First molecular detection and identification of a potyvirus associated with severe mosaic disease of amaryllis (Hippeastrum hybridum Hort.) in India
S. K. Raj A B , S. K. Snehi A , S. Kumar A and M. S. Khan AA Plant Molecular Virology, National Botanical Research Institute, Rana Pratap Marg, Lucknow 226 001 (U.P.), India.
B Corresponding author. Email: skraj2@rediffmail.com
Australasian Plant Disease Notes 4(1) 50-53 https://doi.org/10.1071/DN09021
Submitted: 27 August 2008 Accepted: 4 May 2009 Published: 15 June 2009
Abstract
A potyvirus associated with mosaic disease of amaryllis was detected and identified by reverse-transcription and polymerase chain reaction using degenerate primers specific to the core region of coat protein gene of potyviruses followed by cloning and sequencing. The sequence shared highest (98–95%) amino acid identities with Amaryllis potyvirus that is not yet recognised by the International Committee on Taxonomy of Viruses, Amazon lily mosaic virus (tentative potyvirus) and Hippeastrum mosaic virus (recognised potyvirus). Screening bulbs of elite amaryllis cultivars for potyvirus infection would be useful to establish virus-free propagating material for the floriculture industry in India.
Amaryllis (Hippeastrum hybridum Hort. family Amaryllidaceae) is a popular bulbous ornamental plant grown in gardens and pots for beautiful blooms of various colours. It has been grown on a commercial scale in The Netherlands, South Africa and USA (Meerow et al. 1991) and is now emerging as an important cut flower for the floriculture industry in India. An extensive program was initiated within the botanic gardens of the floriculture division of the National Botanical Research Institute, Lucknow, to develop novel Hippeastrum varieties through conventional breeding resulting in 21 varieties with attractive colour combinations (Datta and Gupta 2000; Datta et al. 2004).
During April–May, 2007, a mosaic disease in ~15% H. hybridum was observed in house gardens in Lucknow, India. The diseased plants exhibited severe mosaic symptoms accompanied with light-green to yellow striped symptoms on leaves (Fig. 1) and affected plants bore small-sized flowers. Since vegetative propagation through amaryllis stock bulbs is commonly used and will likely lead to large-scale dissemination of systemic viruses that may affect the quality of blooms, virus screening of propagation materials is essential.

|
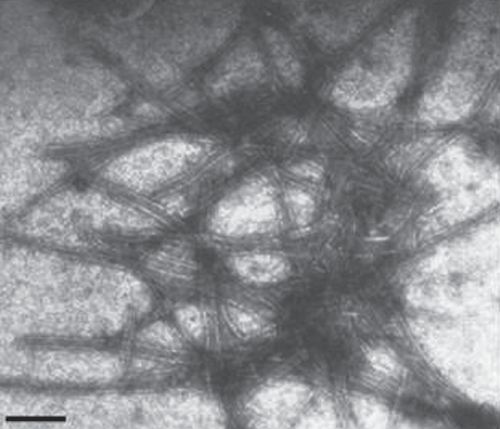
Mechanical inoculations using sap from infected H. hybridum leaf failed to produce any local or systemic symptoms on Chenopodium amaranticolor, Nicotiana glutinosa, N. rustica and N. benthamiana. Purification of the virus from sap of infected young leaves of H. hybridum showing mosaic symptoms, extracted in 0.1 molar sodium phosphate buffer, pH 7.2 was attempted by a protocol described earlier by Aminuddin et al. (1999). The sap was clarified by 15% n-butanol and virus was precipitated with 6% polyethylene glycol (8000) supplemented with 1.5% sodium chloride, followed by sedimentation through 15% sucrose pad containing 0.1% Triton X-100 at 95 868.5g for 2 h in a Beckman Ti 45 rotor at 4°C. The purified preparation was observed under transmission electron microscope (Philips 420, USA) using 2% uranyl acetate, pH 4.2 as a negative strain which revealed the presence of flexuous rod-shaped particles of 730 × 12 nm (Fig. 2). Morphology and size of the particle of the virus isolate was similar to potyviruses (Harrison et al. 1971).
|
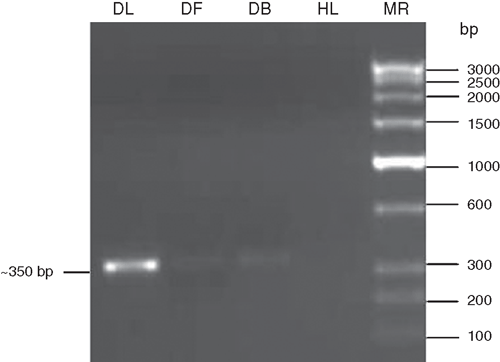
Further evidence of a potyvirus associated with the mosaic disease of H. hybridum was carried out by reverse transcription-polymerase chain reaction (RT–PCR) using coat protein core region specific degenerate primers of potyviruses followed by sequence analysis of RT–PCR amplicons. Total RNA was extracted from 100 mg infected amaryllis leaf, flower and bulb samples and healthy leaf (as negative control) following the method described by Pawlowski et al. (1994). RT–PCR was performed using the degenerate forward primer ‘oligo1n’ encoding the MVWCIEN motif and reverse primer ‘oligo2n’ corresponding to the reverse complimentary of the nucleotide sequence encoding the QMKAAA motif in the core of the coat protein gene of potyviruses (Langeveld et al. 1991; Marie-Jeanne et al. 2000). First strand cDNA synthesis was performed in a 20 µL reaction mixture containing: total RNA as a template (1µg, 6.0 µL); reverse primer ‘oligo2n’ (25 p.m., 1.0 µL); RNA guard (25 units 1.0 µL); RT-buffer (5X, 4.0 µL); AMV-reverse transcriptase (20 units, 1.0 µL, Genei Pvt. Ltd., Bangalore, India); dNTPs (10 mmol/L of each, 2.0 µL); and water to make up reaction volume and reaction was incubated at 42°C for 90 min. PCR was performed in a 50-µL reaction mixture containing: cDNA (3.0 µL); Pfu DNA polymerase buffer (10X, 5.0 µL); MgCl2 (25 mmol/L, 3.0 µL); reverse ‘oligo2n’ and forward ‘oligo1n’ primers (25 p.m., 1.0 µL each); dNTPs (10 mmol/L each, 1.0 µL) and Pfu DNA polymerase (3 U, 1.0 µL, Genei Pvt. Ltd.) and water to make up the reaction volume. The PCR conditions were: initial denaturation at 94°C for 3 min followed by denaturation at 94°C for 30 s, annealing at 50°C for 2 min and extension at 72°C for 50 s. After completing 30 cycles, there was final extension at 72°C for 7 min. The amplified PCR products were electrophoresed with a known low-range DNA ruler (Genei Pvt. Ltd.) on 1.5% agarose gel to assess the size of amplicons. The electrophoresis of PCR products resulted in the expected size (~350 bp) amplicons derived from leaf, flower and bulb samples of diseased H. hybridum but not from healthy plants (Fig. 3).
|
The amplicon obtained from H. hybridum leaf was cloned in pGEM-T easy cloning vector (Promega, Madison, WI, USA), transformed into Escherichia coli strain DH5α and clones were selected on Luria agar plates containing ampicillin (100 mg/L), X-gal (80 µg/mL) and Isopropyl β-D-1 thiogalactopyranosides (0.5 mmol/L). Three positive clones were sequenced, resulting in a consensus sequence with no ambiguities. The sequence data were deposited in NCBI GenBank under accession number EU249375. Basic local alignment search tool (BLAST) analysis of deposited nucleotide sequence revealed that it shared 95% sequence identity with a sequence not yet recognised by the International Committee on Taxonomy of Viruses (ICTVdB Management 2006), referred to as Amaryllis potyvirus (Accession AY566239). The sequence also showed 90–91% identity with Hippeastrum mosaic virus (Accession EF203685) and a tentative member of the Potyviridae Amazon lily mosaic virus (Accession AY590143), 69–68% with Plum pox virus (Accession AF440746) and Nerine yellow stripe virus (Accession EU042758), and 65% with Amazon lily mosaic virus (Accession AB158523, Fuji et al. 2004). The putative amino acid sequence (108 aa) of virus isolate’s core coat protein sequence revealed by BLAST analysis 95–98% identities with the sequence for the Amaryllis potyvirus (Accession AAT67221), Amazon lily mosaic virus (Accession AAT01303) and Hippeastrum mosaic virus (Accession ABM97616) and less than 70% identities with Nerine yellow stripe virus (Accession ABN45757). Earlier, Shukla and Ward (1989) clearly demonstrated that species of potyviruses have less than 80% amino acid homology with other species, whereas strains of a virus show more than 85% amino acid homology. Based on 95–98% amino acid identities with the sequence of the Amaryllis potyvirus (Acc. AAT67221, not recognised by ICTV), Amazon lily mosaic virus (Acc. AAT01303, tentative potyvirus) and Hippeastrum mosaic virus (Acc. ABM97616, recognised potyvirus); and the lack of clarity for the nomenclature and sequence of Amazon lily mosaic virus (two accessions BAD27401 and BAD27400 that have only 62–65% amino acid identity) (Hamilton et al. 1981), the virus under study may therefore be considered a strain of Hippeastrum mosaic virus.
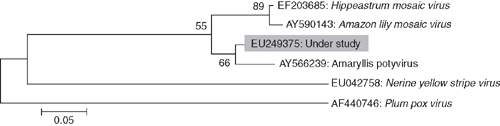
Phylogenetic analysis of nucleotide sequence of the core of the coat protein of the virus isolate with the most related potyviruses or potyvirus-like sequences was performed using molecular evolutionary genetics analysis (MEGA) version 4.0 tool (Tamura et al. 2007). The analyses revealed closest relationships of the virus isolate with the sequence referred to as Amaryllis potyvirus (Accession AY566239) (Fig. 4). On the basis of highest nucleotide (95%) and amino acid (98%) sequence identities at the core region of coat protein gene and closest phylogenetic relationship with Amaryllis potyvirus (Accession AY566239, FL Chiang, YJ Lin, CC Chen, CA Chang, 2005 unpublished report from Taiwan), the virus associated with mosaic disease of H. hybridum was identified as a strain of Hippeastrum mosaic virus.
|
Amaryllis (Hippeastrum species) is known to be infected by Amaryllis mosaic virus (synonym of Hippeastrum mosaic virus, Brants and van den Heuvel 1965; Pirone 1978), Nerine latent virus (Schubert and Herwiing 1980), Tomato spotted wilt virus (Brunt et al. 1996), Cucumber mosaic virus (Gutiérrez-Villegas et al. 2004) and Tobacco mosaic virus (De Leeuw 1972) in Asia, Europe and America where it is being cultivated commercially. A report of a Alphacryptovirus (of family Partitiviridae) in amaryllis from India, based on enzyme-linked immunosorbent assay (ELISA) test, exists in the literature (Raizada et al. 1983). However, molecular detection and identification of a strain of Hippeastrum mosaic virus associated with severe mosaic disease of Hippeastrum hybridum is a first report from India.
Since H. hybridum is propagated through bulbs and the mother stock once infected, act as a source for disease spread in successive generations, detection of potyviruses in bulbs of elite amaryllis cultivars through RT–PCR (a sensitive procedure for detection of RNA viruses) would be of high practical value for establishing virus-free propagation material for the emerging floriculture industry and for quarantine.
Acknowledgements
The authors express their gratitude to the Director, National Botanical Research Institute, Lucknow for facilities and to Director, Central Institute of Medicinal and Aromatic Plants for electron microscope facilities.
Aminuddin
,
Khan JA, Raj SK
(1999) Association of an unknown potyvirus isolate with severe mosaic of Narcissus tazetta L. Indian Journal of Experimental Biology 37, 1034–1036.

Brants DH, van den Heuvel J
(1965) Investigation of Hippeastrum mosaic virus in Hippeastrum hybridum. Netherlands Journal of Plant Pathology 71, 145–151.
| Crossref | GoogleScholarGoogle Scholar |

Datta SK, Gupta VN
(2000)
Hippeastrum-an excellent bulbous plant at National Botanical Research Institute, Lucknow, India. Applied Botany Abstract 20, 248–260.

De Leeuw GTN
(1972) Tobacco mosaic virus in Hippeastrum hybridum. European Journal of Plant Pathology 78, 69–71.

Fuji S,
Terami F,
Furuya H,
Naito H, Fukumoto F
(2004) Nucleotide sequence of the coat protein genes of Alstroemeria mosaic virus and Amazon lily mosaic virus, a tentative species of genus Potyvirus. Archives of Virology 149, 1843–1849.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Gutiérrez-Villegas C,
Ruiz-Medrano R,
Piedra-Ibarra E, de la Torre-Almaráz R
(2004) Characterization of a Cucumber mosaic virus strain associated with yellow mottle symptoms of amaryllis (Hippeastrum × hybridum Leopoldii) in México. Agrociencia (Montecillo) 38, 343–354.

Hamilton RI,
Edwardson JR,
Francki RIB,
Hsu HT,
Hall R,
Koenig R, Milne RG
(1981) Guidelines for the identification and characterization of plant viruses. The Journal of General Virology 54, 223–241.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Harrison BD,
Finch JT,
Gibbs AJ,
Hollings M,
Shepard RJ,
Valena V, Wetter C
(1971) Sixteen group of plant viruses. Virology 45, 356–363.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Langeveld SA,
Doe JM,
Memelink J,
Derks AFLM,
van der Vlugt CIM,
Asjes CJ, Bol JF
(1991) Identification of potyviruses using the polymerase chain reaction with degenerate primers. The Journal of General Virology 72, 1531–1541.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Marie-Jeanne V,
Ioos R,
Peyre J,
Alliot B, Signoret P
(2000) Differentiation of Poaceae potyviruses by reverse transcription-polymerase chain reaction and restriction analysis. Journal of Phytopathology 148, 141–151.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Meerow AW,
Broschat TK, Kane ME
(1991)
Hippeastrum breeding at University of Florida. Herbertia 47, 4–10.

Pawlowski K,
Kunze R,
Vries SD, Bisseling T
(1994) Isolation of total, poly(A) and polysomal RNA from plant tissues. Plant Molecular Biology Manual D5, 1–13.

Raizada RK,
Srivastava KM, Singh BP
(1983) Detection of Amaryllis cryptic virus by Enzyme linked immunosorbent assay. Indian Journal of Experimental Biology 21, 299–300.

Shukla DD, Ward CW
(1989) Identification and classification of potyvirus on the basis of coat protein sequence data and serology. Archives of Virology 106, 171–200.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Tamura K,
Dudley J,
Nei M, Kumar S
(2007) MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |



