Apricot is a new host of Apple scar skin viroid
Ying Zhao A and Jianxin Niu A BA Horticultural Department, Agricultural College of Shihezi University, Shihezi 832003, China.
B Corresponding author. Email: njx105@163.com
Australasian Plant Disease Notes 3(1) 98-100 https://doi.org/10.1071/DN08039
Submitted: 7 March 2008 Accepted: 8 July 2008 Published: 17 July 2008
Abstract
Apple scar skin viroid (ASSVd), a pathogenic RNA viroid, infects apple and pear trees. To determine if ASSVd can also infect apricot, we performed reverse transcription–polymerase chain reaction (RT-PCR), spot hybridisation and in situ RT-PCR and detected ASSVd-specific sequences from apricot. We cloned and sequenced the apricot-derived ASSVd and registered it in GenBank. In situ RT-PCR further confirmed the existence of ASSVd in apricot leaf tissues. Our data indicate that apricot is a novel host of ASSVd. These findings provide the basis for the rapid identification of ASSVd in apricot.
Viroids are the smallest pathogens discovered to date, consisting of a single-stranded RNA of 246–574 nucleotides in an unencapsulated, covalently closed-circular conformation. Viroids do not encode any proteins and they depend on the host for replication (Guo 2006). Viroids are classified into two families, Pospiviroidae and Avsunviroidae, based on homology in primary sequences, secondary structures and replication characteristics. Apple scar skin viroid (ASSVd), a member of the Pospiviroidae that replicates in the nucleus through an asymmetrical rolling circle mechanism, usually comprises 330 nucleotides, and possesses no nuclease activity.
Koganezawa et al. (1983) first reported the detection of ASSVd RNA in diseased apple fruits. Subsequently, Chen et al. (1986) isolated ASSVd from diseased branches, determined the sequence of the RNA, and proved that apple scar skin was a viroid disease. Hashimoto and Koganezawa (1987) first reported the entire sequence of ASSVd. Subsequently, Guo et al. (2005) and Zhao and Niu (2006) have cloned and analysed its sequence. ASSVd can infect apple and pear (Hadidi et al. 1991; Yang et al. 1992; Koganezawa et al. 2003; Kyriakopoulou et al. 2003), but previously it was not known if ASSVd could also infect apricot. In this study, we detected ASSVd by reverse transcription–polymerase chain reaction (RT-PCR), spot hybridisation and in situ RT-PCR in apricot trees in Xinjiang, China. Thus, for the first time, our study showed that ASSVd is also a pathogen of apricot trees, suggesting that ASSVd may have a wider impact in agricultural disease.
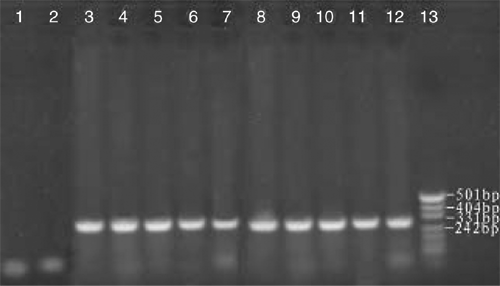
Tender leaves and shoots used for RNA extraction were collected from a 50-year-old orchard in Yanqi County, Xinjiang, China, growing many species of fruit trees, including apple, pear, peach and apricot. The infected apricot tree showed the symptoms of leaf epinasty and distortion. Biologically characterised ASSVd samples numbered AP1 to AP10 were thus obtained from symptomatic trees. Samples AP1 to AP10 were collected from 10 different trees that had symptomatic leaves. Spot hybridisation confirmed the presence of ASSVd in each of the samples. RNA extraction was performed using ~250 mg of fresh or frozen tender leaf tissue, which was ground to a fine powder in liquid nitrogen and extracted using the method of Zhao and Niu (2006). The isolated total RNA (0.2 µg) from each sample was used as template for RT. Primers were designed based on the reported sequences by The National Center for Biotechnology Information of USA (NCBI, GenBank accession number X17696) (Puchta et al. 1990): ASSVd1: 5′-CAGCACCACAGGAACCTCACGG-3′ (antisense, 10–32); ASSVd2: 5′-CTCGTCGTCGACGAAGG–3′ (sense, 80–97) and were synthesised by Shanghai Biological Engineering Technology and Service Corp. The antisense primer ASSVd1(20 µM) was used as a primer for RT by Moloney murine leukemiavirus (M-MLV) reverse transcriptase (Shanghai Biological Engineering Technology & Service Corp.) at 42°C for 1 h in 50 mM Tris-Cl pH 8.3, 75 mM KCl, 3 mM MgCl2, and 10 mM DDT. PCR was performed in 10 mM dNTPs, 2 mM Mg2+, antisense and sense primers (ASSVd1 and ASSVd2 at 20 µM), cDNA (3 µg) and 2.5 U Taq DNA polymerase (Shanghai Biological Engineering Technology & Service Corp.). Samples were amplified for 32 cycles as follows: denaturation at 95°C for 3 min, 62°C for 45 s and 72°C for 60 s, with the final extensions at 72°C for 7 min. The amplified products were separated by electrophoresis on 2% (w/v) non-denaturing agarose gel in 1 × TBE buffer at 120 V for 40 min, and visualised by staining with Gold Viewer Dye (Beijing Liuyi Factory, China). PCR markers (Hua Mei, China) were used to determine the sizes of the amplified fragments. Expected fragments of ~330 bp were obtained through RT-PCR using specific primers for ASSVd (Fig. 1).
|
To identify the sequence of each amplified product from RNA isolated from each sample of AP1 to AP10, the amplified products were separated by electrophoresis on a 2% agarose gel in 1 × TAE buffer, excised from the gel under ultraviolet light and recovered using UNIQ-10 Gel Extraction Kit (Sangon) according to manufacturer’s protocol. The recovered fragments were cloned into pUCm-T vector (Shanghai Biological Engineering Technology and Service Corp.). A representative clone from each sample was submitted to Shanghai Biological Engineering Technology and Service Corp for sequencing. The 10 sequences obtained in this study showed at least 98% sequence identity to each other and were submitted to GenBank (accession numbers EU031487 to EU031496). Sequence analysis of the selected clones revealed that the full-length of viroid in apricot was 330 nt and exhibited the highest homology with the ASSVd (accession number X17696) using Basic Local Alignment Search Tool (BLAST).
Three full length apple isolates of ASSVd from different regions of China (Liaoning isolate (accession number AY972082, Guo et al. 2005), China isolate (accession numbers NC_001340 and X17696, Puchta et al. 1990) and Xinjiang isolate, Zhao et al. 2006)) showed no less than 99.7% homology with each other demonstrating low sequence variation across geographical regions. The ASSVd sequences obtained in this study from an apricot tree grown in Yanqi County, Xinjiang are 98% identical to these apple isolates. This finding suggests that the apricot isolate of ASSVd has been transmitted from an infected apple tree in this region.
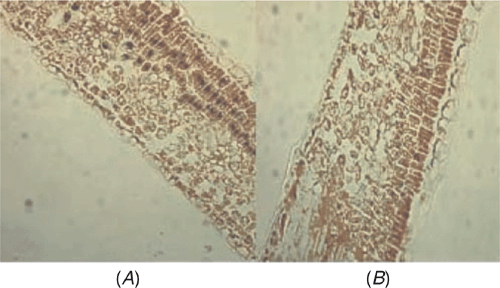
The distribution of ASSVd within apricot leaf tissues was investigated by in situ RT-PCR. Paraffin sections of apricot leaves were prepared according to the method of Niu et al. (2007). Complementary DNA synthesis was performed according to the method of Zhao and Niu (2006) and in situ RT-PCR amplification and immunological detection used the methods of Niu et al. (2007). Positive signals (Fig. 2A) were detected in palisade cells while it was not detected when no RT was included in the protocol (Fig. 2B). These data demonstrate that ASSVd does indeed infect apricot cells.
|
Previously it has been shown that ASSVd can infect apple and pear trees (Puchta et al. 1990; Zhu et al. 1995; Guo et al. 2005; Zhao and Niu 2006, 2007) but there was no report that ASSVd has been discovered in trees that bear drupes. We describe the amplification, cloning and sequencing of full-length ASSVd from apricot grown in China (GenBank accessions EU031477 to EU031486) and the existence of ASSVd in apricots was reconfirmed by in situ RT-PCR. Our study showed that ASSVd was detectable in apricot and provides the basis for the rapid identification of ASSVd in apricot.
Acknowledgements
This work was supported by the National Natural Science Foundation of China program (30360066) and the National Key Technologies R&D Program (2003BA546C).
Chen W,
Tian P,
Jin LP,
Wang GP, Liu FC
(1986) Study of viroid RNA isolated from apple scar skin disease tissues. Chinese Journal of Virology 2, 79–84.

Guo R,
Li SF,
Dong YF, Cheng ZM
(2005) Occurrence of Apple scar skin disease and sequence analysis of Apple scar skin viroid in Liaoning. Acta Phytopathologica Sinica 35, 472–474.

Hadidi A,
Hansen AJ,
Parish CJ, Yang X
(1991) Scar skin and dapple apple viroids are seed-borne and persistent in infected apple trees. Research in Virology 142, 289–296.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hashimoto J, Koganezawa H
(1987) Nucleotide sequence and secondary structure of Apple scar skin viroid. Nucleic Acids Research 15, 7045–7052.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Koganezawa H,
Yanase H, Sakuma T
(1983) Viroid-like RNA associated with apple scar skin (or dapple apple) disease. Acta Horticulturae 130, 193–197.

Niu JX,
Zhou MS,
Ma BG,
Zhao Y, Liu H
(2007) Detection of Apple chlorotic leaf sopt virus in pear by in situ RT-PCR. Acta Horticulturae Sinica 34, 53–58.

Puchta H,
Luckinger R,
Yang XC,
Hadidi A, Saenger HL
(1990) Nucleotide sequence and secondary structure of Apple scar skin viroid (ASSVd) from China. Plant Molecular Biology 14, 1065–1067.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Yang X,
Hadidi A, Hammond RW
(1992) Nucleotide sequence of Apple scar skin viroid reverse transcribed in host extracts and amplified by the polymerase chain reaction. Acta Horticulturae 309, 305–309.

Zhao Y, Niu JX
(2006) Cloning and sequencing of Sinkiang isolate of Apple scar skin viroid (ASSVd). Journal of Fruit Science 23, 896–898.

Zhao Y, Niu JX
(2007) Detection of Apple scar skin viroid in pear cultivars by RT-PCR and spot hybridization in Xinjiang area. Journal of Fruit Science 24, 761–764.

Zhu SF,
Hadidi A, Hammond RW
(1995) Nucleotide sequence and secondary structure of pome fruit viroids from apple disease apples, pear rusty skin diseased pears and apple scar skin symptomless pear. Acta Horticulturae 386, 554–559.



