crop plants. Soil Science Society of America Journal 13, 224-226.
| Crossref | Google Scholar |
Bafoil M, Le Ru A, Merbahi N, Eichwald O, Dunand C, Yousfi M (2019) New insights of low-temperature plasma effects on germination of three genotypes of Arabidopsis thaliana seeds under osmotic and saline stresses. Scientific Reports 9(1), 8649.
| Crossref | Google Scholar |
Bellati J, Champeyroux C, Hem S, Rofidal V, Krouk G, Maurel C, Santoni V (2016) Novel aquaporin regulatory mechanisms revealed by interactomics. Molecular & Cellular Proteomics 15(11), 3473-3487.
| Crossref | Google Scholar |
Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, Le BH, Drews GN, Brady SM, Goldberg RB, Harada JJ (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proceedings of the National Academy of Sciences 110(5), E435-E444.
| Crossref | Google Scholar |
Bewley JD (1997) Seed germination and dormancy. The Plant Cell 9(7), 1055-1066.
| Crossref | Google Scholar |
Bewley JD, Bradford KJ, Hilhorst HWM, Nonogaki H (2013) ‘Seeds: physiology of development, germination and dormancy.’ 3rd edn. (Springer: New York, NY, USA) doi:10.1007/978-1-4614-4693-4
Bienert GP, Chaumont F (2014) Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochimica et Biophysica Acta (BBA) – General Subjects 1840(5), 1596-1604.
| Crossref | Google Scholar |
Byrt CS, Zhao M, Kourghi M, Bose J, Henderson SW, Qiu J, Gilliham M, Schultz C, Schwarz M, Ramesh SA, Yool A, Tyerman S (2017) Non-selective cation channel activity of aquaporin AtPIP2;1 regulated by Ca2+ and pH. Plant, Cell & Environment 40(6), 802-815.
| Crossref | Google Scholar |
Ceciliato PHO, Zhang J, Liu Q, Shen X, Hu H, Liu C, Schäffner AR, Schroeder JI (2019) Intact leaf gas exchange provides a robust method for measuring the kinetics of stomatal conductance responses to abscisic acid and other small molecules in Arabidopsis and grasses. Plant Methods 15(1), 38.
| Crossref | Google Scholar |
Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R (2001) Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiology 125(3), 1206-1215.
| Crossref | Google Scholar |
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16(6), 735-743.
| Crossref | Google Scholar |
Coolbear P, Francis A, Grierson D (1984) The effect of low temperature pre-sowing treatment on the germination performance and membrane integrity of artificially aged tomato seeds. Journal of Experimental Botany 35(11), 1609-1617.
| Crossref | Google Scholar |
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133(2), 462-469.
| Crossref | Google Scholar |
Czechowski T, Stitt M, Altmann T, Udvardi M, Scheible W-R̈ (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139(1), 5-17.
| Crossref | Google Scholar |
Da Ines O, Graf W, Franck KI, Albert A, Winkler JB, Scherb H, Stichler W, Schaffner AR (2010) Kinetic analyses of plant water relocation using deuterium as tracer – reduced water flux of Arabidopsis pip2 aquaporin knockout mutants. Plant Biology 12, 129-139.
| Crossref | Google Scholar |
Daszkowska-Golec A (2011) Arabidopsis seed germination under abiotic stress as a concert of action of phytohormones. OMICS: A Journal of Integrative Biology 15(11), 763-774.
| Crossref | Google Scholar |
do Espirito Santo Pereira A, Caixeta Oliveira H, Fernandes Fraceto L, Santaella C (2021) Nanotechnology potential in seed priming for sustainable agriculture. Nanomaterials 11(2), 267.
| Crossref | Google Scholar |
Duan L, Dietrich D, Ng CH, Chan PMY, Bhalerao R, Bennett MJ, Dinneny JR (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. The Plant Cell 25(1), 324-341.
| Crossref | Google Scholar |
Dynowski M, Schaaf G, Loque D, Moran O, Ludewig U (2008) Plant plasma membrane water channels conduct the signalling molecule H2O2. Biochemical Journal 414(1), 53-61.
| Crossref | Google Scholar |
Fenner M, Thompson K (2005) ‘The ecology of seeds.’ (Cambridge University Press: Cambridge, UK) doi:10.1017/CBO9780511614101.007
Footitt S, Clewes R, Feeney M, Finch-Savage WE, Frigerio L (2019) Aquaporins influence seed dormancy and germination in response to stress. Plant, Cell & Environment 42, 2325-2339.
| Crossref | Google Scholar |
Gattolin S, Sorieul M, Frigerio L (2011) Mapping of tonoplast intrinsic proteins in maturing and germinating Arabidopsis seeds reveals dual localization of embryonic TIPs to the tonoplast and plasma membrane. Molecular Plant 4(1), 180-189.
| Crossref | Google Scholar |
Grondin A, Rodrigues O, Verdoucq L, Merlot S, Leonhardt N, Maurel C (2015) Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. The Plant Cell 27(7), 1945-1954.
| Crossref | Google Scholar |
Groszmann M, Osborn HL, Evans JR (2017) Carbon dioxide and water transport through plant aquaporins. Plant, Cell & Environment 40(6), 938-961.
| Crossref | Google Scholar |
Groszmann M, De Rosa A, Chen W, Qiu J, McGaughey SA, Byrt CS, Evans JR (2021) Permeability profiling of all 13 Arabidopsis PIP aquaporins using a high throughput yeast approach [Preprint]. bioRxiv
| Crossref | Google Scholar |
Guha T, Das H, Mukherjee A, Kundu R (2021) Elucidating ROS signaling networks and physiological changes involved in nanoscale zero valent iron primed rice seed germination sensu stricto. Free Radical Biology and Medicine 171, 11-25.
| Crossref | Google Scholar |
Han C-s, Kim S, Lee S-e, Choi S, Kim S-h, Yoon Is, Hwang Y-s (2018) Cross-talk between ABA and sugar signaling is mediated by the ACGT core and CE1 element reciprocally in OsTIP3;1 promoter. Journal of Plant Physiology 224-225, 103-111.
| Crossref | Google Scholar |
Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2(1), 19.
| Crossref | Google Scholar |
Hoai PTT, Tyerman SD, Schnell N, Tucker M, McGaughey SA, Qiu J, Groszmann M, Byrt CS (2020) Deciphering aquaporin regulation and roles in seed biology. Journal of Experimental Botany 71(6), 1763-1773.
| Crossref | Google Scholar |
Hooijmaijers C, Rhee JY, Kwak KJ, Chung GC, Horie T, Katsuhara M, Kang H (2012) Hydrogen peroxide permeability of plasma membrane aquaporins of Arabidopsis thaliana. Journal of Plant Research 125(1), 147-153.
| Crossref | Google Scholar |
Hu Y, Omary M, Hu Y, Doron O, Hoermayer L, Chen Q, Megides O, Chekli O, Ding Z, Friml J, Zhao Y, Tsarfaty I, Shani E (2021) Cell kinetics of auxin transport and activity in Arabidopsis root growth and skewing. Nature Communications 12(1), 1657.
| Crossref | Google Scholar |
Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiology 126(4), 1358-1369.
| Crossref | Google Scholar |
Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. The Plant Journal : for Cell and Molecular Biology 88(6), 1058-1070.
| Crossref | Google Scholar |
Kourghi M, Nourmohammadi S, Pei JV, Qiu J, McGaughey S, Tyerman SD, Byrt CS, Yool AJ (2017) Divalent cations regulate the ion conductance properties of diverse classes of aquaporins. International Journal of Molecular Sciences 18(11), 2323.
| Crossref | Google Scholar |
Li X, Pan Y, Chang B, Wang Y, Tang Z (2016) NO promotes seed germination and seedling growth under high salt may depend on EIN3 protein in Arabidopsis. Frontiers in Plant Science 6, 1203.
| Crossref | Google Scholar |
Liu C (2015) The PIP1 protein expression is positively regulated by PIP2;1 and PIP2;2 in Arabidopsis thaliana. Dissertation, Ludwig-Maximilians-Universität München. Available at https://edoc.ub.uni-muenchen.de/18857/1/Liu_Chen.pdf
Liu H-Y, Yu X, Cui D-Y, Sun M-H, Sun W-N, Tang Z-C, Kwak S-S, Su W-A (2007) The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Research 17, 638-649.
| Crossref | Google Scholar |
Liu C, Fukumoto T, Matsumoto T, Gena P, Frascaria D, Kaneko T, Katsuhara M, Zhong S, Sun X, Zhu Y, Iwasaki I, Ding X, Calamita G, Kitagawa Y (2013a) Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination. Plant Physiology and Biochemistry 63, 151-158.
| Crossref | Google Scholar |
Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang H-Q, Luan S, Li J, He Z-H (2013b) Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proceedings of the National Academy of Sciences 110(38), 15485-15490.
| Crossref | Google Scholar |
Liu X-M, Nguyen XC, Kim KE, Han HJ, Yoo J, Lee K, Kim MC, Yun D-J, Chung WS (2013c) Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochemical and Biophysical Research Communications 430(3), 1054-1059.
| Crossref | Google Scholar |
Lutts S, Benincasa P, Wojtyla L, Szymon Kubala S, Pace R, Lechowska K, Quinet M, Garnczarska M (2016) Seed priming: new comprehensive approaches for an old empirical technique. In ‘New challenges in seed biology – basic and translational research driving seed technology’. (Eds S Araujo, A Balestrazzi) (pp. 1–30). (InTech: London, UK) doi:10.5772/64420
Mahdieh M, Mostajeran A, Horie T, Katsuhara M (2008) Drought stress alters water relations and expression of PIP-type aquaporin genes in Nicotiana tabacum plants. Plant and Cell Physiology 49(5), 801-813.
| Crossref | Google Scholar |
Manz B, Muller K, Kucera B, Volke F, Leubner-Metzger G (2005) Water uptake and distribution in germinating tobacco seeds investigated in vivo by nuclear magnetic resonance imaging. Plant Physiology 138(3), 1538-1551.
| Crossref | Google Scholar |
Matsunami M, Hayashi H, Murai-Hatano M, Ishikawa-Sakurai J (2022) Effect of hydropriming on germination and aquaporin gene expression in rice. Plant Growth Regulation 97, 263-270.
| Crossref | Google Scholar |
Maurel C, Chrispeels M, Lurin C, Tacnet F, Geelen D, Ripoche P, Guern J (1997) Function and regulation of seed aquaporins. Journal of Experimental Botany 48(Special_Issue), 421-430.
| Crossref | Google Scholar |
McDowell SC, Akmakjian G, Sladek C, Mendoza-Cozatl D, Morrissey JB, Saini N, Mittler R, Baxter I, Salt DE, Ward JM, Schroeder JI, Guerinot ML, Harper JF (2013) Elemental concentrations in the seed of mutants and natural variants of Arabidopsis thaliana grown under varying soil conditions. PLoS ONE 8(5), e63014.
| Crossref | Google Scholar |
McGaughey SA, Qiu J, Tyerman SD, Byrt CS (2018) Regulating root aquaporin function in response to changes in salinity. Annual Plant Reviews online 1(2), 1-36.
| Crossref | Google Scholar |
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annual Review of Plant Biology 59(1), 651-681.
| Crossref | Google Scholar |
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15(3), 473-497.
| Crossref | Google Scholar |
Narsai R, Gouil Q, Secco D, Srivastava A, Karpievitch YV, Liew LC, Lister R, Lewsey MG, Whelan J (2017) Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biology 18, 172.
| Crossref | Google Scholar |
Nasri N, Maatallah S, Kaddour R, Lachaal M (2016) Effect of salinity on Arabidopsis thaliana seed germination and acid phosphatase activity. Archives of Biological Sciences 68(1), 17-23.
| Crossref | Google Scholar |
Niemann S, Burghardt M, Popp C, Riederer M (2013) Aqueous pathways dominate permeation of solutes across Pisum sativum seed coats and mediate solute transport via diffusion and bulk flow of water. Plant, Cell & Environment 36(5), 1027-1036.
| Crossref | Google Scholar |
Obroucheva NV (2013) Aquaporins in seeds. Seed Science Research 23(4), 213-216.
| Crossref | Google Scholar |
Paul S, Dey S, Kundu R (2022) Seed priming: an emerging tool towards sustainable agriculture. Plant Growth Regulation 97(2), 215-234.
| Crossref | Google Scholar |
Péret B, Li G, Zhao J, Band LR, Voß U, Postaire O, Luu D-T, Da Ines O, Casimiro I, Lucas M (2012) Auxin regulates aquaporin function to facilitate lateral root emergence. Nature Cell Biology 14(10), 991-998.
| Crossref | Google Scholar |
Prado K, Cotelle V, Li G, Bellati J, Tang N, Tournaire-Roux C, Martinière A, Santoni V, Maurel C (2019) Oscillating aquaporin phosphorylation and 14-3-3 proteins mediate the circadian regulation of leaf hydraulics. The Plant Cell 31(2), 417-429.
| Crossref | Google Scholar |
Qing D, Yang Z, Li M, Wong WS, Guo G, Liu S, Guo H, Li N (2016) Quantitative and functional phosphoproteomic analysis reveals that ethylene regulates water transport via the C-terminal phosphorylation of aquaporin PIP2;1 in Arabidopsis. Molecular Plant 9(1), 158-174.
| Crossref | Google Scholar |
Qiu J, McGaughey SA, Groszmann M, Tyerman SD, Byrt CS (2020) Phosphorylation influences water and ion channel function of AtPIP2;1. Plant, Cell & Environment 43(10), 2428-2442.
| Crossref | Google Scholar |
Ravindran P, Yong SY, Mohanty B, Kumar PP (2020) An LRR-only protein regulates abscisic acid-mediated abiotic stress responses during Arabidopsis seed germination. Plant Cell Reports 39(7), 909-920.
| Crossref | Google Scholar |
Reid DA, Lott JNA, Attree SM, Fowke LC (1999) Imbibition of white spruce seeds and somatic embryos: a study of morphological changes in an environmental scanning electron microscope and potassium leakage. In Vitro Cellular & Developmental Biology – Plant 35(4), 303-308.
| Crossref | Google Scholar |
Rodrigues O, Reshetnyak G, Grondin A, Saijo Y, Leonhardt N, Maurel C, Verdoucq L (2017) Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proceedings of the National Academy of Sciences 114(34), 9200-9205.
| Crossref | Google Scholar |
Rosental L, Nonogaki H, Fait A (2014) Activation and regulation of primary metabolism during seed germination. Seed Science Research 24(1), 1-15.
| Crossref | Google Scholar |
Shabala S, Shabala L, Volkenburgh EV (2003) Effect of calcium on root development and root ion fluxes in salinised barley seedlings. Functional Plant Biology 30(5), 507-514.
| Crossref | Google Scholar |
Simon EW, Harun RMR (1972) Leakage during Seed Imbibition. Journal of Experimental Botany 23(4), 1076-1085.
| Crossref | Google Scholar |
Su Y, Liu Z, Sun J, Wu C, Li Y, Zhang C, Zhao L (2022) Genome-wide identification of maize aquaporin and functional analysis during seed germination and seedling establishment. Frontiers in Plant Science 13, 831916.
| Crossref | Google Scholar |
Takahashi H, Rai M, Kitagawa T, Morita S, Masumura T, Tanaka K (2004) Differential localization of tonoplast intrinsic proteins on the membrane of protein body type II and aleurone grain in rice seeds. Bioscience, Biotechnology, and Biochemistry 68(8), 1728-1736.
| Crossref | Google Scholar |
Tian S, Wang X, Li P, Wang H, Ji H, Xie J, Qiu Q, Shen D, Dong H (2016) Plant aquaporin AtPIP1;4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiology 171(3), 1635-1650.
| Crossref | Google Scholar |
Tuan PA, Nguyen T-N, Jordan MC, Ayele BT (2021) A shift in abscisic acid/gibberellin balance underlies retention of dormancy induced by seed development temperature. Plant, Cell & Environment 44(7), 2230-2244.
| Crossref | Google Scholar |
Tyerman SD, McGaughey SA, Qiu J, Yool AJ, Byrt CS (2021) Adaptable and multifunctional ion-conducting aquaporins. Annual Review of Plant Biology 72(1), 703-736.
| Crossref | Google Scholar |
Utsugi S, Shibasaka M, Maekawa M, Katsuhara M (2015) Control of the water transport activity of barley HvTIP3;1 specifically expressed in seeds. Plant and Cell Physiology 56(9), 1831-1840.
| Crossref | Google Scholar |
Vander Willigen C, Postaire O, Tournaire-Roux C, Boursiac Y, Maurel C (2006) Expression and inhibition of aquaporins in germinating Arabidopsis seeds. Plant and Cell Physiology 47(9), 1241-1250.
| Crossref | Google Scholar |
Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136(6), 1005-1016.
| Crossref | Google Scholar |
Van Steveninck J, Ledeboer AM (1974) Phase transitions in the yeast cell membrane the influence of temperature on the reconstitution of active dry yeast. Biochimica et Biophysica Acta (BBA) – Biomembranes 352(1), 64-70.
| Crossref | Google Scholar |
Verdoucq L, Rodrigues O, Martiniere A, Luu DT, Maurel C (2014) Plant aquaporins on the move: reversible phosphorylation, lateral motion and cycling. Current Opinion in Plant Biology 22, 101-107.
| Crossref | Google Scholar |
Wang Y, Tajkhorshid E (2010) Nitric oxide conduction by the brain aquaporin AQP4. Proteins: Structure, Function, and Bioinformatics 78(3), 661-670.
| Crossref | Google Scholar |
Wang C, Hu H, Qin X, Zeise B, Xu D, Rappel W-J, Boron WF, Schroeder JI (2016) Reconstitution of CO2 regulation of SLAC1 anion channel and function of CO2-permeable PIP2;1 aquaporin as CARBONIC ANHYDRASE4 interactor. The Plant Cell 28(2), 568-582.
| Crossref | Google Scholar |
Weitbrecht K, Müller K, Leubner-Metzger G (2011) First off the mark: early seed germination. Journal of Experimental Botany 62(10), 3289-3309.
| Crossref | Google Scholar |
Wilson RL, Kim H, Bakshi A, Binder BM (2014) The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiology 165(3), 1353-1366.
| Crossref | Google Scholar |
Wojtyla L, Lechowska K, Kubala S, Garnczarska M (2016) Different modes of hydrogen peroxide action during seed germination. Frontiers in Plant Science 7, 66.
| Crossref | Google Scholar |
Zhang W-H, Zhou Y, Dibley KE, Tyerman SD, Furbank RT, Patrick JW (2007) Review: Nutrient loading of developing seeds. Functional Plant Biology 34(4), 314-331.
| Crossref | Google Scholar |
Zhang S, Feng M, Chen W, Zhou X, Lu J, Wang Y, Li Y, Jiang C-Z, Gan S-S, Ma N, Gao J (2019) In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nature Plants 5(3), 290-299.
| Crossref | Google Scholar |
Zhou Y, Setz N, Niemietz C, Qu H, Offler CE, Tyerman SD, Patrick JW (2007) Aquaporins and unloading of phloem-imported water in coats of developing bean seeds. Plant, Cell & Environment 30(12), 1566-1577.
| Crossref | Google Scholar |


