Plant species, nitrogen status and endophytes are drivers of soil microbial communities in grasslands
Susanne Rasmussen A , Anthony J. Parsons A , Julia Russell B , Daniel A. Bastías C and Qianhe Liu
C and Qianhe Liu  C *
C *
A
B
C
Abstract
There is concern that the introduction of ‘novel’ plant germplasm/traits could outpace our capacity to measure and so assess their impacts on soil microbial communities and function.
This study aimed to investigate the effects of plant species/functional traits, nitrogen (N) fertilisation and endophyte infection on grassland soil microbial communities within a short time span of 2 years.
Two field experiments with monoculture plots were conducted in a common soil. Experiment 1 compared grasses and legumes, using two cultivars of perennial ryegrass (Lolium perenne) that varied in fructan content, along with the legumes white clover (Trifolium repens) and bird’s-foot trefoil (Lotus pedunculatus) that varied in tannin content. Grass treatments received high and low N application levels. Experiment 2 compared the presence/absence of Epichloë strains in ryegrass, tall fescue (Schedonorus phoenix) and meadow fescue (Schedonorus pratensis). Soil microbial communities were analysed by using high-throughput sequencing of DNA isolated from bulk soil cores.
Higher abundance of ligninolytic fungi was found in grass soils and pectinolytic fungi in legume soils. Levels of N fertilisation and fructan in ryegrass had only minor effects on soil fungal communities. By contrast, N fertilisation or fixation had a strong effect on bacterial communities, with higher abundance of nitrifiers and denitrifiers in high-N grass soils and in legume soils than in low-N grass soils. Epichloë affected soil microbiota by reducing the abundance of certain fungal phytopathogens, increasing mycorrhizal fungi and reducing N-fixing bacteria.
Chemical composition of plant cell walls, which differs between grasses and legumes, and presence of Epichloë in grasses were the main drivers of shifts in soil microbial communities.
Impacts of farming practices such as mono- or poly-culture, N fertilisation and presence of Epichloë in grasses on soil microbial communities should be considered in pasture management.
Keywords: bulk soil, Epichloë, legumes, microbiota, nitrogen, pastures, soil microbial community, temperate grasslands.
Introduction
The myriad soil microbial species perform a diverse array of metabolic functions and are central to global carbon (C) and nitrogen (N) sequestration and cycling. Interactions and feedback between above- and belowground organisms are increasingly seen to control terrestrial ecosystem processes and services (Wardle et al. 2004; De Deyn et al. 2008; van der Heijden et al. 2008; Zak et al. 2011; Tomazelli et al. 2023). Sustaining those services, with any degree of certainty, faces increasing challenge.
Managed temperate grasslands occupy 25% of the world’s land area, which is 70% of global agricultural land (Ghahramani et al. 2019). Those grasslands are a major source or sink for C and N (Oenema et al. 2005; McSherry and Ritchie 2013), and their management has significant implications for greenhouse gas emissions (Jones and Donnelly 2004; Reisinger and Ledgard 2013; Parsons et al. 2016). In grassland ecosystems, even simple management changes such as N fixation or fertilisation can substantially affect the scale of C and N inputs, as well as the C:N ratio balance, not least because grazing ruminants (as major aboveground heterotrophs) effectively ‘uncouple’ the C and N cycles (Parsons et al. 2013; Soussana and Lemaire 2014). Over the past few decades, pressure for greater food production has led to substantially increased N fertiliser usage and a decline in reliance on legume N fixation (Brisson et al. 2010). There have been increased calls for plants with novel functional traits, such as grasses with high C:N ratios (Parsons et al. 2011) and white clover (Trifolium repens L.) with high condensed tannins (Hancock et al. 2012), or novel forage plant germplasm, for example ‘extended phenotypes’ of endophyte-infected grasses (Schardl et al. 2013), to meet human demands for food. Any introduced desirable traits or extended phenotypes have a substantial impact on some aspect of ecosystem functioning; however, the potential effects of these introductions on soils have received little assessment. Indeed, many of the forage traits targeted by industry directly affect C and N cycling in plants, animals and soils (Parsons et al. 2011).
Conventional assessments of the impacts and outcomes on soil C:N states and organic matter, from even simple changes in farm practices, contain a high degree of uncertainty (Kelliher et al. 2013) and can require extensive re-measurement across sites and time, for example of some 30+ years (Schipper et al. 2010; Mudge et al. 2017). Impacts on soil function will be related to, and likely prefaced by, changes in the soil microbial community. Therefore, a first step is to assess whether plant species and their functional traits, N fertilisation and plant extended phenotypes lead to changes that are detectable via a detailed analysis of soil microbial communities, and whether any effects are significant on a short time scale. With the advent of high-throughput next-generation sequencing methods, more detailed studies became possible for analysis of effects of factors such as N fertilisation (Nemergut et al. 2008; Nacke et al. 2011), global localisation (Prober et al. 2015), elevation gradients (Pellissier et al. 2014) or endophyte infection (Rojas et al. 2016; Mahmud et al. 2021; Chen et al. 2022) on grassland soil microbial community composition and diversity. Whereas, soil types and their physico-chemical properties, as well as plant species and their associated management, have major impacts on microbial composition (Bezemer et al. 2006; Young and Crawford 2004), most studies have been performed across a range of different soils and with limited knowledge about past or present plant cover and management practices.
Changes in the number of species present per se might not be the most relevant measure for deducing any change in soil function (van der Wal et al. 2013). In addition, studies that attempt to identify and catalogue the wide range of soil organisms present face the difficulty that many of the soil microbial species, fungi in particular, have not been sequenced, let alone functionally classified. In the present study, we attempt to overcome these limitations by using a series of systematically different experimental treatments, analysing only for significant differences between treatments with respect to the microbial communities they foster in what was previously a common soil mineralogy and community. This approach allows us to identify changes in community composition, even when many of the organisms present remain ‘unnamed’ or where the bulk of the soil community may be unaltered. Specifically, we performed two concurrently run experiments in a common soil, each with a random spatial distribution of treatments and replications, for effects on soil microbial communities. In Experiment 1 (plant species/traits, N), we tested for differential impacts of two major forage legume species (white clover, Trifolium repens L.; and bird’s-foot trefoil, Lotus pedunculatus Cav.) and two cultivars of perennial ryegrass (Lolium perenne L.) associated with various N application levels. In Experiment 2 (grass extended phenotype), we tested for differential impacts of grasses containing various strains of Epichloë fungal endophytes. Both fungal and bacterial communities were analysed using high-throughput sequencing of DNA isolated from bulk soil cores harvested 2 years after imposing the above treatments. The study focused on implications for C and N cycling, and whether any changes were detectable within a short time span of 2 years.
Materials and methods
Experimental design
All experimental field plots were established on Paparua sandy loam soil at the Lincoln University Research Dairy Farm, Lincoln, New Zealand. The whole experimental area had been previously used for cereal cropping and was ploughed prior to sowing forage plants as described below. For a detailed analysis of soil physico-chemical properties, see Cripps et al. (2013). Two experiments were set up in April 2010; in both, four experimental replicates (plots) of each treatment were sown in a randomised block design. Fertiliser N in the form of urea was applied to all plots at a rate of 40 kg N ha−1 every 4 months, except for the legume and low-N treatments in Experiment 1 (see Table 1), which were fertilised with 10 kg N ha−1. The area surrounding the experimental plots was sown with L. perenne cv. Alto (containing AR1 endophyte). Individual plot areas were 2.1 m by 6 m in Experiment 1, and 2.1 m by 3 m in Experiment 2. Plots were grazed and mown as described by Cripps et al. (2013).
| Experiment 1: plant species/trait and nitrogen level | |||||
|---|---|---|---|---|---|
| Plant type | Species | Common name and cultivar | Nitrogen applied | Treatment abbreviation | |
| Legume | Trifolium repens | White clover cv. Kopu | 10 kg N ha−1 | WCl | |
| Lotus pedunculatus | Bird’s-foot trefoil cv. Barsille | 10 kg N ha−1 | LoP | ||
| Grass | Lolium perenne | Perennial ryegrass cv. Impact (normal sugar variety) | 10 kg N ha−1 | LN RG | |
| 40 kg N ha−1 | HN RG | ||||
| Perennial ryegrass cv. AberDart (high sugar variety) | 10 kg N ha−1 | LN HSG | |||
| 40 kg N ha−1 | HN HSG | ||||
| Experiment 2: grass extended phenotype (i.e. endophyte infection) | |||||
|---|---|---|---|---|---|
| Grass common name | Species and cultivar | Endophyte species | Endophyte treatment/strain | Treatment abbreviation | |
| Perennial ryegrass | Lolium perenne cv. Alto | Epichloë festucae var. lolii | Uninfected | RG Nil | |
| Wild type | RG WT | ||||
| NEA2 | RG NEA2 | ||||
| AR1 | RG AR1 | ||||
| Epichloë sp. LpTG-3 | AR37 | RG AR37 | |||
| Tall fescue | Schedonorus phoenix cv. Grassland Advance | Epichloë coenophiala | Uninfected AR542 | TF Nil TF AR542 | |
| Meadow fescue | Schedonorus pratensis | Epichloë uncinata | Uninfected | MF Nil | |
| U2 | MF U2 | ||||
There were four replicates of each treatment. In Experiment 1, soil bacterial communities were analysed for only three replicates.
Experiment 1, examining plant species/traits and levels of N, comprised four replicates of six treatments: two legumes, white clover cv. Kopu sown at 4 kg seed ha−1 (WCl) and bird’s-foot trefoil cv. Barsille sown at 4 kg seed ha−1 (LoP); and two Epichloë-free perennial ryegrass cultivars, normal sugar cv. Impact (RG) and high sugar cv. AberDart (HSG), both sown at 20 kg seed ha−1, and both with a low N (LN, 10 kg N ha−1) and a high N (HN, 40 kg N ha−1) fertilisation rate treatment. The two legume species differ substantially in their ability to accumulate antimicrobial, protein-binding condensed tannins (Terrill et al. 1992), and the two ryegrass cultivars differ significantly in their ability to produce fructans in leaf blades (Parsons et al. 2011). A summary of the experiments is shown at Table 1.
Experiment 2, examining grass extended phenotypes, comprised four replicates of nine treatments: five treatments with perennial ryegrass cv. Alto (RG), four of which contained one of three different strains of Epichloë spp. (AR1, AR37 and the wild-type strain, WT) or a strain blend of NEA2 and NEA6 (branded ‘NEA2’, Eady 2021), and one that was endophyte-free (nil); two treatments with tall fescue (Schedonorus phoenix (Scop.) Holub) cv. Grasslands Advance (TF), one containing Epichloë coenophiala (strain AR542) and one endophyte-free; and two treatments with meadow fescue (Schedonorus pratensis (Huds.) P.Beauv.) (MF), one containing Epichloë uncinata (strain U2) and one endophyte-free (see Table 1). Each grass treatment was sown at a rate of 15 kg seed ha−1. All grass–endophyte combinations were assessed for the presence of endophyte in January 2011 by microscopic examination of leaf sheath tissue stained with lactophenol cotton blue. Endophyte-containing grasses ranged from 85% to 100% infected, and the endophyte-free grasses were 0–5% infected (for further details, see Cripps et al. 2013). The commonly used endophytic Epichloë festucae var. lolii strains (AR1, NEA2 and WT) and Epichloë sp. LpTG-3 strain AR37 differ in their ability to produce certain fungal alkaloids.
Soil sampling and DNA extraction
In April 2012, soil was collected to a depth of 7.5 cm with a metal corer (diameter 3 cm). In Experiment 1 six cores and in Experiment 2 four cores were sampled randomly from each plot. Soil cores from each individual plot were mixed and passed through a 4-mm metal sieve to remove larger stones and plant root parts, bagged, immediately frozen in liquid N2 for transport and stored at −80°C until analysis. The above procedures of collection and treatment for each soil sample were completed within <15 min.
Genomic DNA was extracted from soil by means of the PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA) following the manufacturer’s instructions. DNA concentrations were measured with a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA). DNAs (~25 ng μL−1) were used for polymerase chain reaction (PCR) as detailed below.
Analysis of soil fungal community composition: 454 pyrosequencing
Soil fungal community composition was analysed by 454 pyrosequencing on the GS-FLX 454 System (454 Life Sciences/Roche Applied Biosystems, Nutley, NJ, USA) at Waikato University, Hamilton, NZ. DNA samples were amplified using general primers linked to 454 pyrosequencing adapters A and B, and a barcode (multiplex identifier, MID) for each sample, as recommended for the 454 GS-FLX One-Way Read method. The primer pair used in this study, NSI1mod (modified NSI1 primer) and 5.8A2R (Martin and Rygiewicz 2005), was designed to target fungal DNA in the partial 18S SSU and the variable ITS1 region of the non-coding nuclear rDNA ITS region, which is recognised as the universal DNA barcode marker for fungi (Schoch et al. 2012). We modified the NSI1 primer, which targets primarily Ascomycota and Basidiomycota, with a degenerate base at the 3′ end in order to include potential Glomeromycota sequences. Thus, the composite forward primer was NSI1mod 5′-CCATCTCATCCTGCGTGTCTCCGACTCAG xxx GATTGAATGGCTTAGTGAGK-3′ and the reverse primer was 5.8A2R 5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCTGCGTTCTTCATCGAT-3′, where manufacturer adapters are underlined, barcode is indicated by xxx, and the general primers NSI1mod and 5.8A2R are shown in italics.
For each sample, a PCR mix was prepared containing 1 μL FastStart High Fidelity Taq polymerase (Roche Molecular Systems, Pleasanton, CA, USA), 10 μL reaction buffer 10X, 2 μL PCR grade nucleotide mix, and 0.8 μM each primer in a total volume of 100 μL. Three replicate PCRs per DNA sample were performed by adding 1 μL template DNA to 25 μL reaction mix and using the following cycling conditions: 94°C for 4 min; 30 cycles of 30 s at 94°C, 50°C for 1 min and 72°C for 1.5 min; followed by 7 min at 72°C. Replicate PCR products were pooled and separated on 0.8% agarose gel in 1× TAE (Tris-acetate-EDTA) buffer. PCR products were purified from the gel by using the ISOLATE Gel Purification Kit (Bioline, Alexandria, NSW, Australia), and DNA concentrations in the purified products (amplicon libraries) were measured with a QuBit fluorometer (Invitrogen, Eugene, OR, USA). Aliquots (200 pg μL−1) of each sample were run on a Bioanalyser (Agilent Technologies, Santa Clara, CA, USA) and the number of molecules in each sample was calculated. Amplicon libraries were mixed at equimolar concentrations to make a solution of 1 × 109 molecules DNA in 200 μL.
A 1-μL sample of the combined library solution was subjected to pyrosequencing and the datasets were processed using mothur v.1.32 (Schloss et al. 2009). Reads were subjected to denoising, quality trimming and chimera removal using AmpliconNoise v.1.28 (Quince et al. 2011). Sequences shorter than 400 bp after quality trimming were discarded. Sequences were clustered into molecular operational taxonomic units (OTUs) using mothur v.1.32. Distance was calculated using ESPRIT-Tree (pair-wise alignment) (Cai and Sun 2011); of the four stringencies (unique, 1%, 3%, 5% distance) tested with nearest, average and furthest neighbour strategy, we chose the 3% nearest neighbour stringency with a 97% sequence identity threshold. Following OTU clustering, a reference sequence from each OTU was mathematically determined and used for taxonomic identification of the OTU. Representative sequences for each OTU were queried against the non-redundant GenBank database using the BLASTN algorithm.
Analysis of soil bacterial community composition: MiSeq Illumina Sequencing
Bacterial community composition was investigated via the MiSeq sequencing platform (Illumina, San Diego, CA, USA) at the Massey Genome Service (Massey University, Palmerston North, NZ). Genomic DNA (10 ng μL−1) was prepared from three replicates of the six treatments from Experiment 1 and from all 36 samples from Experiment 2 according to the specifications of the Massey Genome Service. Amplicon libraries were prepared using a single-step PCR; libraries were purified by the Massey Genome Service. A set of index-tagged PCR primers containing the Illumina complementary adapter sequence for binding to the flowcell, which flanks the 16S fourth hypervariable (V4) region, was used for amplification. The amplicon libraries were pooled at equal molarity. To control for uneven representation of bases at each cycle with 16S libraries, the Illumina PhiX control library was added at ~10% volume before loading onto the Illumina MiSeq instrument.
All bioinformatics analysis steps were performed using mothur (Schloss et al. 2009). Sample reads were combined to generate ~250 bp reads (contigs), using FLASH after preliminary quality-control steps. Raw sequence data were filtered by removing any sequences that contained ambiguous call ‘N’, lengths >275 bp, and low-quality regions. Data were aligned to 16s rRNA reference database (RDP) v.9 and unaligned data were removed. Chimeras were detected using UCHIME (Edgar et al. 2011) implemented in mothur. All reads were taxonomically classified using the RDP classifier (Wang et al. 2007); reads that were classified as chloroplast, mitochondria, unknown, Archaea and Eukaryota were removed.
Statistical analyses
Prior to statistical analyses, sequences of very low abundance were removed from the datasets; we used fungal or bacterial sequences of <10 reads on average per treatment as an arbitrary cut-off, i.e. an OTU had to be present at least 60 times in the sample set from Experiment 1 and 90 times in the sample set from Experiment 2. We used partial least square discrimination analysis (PLSDA) to identify microbial genera (i.e. OTUs) contributing the most to a segregation of treatment groups. Those genera with a VIP (variable importance in projection) score >1.0 on each of the relevant PLSDA axes were subsequently selected for analysis of variance (ANOVA). In Experiment 1, one-way ANOVA was used to separate treatment effects (i.e. plant type, plant species, plant cultivar, N applied). In Experiment 2, two-way ANOVA was used to separate effects of endophyte with grass species as factor 1 and endophyte status (endophyte or nil) as factor 2; and one-way ANOVA was used to separate strain dependency within ryegrass group, with nil, WT, AR1, AR37 and NEA2 as treatment groups. All statistical analyses were performed using XLSTAT v.2015.6.01.25106 (Addinsoft, Paris, France). To homogenise the variance, all data were Box–Cox transformed prior to analyses.
Results
Experiment 1: impacts of plant species/traits and nitrogen fertilisation level on soil microbial communities
In total, 6648 fungal and 512 bacterial OTUs were identified in soils from Experiment 1 across all treatments, with the most abundant fungal OTU (Mycena sp.) being amplified 12 796 times and the most abundant bacterial OTU (unclassified Actinobacteria_Gp6) being amplified 6691 times. The top 3.8% of fungal OTUs (235 OTUs, total reads 107 406) and 31.25% of bacterial OTUs (160 OTUs, total reads 145 902) were selected for further analysis.
The soil fungal microbiota was dominated by phyla Ascomycota (44%), Basidiomycota (34%), Glomeromycota (6%), Zygomycota (2%), Chytridiomycota (0.4%) and unknown ectomycorrhizal fungi (1%). Approximately 12% of the OTUs aligned to unknown fungi. The abundance was significantly different among treatments for Glomeromycota (arbuscular mycorrhizae; P = 0.013) and Chytridiomycota (saprotrophs; P < 0.001). Glomeromycota represented by the Glomeraceae family was most abundant in WCl soils and least abundant in grass (both HSG and RG) soils at low N, whereas the Chytridiomycota represented by the Olpidiaceae family was more abundant in RG soils at both N levels than in legume soils. Within the Basidiomycota, the class of Agaricomycetes was more abundant in HSG (both N levels) than WCl soils (P = 0.004); and within the Ascomycota, the class Leotiomycetes was more abundant in grass than legume soils (P < 0.001). The soil bacterial microbiota was dominated by phyla Proteobacteria (25%), Acidobacteria (12%) and Bacteroidetes (12%). Up to 29% of the OTUs were aligned to unclassified bacteria. The abundance was different among treatments for Armatimonadetes (P = 0.008, represented by the Armatimonadaceae family), Bacteroidetes (P = 0.032), and Nitrospirae (P = 0.017, represented by the Nitrospiraceae family).
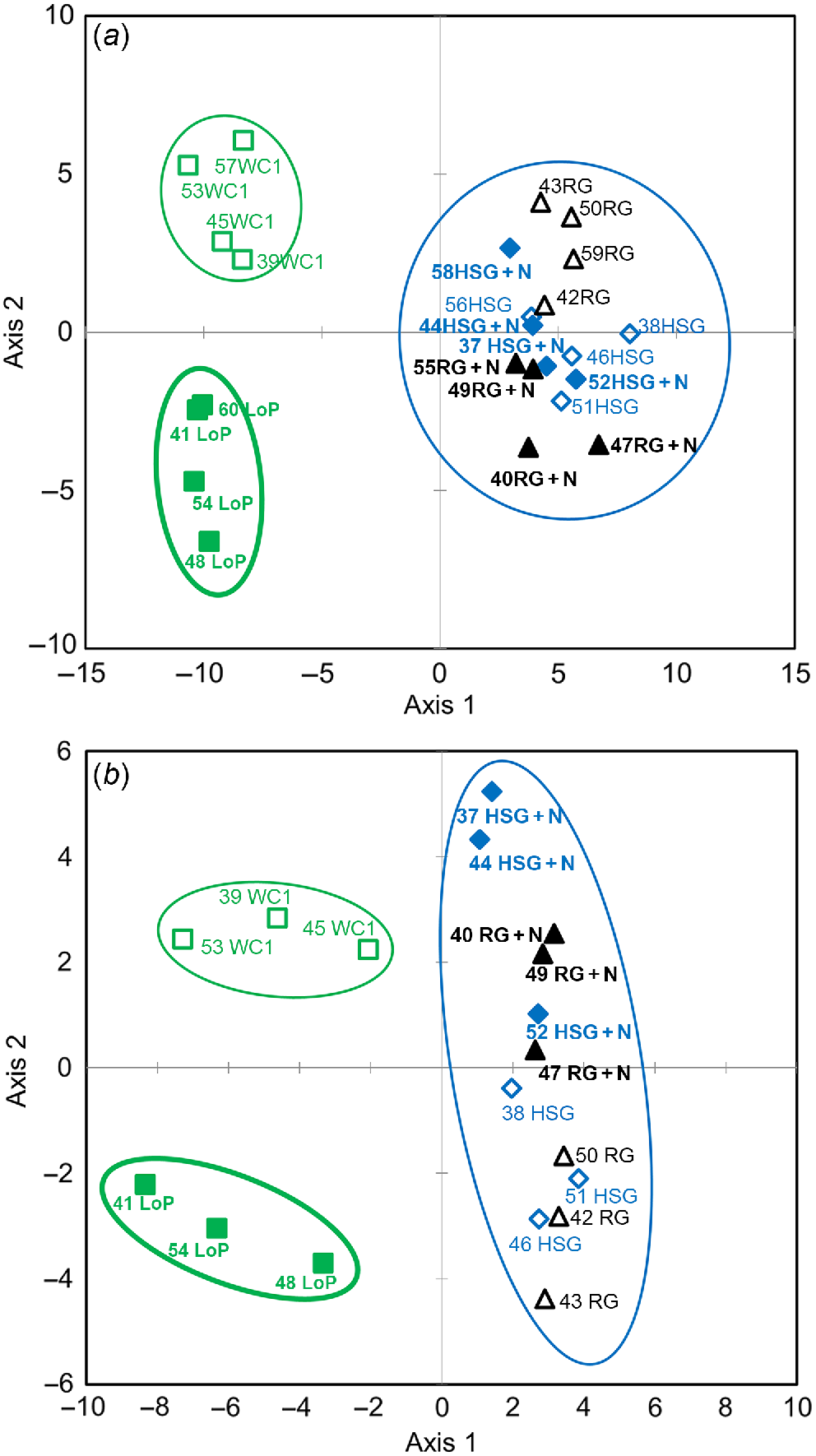
With respect to fungal genera, legume soils segregated from grass soils on PLSDA axis 1, whereas WCl soils segregated from LoP soils on axis 2 (Fig. 1a), and a weak segregation of HSG vs RG soils and low N vs high N soils was observed on axis 3 (Supplementary Fig. S1a). Regarding bacterial genera, similar to fungi, legume soils segregated from grass soils on PLSDA axis 1 and WCl from LoP soils on axis 2 (Fig. 1b), low N segregated from high N soils on axis 3 (Fig. S1b), and there was a weak segregation of HSG from RG soils on axis 4 (Fig. S1b).
Partial least square discrimination analysis (PLSDA) of impacts of plant species/traits and N fertilisation level on the abundance of (a) fungal and (b) bacterial genera. Blue open diamonds, HSG (high sugar perennial ryegrass cv. AberDart) (low N); blue closed diamonds, HSG + N (high N); black open triangles, RG (normal sugar perennial ryegrass cv. Impact) (low N); black closed triangles, RG + N (high N); green open squares, WCl (white clover); green closed squares, LoP (bird’s-foot trefoil). Abundances were Box–Cox transformed prior to PLSDA. Groups showing major spatial separations are confined within ellipses.
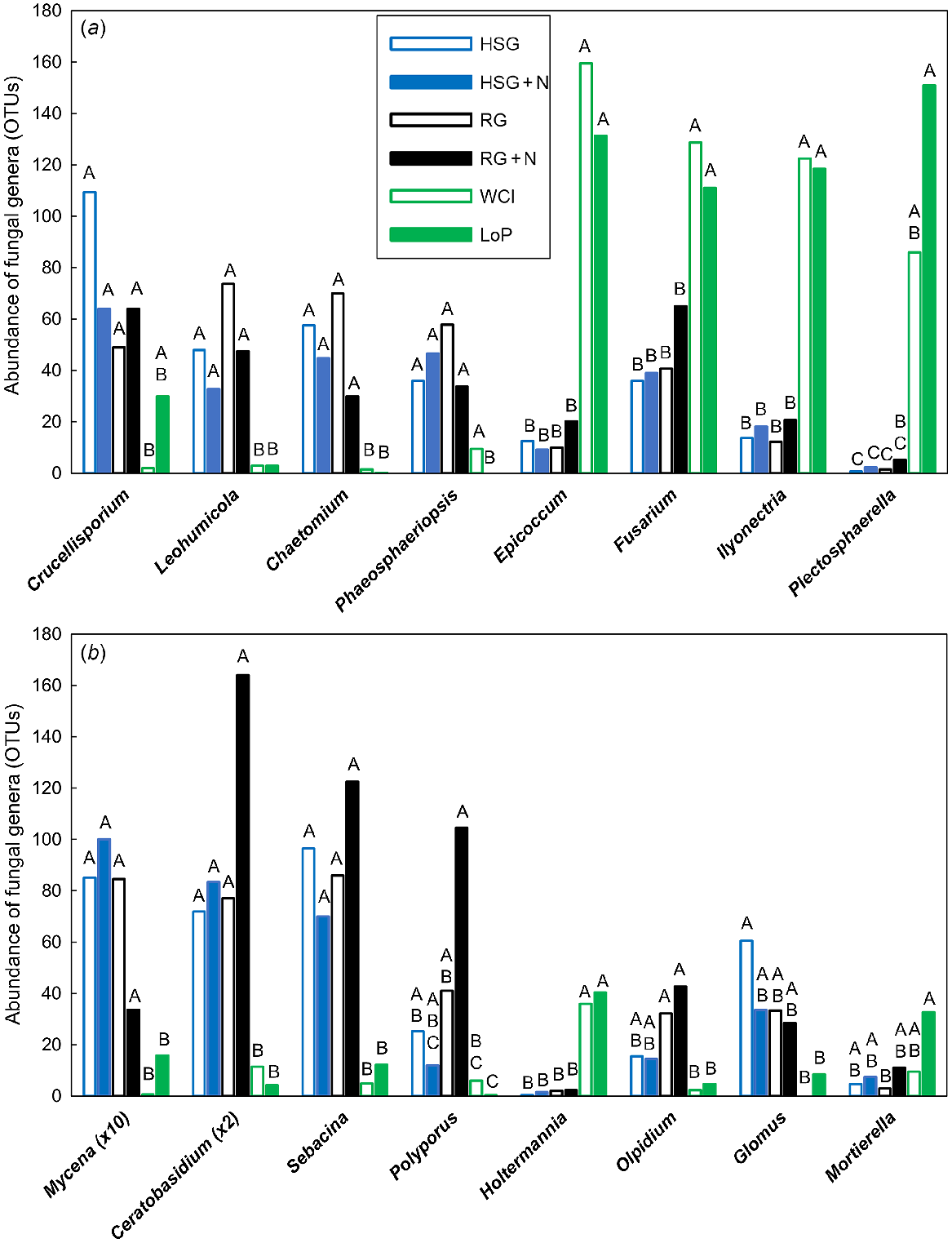
In total, 75 fungal OTUs were significantly different (VIP >1.0), with 51 of these being most important for segregation on axis 1, nine on axis 2 and 15 on axis 3 (data not shown). Genera that were significantly affected by treatment are shown in Fig. 2. The Ascomycota Leohumicola sp. (root endophyte) and Chaetomium sp. (mould) were more abundant in grass soils than in legume soils (Fig. 2a). Crucellisporium sp. was also more abundant in grass soils but only compared with WCl (not LoP) soil, whereas Phaeosphaeriopsis sp. (root endophyte) was more abundant in grass and WCl soils than in LoP soil. The Ascomycota Epicoccum sp. (humic acid/melanin forming), Fusarium sp. (plant pathogen), Ilyonectria sp. (plant pathogen) and Plectosphaerella sp. (plant pathogen) were substantially more abundant in legume soils than in grass soils (Fig. 2a). The abundance of Xylariales fungi (e.g. Pestalotiopsis sp., Monographella sp. and Anthostomella sp.) appeared to be low in legume soils (data not shown). The Basidiomycota Mycena sp. (white-rot fungus), Ceratobasidium sp. (saprotrophic fungus) and Sebacina sp. (mycorrhizal fungus) were more abundant in grass soils than in legume soils, whereas Holtermannia sp. (fungal pathogen) was less abundant in grass soils than in legume soils (Fig. 2b). The white-rot fungus Polyporus sp. was also more abundant in grass soils, except in HSG soil at high N, but only compared with LoP (not WCl) soil. The Chytridiomycota Olpidium sp. (root pathogen) was more abundant in RG soils (both N treatments) than in legume soils, and the Glomeromycota Glomus sp. (arbuscular mycorrhizal fungus) was more abundant in HSG soil at low N than in legume soils (Fig. 2b). By contrast, the Zygomycota Mortierella sp. (saprotrophic fungus) was less abundant in RG soil at low N than in LoP soil.
Impacts of plant species/traits and N fertilisation level on the abundance of influential fungal genera within the PLSDA related to Experiment 1. Fungal genera are shown according to the associated phyla within (a) Ascomycota and (b) Basidiomycota. Only those genera that featured strongly as VIP >1 were analysed further using one-way ANOVA. Untransformed means of statistically significant different genera (P < 0.05) are shown. Blue open bars, HSG (high sugar perennial ryegrass cv. AberDart) (low N); blue closed bars, HSG + N (high N); black open bars, RG (normal sugar perennial ryegrass cv. Impact) (low N); black closed bars, RG + N (high N); green open bars, WCl (white clover); green closed bars, LoP (bird’s-foot trefoil). Abundances were Box–Cox transformed prior to one-way ANOVA; within each genus, treatment means with the same letter are not significantly different according to Tukey’s HSD at P = 0.05.
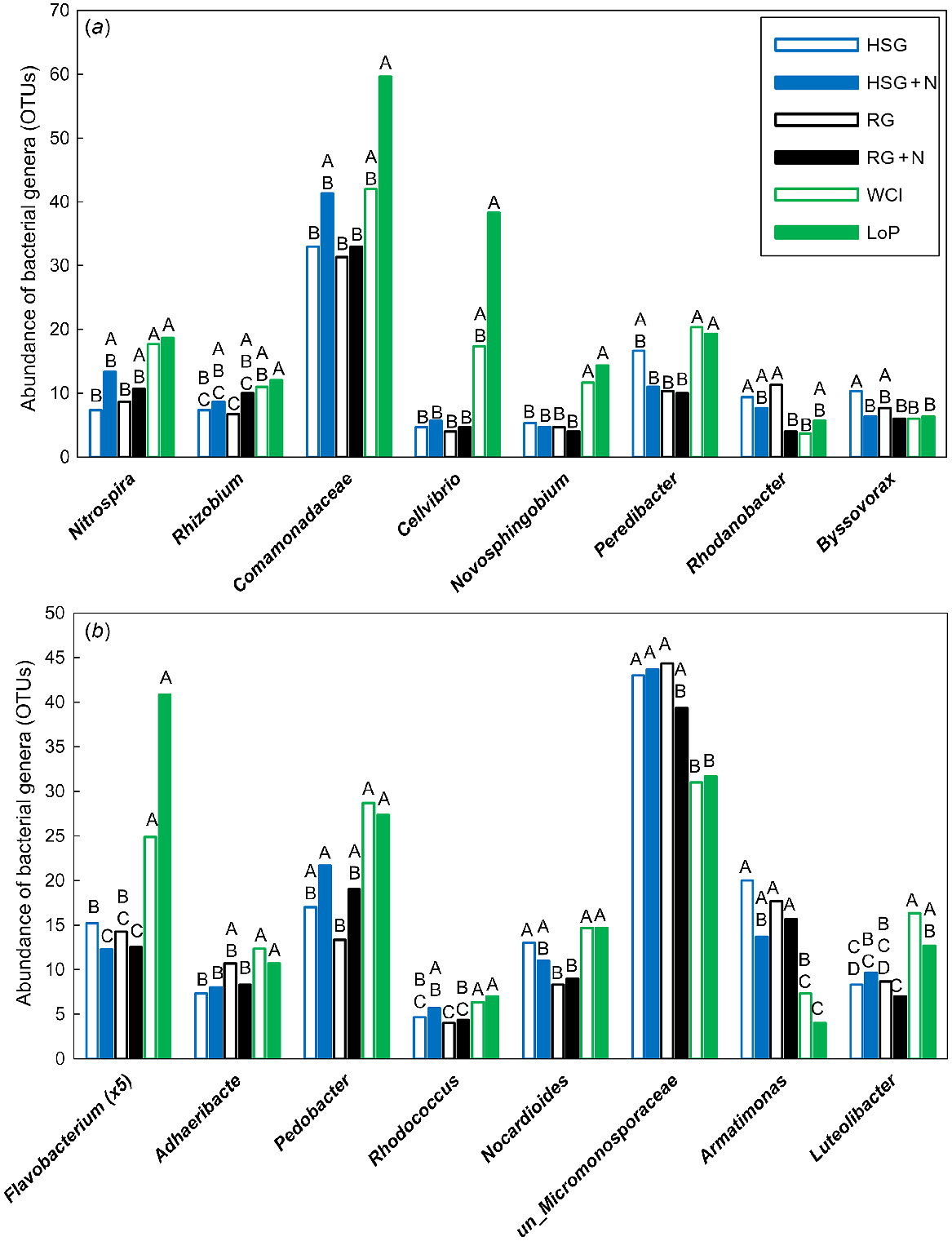
In total, 27 bacterial OTUs were significantly different (VIP >1.0), with 18 of these being most important for segregation on axis 1, two on axis 2, four on axis 3, and three on axis 4 (data not shown). Genera that were significantly affected by treatment are shown in Fig. 3. The nitrifying Nitrospira sp. and the N-fixing Rhizobium sp. were less abundant in grass soils at low N than in both legume soils (Nitrospira sp.), and in LoP soil only (Rhizobium sp.) (Fig. 3a). The denitrifying unclassified Comamonadaceae were also less abundant in HSG soil at low N and RG soils at both N levels than in LoP soil (Fig. 3a). The cellulolytic Cellvibrio sp. and the aromatic-degrading Novosphingobium sp. were less abundant in all grass soils than in LoP soil (Cellvibrio sp.), and in both legume soils (Novosphingobium sp.) (Fig. 3a). Peredibacter sp., a bacterium predating on other bacteria, was less abundant in grass soils (except HSG soil at low N) than in legume soils. The denitrifying Rhodanobacter sp. was most abundant in grass soils at low N and least abundant in RG soil at high N and WCl soil (Fig. 3a). The cellulolytic Byssovorax sp. was more abundant in HSG soil at low N than in all other soils except RG soil at low N (Fig. 3a). The carbohydrate-degrading Flavobacterium sp., Adhaeribacter sp. and Pedobacter sp. were highly abundant in legume soils and generally much less abundant in grass soils (Fig. 3b). Flavobacterium sp. was also affected by N, being less abundant in HSG soil at high N than at low N (Fig. 3b). Generally more abundant in legume soils were Rhodococcus sp. and Nocardioides sp., which are known to be able to degrade phenolics and pesticides. By contrast, the ligninolytic unclassified Micromonosporaceae were highly abundant in all grass soils and, except for the comparison with RG soil at high N, were less abundant in both legume soils (Fig. 3b). The chemoheterotrophic Armatimonas sp. was more abundant in grass soils than in legume soils, whereas Luteolibacter sp. was generally less abundant in grass soils than in legume soils (Fig. 3b).
Impacts of plant species/traits and N fertilisation level on the abundance of influential bacterial genera within the PLSDA related to Experiment 1. Bacterial genera were split into (a) and (b) for illustrative purposes. Only those genera that featured strongly as VIP >1 were analysed further using one-way ANOVA. Untransformed means of statistically significant different genera (P < 0.05) are shown. Blue open bars, HSG (high sugar perennial ryegrass cv. AberDart) (low N); blue closed bars, HSG + N (high N); black open bars, RG (normal sugar perennial ryegrass cv. Impact) (low N); black closed bars, RG + N (high N); green open bars, WCl (white clover); green closed bars, LoP (bird’s-foot trefoil). Abundances were Box–Cox transformed prior to one-way ANOVA; within each genus, treatment means with the same letter are not significantly different according to Tukey’s HSD at P = 0.05.
Experiment 2: impacts of plant extended phenotype on soil microbial communities
In total, 5188 fungal and 569 bacterial OTUs were identified in soils from Experiment 2 across all treatments, with the most abundant fungal OTU (Mycena sp.) being amplified 11 941 times, and the most abundant bacterial OTU (Acidobacteria_Gp6) being amplified 15 901 times. The top 3.2% of fungal OTUs (165 OTUs, total reads 120 663) and 19% of bacterial OTUs (108 OTUs, total reads 171 474) were selected for further analysis.
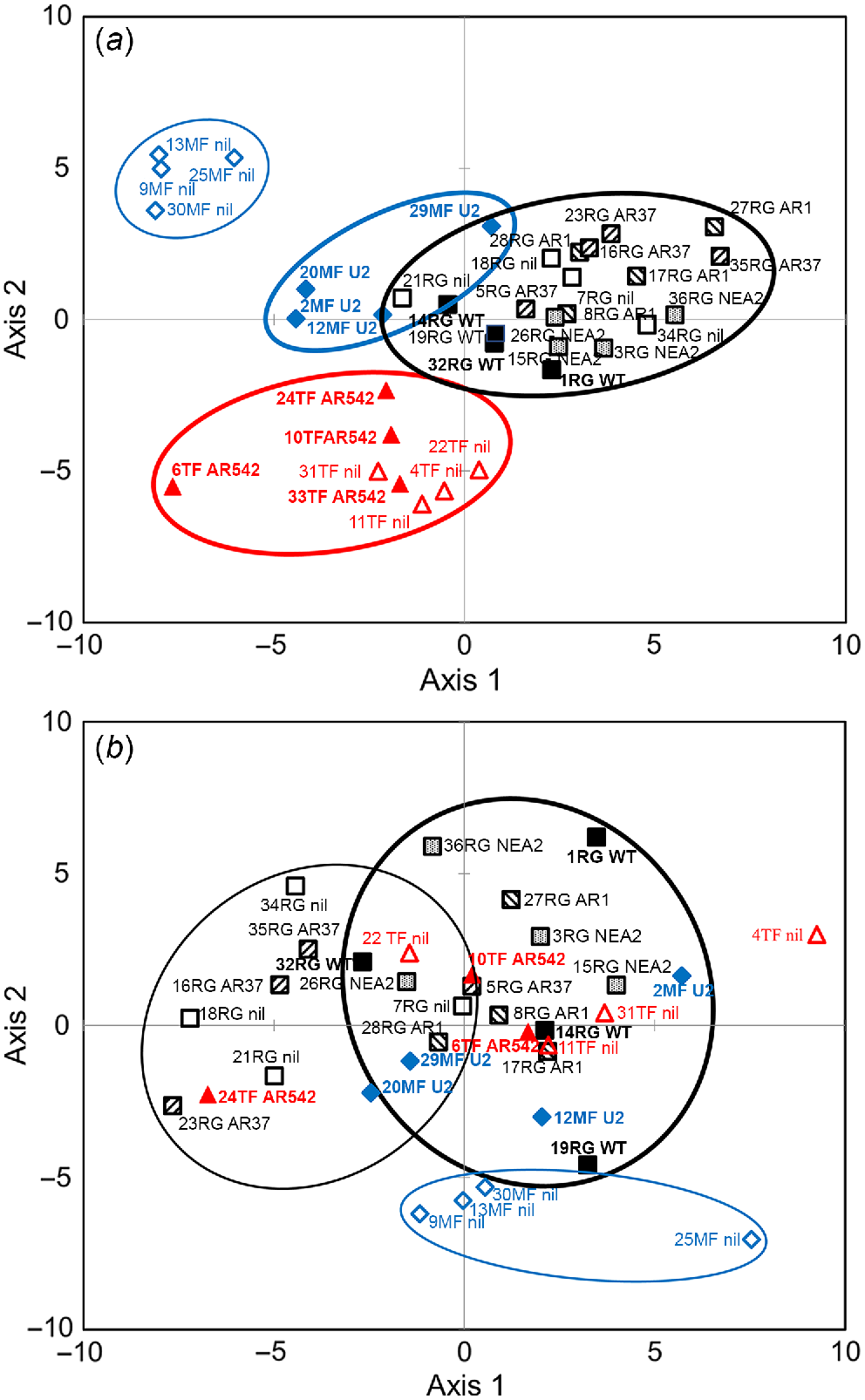
Soil fungal microbiota was dominated by phyla Basidiomycota (34.7%), Ascomycota (34.4%), Glomeromycota (5.4%), Zygomycota (2.1%) and unknown ectomycorrhizal fungi (1%). Up to 22% of the OTUs aligned to unknown fungi. PLSDA for fungal genera showed segregation of MF and TF vs RG soils, and MF_nil vs MF_U2 soils, on axis 1 (Fig. 4a); MF vs TF soils on axis 2 (Fig. 4a); and a weak segregation between RG soils infected with various strains on axis 3 (Fig. S2a). Bacterial phyla in the soils were dominated by Proteobacteria (24%), Acidobacteria (17%), Verrucomicrobia (12%) and Bacteroidetes (11%). Up to 28% of the OTUs aligned to unclassified bacteria. On the level of phyla abundance, differences were found among treatments for Actinobacteria (P = 0.013) and Firmicutes (P = 0.036). Abundance of Actinobacteria was higher in RG_AR37 than in RG_nil soil (P = 0.019), whereas abundance of Firmicutes was higher in RG_nil and TF_AR542 soils than in RG_NEA2 soil (P = 0.046). Bacterial genera in RG_nil and RG_AR37 soils appeared to segregate from the three other endophyte-infected RG groups (WT, AR1 and NEA2), and MF_nil segregated from all other soils on axis 2 (Fig. 4b). TF soils segregated from RG soils on axis 3 and from MF soils on axis 4 (Fig. S2b).
Partial least square discrimination analysis (PLSDA) of impacts of plant extended phenotype (endophyte) and grass species on the abundance of (a) fungal and (b) bacterial genera. Black open squares, RG_nil (ryegrass without endophyte); black closed squares, RG_WT (ryegrass infected with Epichloë festucae var. lolii wild-type); black downward-striped squares, RG_AR1 (ryegrass infected with E. festucae var. lolii AR1); black upward-striped squares, RG_AR37 (ryegrass infected with Epichloë sp. LpTG-3 strain AR37); black stippled squares, RG_NEA2 (ryegrass infected with E. festucae var. lolii NEA2); blue open diamonds, MF_nil (meadow fescue without endophyte); blue closed diamonds, MF_U2 (meadow fescue infected with Epichloë uncinata U2); red open triangles, TF_nil (tall fescue without endophyte); red closed triangles, TF_AR542 (tall fescue infected with Epichloë coenophiala AR542). Groups showing major spatial separations are confined within ellipses.
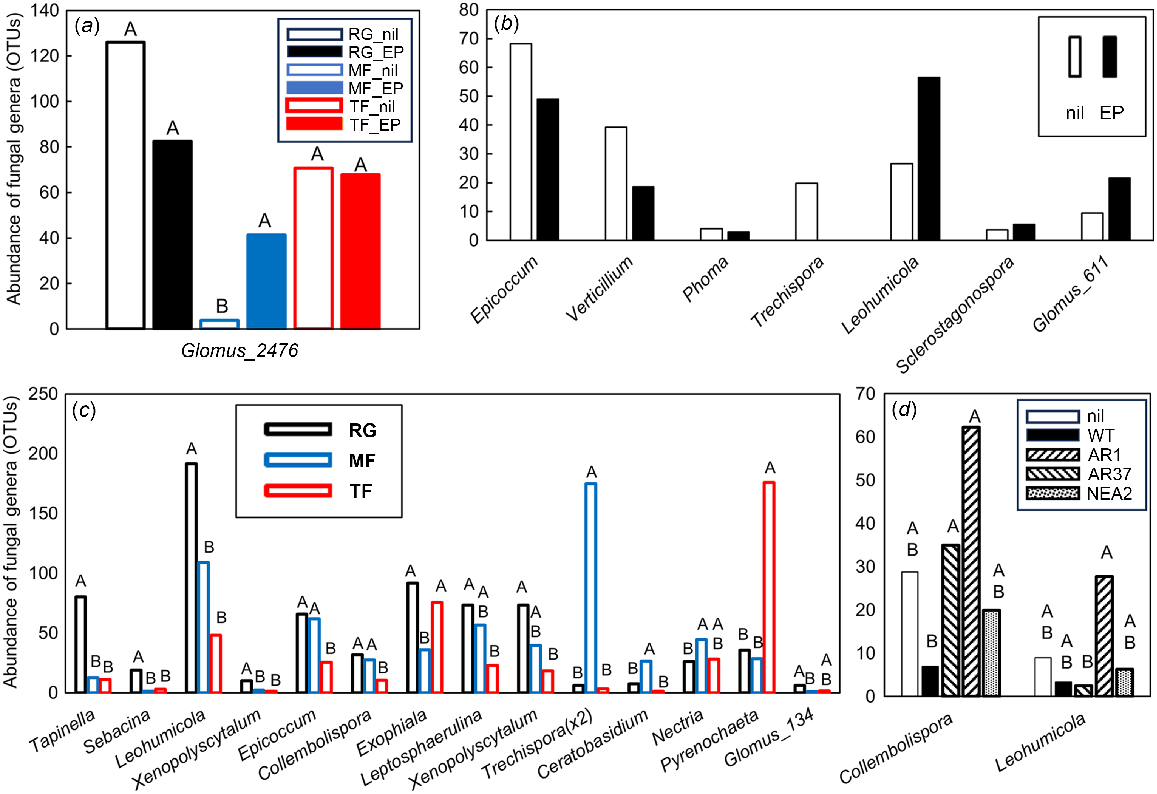
In total, 46 fungal OTUs were significantly different (VIP >1.0), with 26 of these being most important for segregation on axis 1, 13 on axis 2 and seven on axis 3 (data not shown). Five fungal OTUs were significantly affected by the interaction between grass species and endophyte infection; these were the endomycorrhizal Glomus sp. (Fig. 5a), as well as Boletus sp. (detected in MF_nil soils only), an unclassified Ascomycete, Zygomycete and an unknown fungus (data not shown). Nine fungal OTUs were significantly affected by the symbiotic endophyte status of the grass. The Epicoccum sp. (humic acid producer), plant pathogens Verticillium sp. and Phoma sp. and turfgrass pathogen Trechispora sp. were all less abundant in soils from infected grasses, whereas the ectomycorrhizal Leohumicola sp., endophytic Sclerostagonospora sp., and arbuscular mycorrhizal Glomus sp. were more abundant in these soils (Fig. 5b); two OTUs could not be classified. The abundance of 32 fungal genera was affected by grass species. Within these influential genera, the brown-rot fungus Tapinella sp., the mycorrhizal fungi Sebacina sp. and Leohumicola sp., the hemicellulolytic Xenopolyscytalum sp., the humic acid producer Epicoccum sp., the saprotrophic Collembolispora sp. and Exophiala sp., the turfgrass pathogen Leptosphaerulina sp., and the arbuscular mycorrhizal Glomus sp. were all higher in RG soils than in either MF or TF or both fescue soils (Fig. 5c). The turfgrass pathogen Trechispora sp. and the saprotrophic Ceratobasidium sp. and Nectria sp. were highest in MF soils, and the pathogenic Pyrenochaeta sp. was highest in TF soils (Fig. 5c). Five fungal genera were differentially affected by endophytic strains in RG soil. The saprotrophic Collembolispora sp. was higher in RG_AR1 and RG_AR37 soils than in RG_WT soil, whereas the mycorrhizal Leohumicola sp. was highest in RG_AR37 but lowest in RG_AR1 soil (Fig. 5d); three OTUs could not be classified.
Impacts of plant extended phenotype (endophyte) and grass species on the abundance of influential fungal genera within the PLSDA related to Experiment 2. Only those genera that featured strongly as VIP >1 were analysed further using either (a–c) two-way ANOVA or (d) one-way ANOVA. Untransformed means of statistically significant different genera (P < 0.05) are shown. (a) Black open bars, RG_nil (ryegrass without endophyte); black closed bars, RG_EP (ryegrass infected with Epichloë sp.); blue open bars, MF_nil (meadow fescue without endophyte); blue closed bars, MF_EP (meadow fescue infected with Epichloë uncinata U2,); red open bars, TF_nil (tall fescue without endophyte); red closed bars, TF_EP (tall fescue infected with Epichloë coenophiala AR542). (b) Open bars, nil (grasses without endophyte); closed bars, grasses infected with Epichloë sp.. (c) Black bars, RG (ryegrass); blue bars, MF (meadow fescue); red bars, TF (tall fescue). (d) Open bars, RG_nil (ryegrass without endophyte); closed bars, RG_WT (ryegrass infected with E. festucae var. lolii wild-type); downward striped bars, RG_AR1 (ryegrass infected with E. festucae var. lolii AR1); upward striped bars, RG_AR37 (ryegrass infected with Epichloë sp. LpTG-3 strain AR37); stippled bars, RG_NEA2 (ryegrass infected with E. festucae var. lolii NEA2). Abundances were Box–Cox transformed prior to ANOVA; within each genus, treatment means with the same letter are not significantly different according to Tukey’s HSD at P = 0.05.
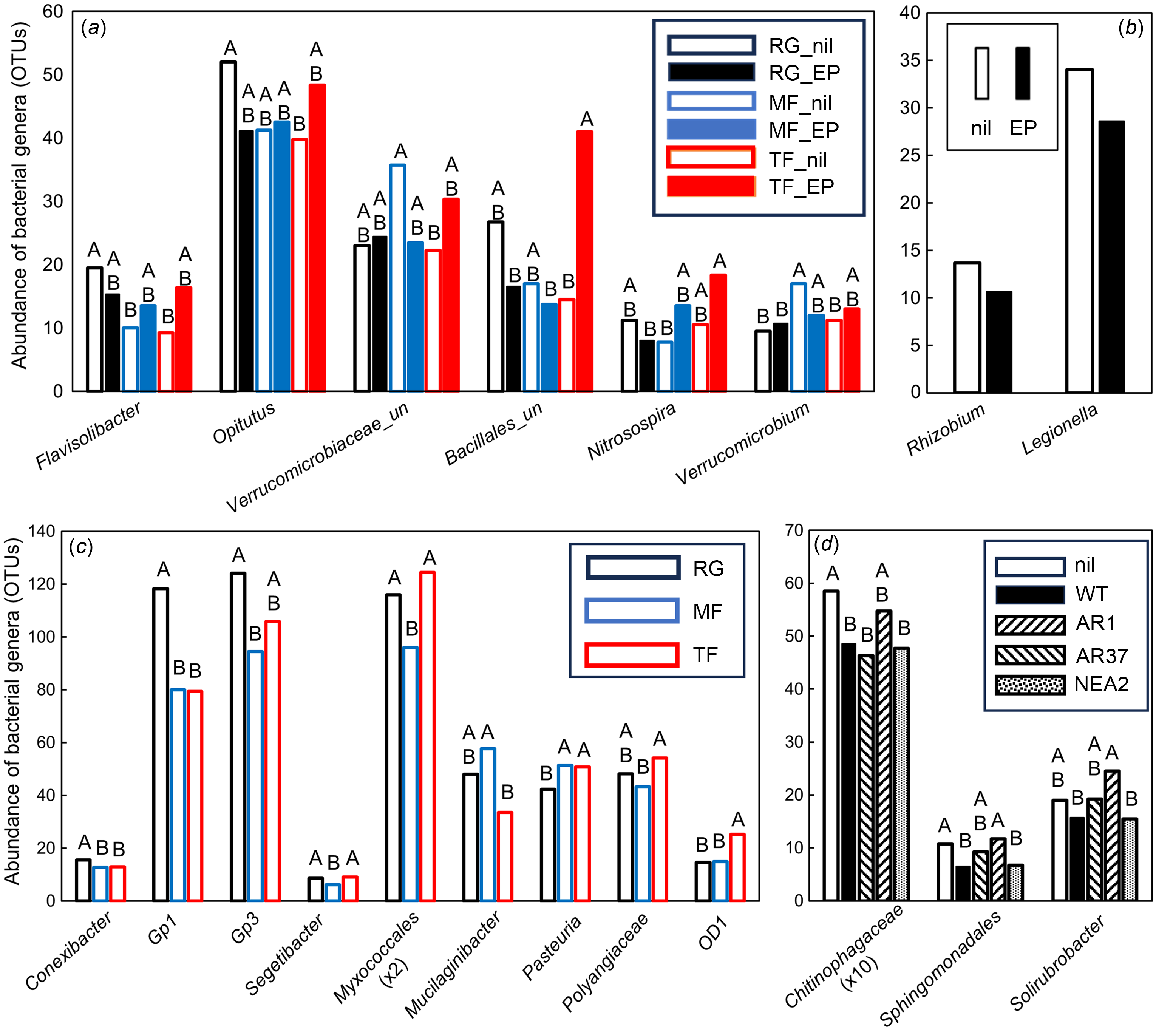
In total, 22 bacterial genera were significantly different (VIP >1.0), with seven of these being most important for segregation on axis 1, seven on axis 2, seven on axis 3, and one on axis 4. Six bacterial OTUs were significantly affected by the interaction between grass species and endophyte infection (Fig. 6a). Flavisolibacter sp. (Bacteroidetes, Chitinophagaceae) and Opitutus sp. (Verrucomicrobia) were more abundant in RG_nil soil than in MF_nil and TF_nil soils (Flavisolibacter sp.), and in TF_nil soil only (Opitutus sp.) (Fig. 6a). An unclassified Verrucomicrobiaceae was more abundant in MF_nil than in TF_nil soil, while an unclassified Bacillales and the nitrite-oxidising nitrifier Nitrospira sp. were most abundant in TF_EP soil. Verrucomicrobium sp. was more abundant in MF_nil soil than in RG and TF_nil soils. Two bacterial OTUs were significantly affected by the endophyte status of the grass species: the N-fixing Rhizobium sp. and the Gammaproteobacteria Legionella sp., both of which were less abundant in soils from endophyte-infected than uninfected grasses (Fig. 6b). Most bacterial genera (14) were affected by grass species and the groups with abundance >15 were further analysed. The Actinobacterium Conexibacter sp. and an Acidobacteria_Gp1 were more abundant in RG than in MF and TF soils, whereas the Acidobacteria_Gp3, the Chitinophagaceae Segetibacter sp. and an unclassified Myxococcales were more abundant in RG and TF than in MF soils (Fig. 6c). The Sphingobacteria Mucilaginibacter sp. was most abundant in RG and MF soils. The Bacillales Mucilaginibacter sp., an unclassified Polyangiaceae and OD1 were all most abundant in TF soils (Fig. 6c). Three bacterial genera were differentially affected by endophytic strain. An unclassified Chitinophagaceae, an unclassified Sphingomonadales and the Actinobacterium Solirubrobacter sp. were all highly abundant in RG_nil and RG_AR37 soils (Fig. 6d).
Impacts of plant extended phenotype (endophyte) and grass species on the abundance of influential bacterial genera within the PLSDA related to Experiment 2. Only those genera that featured strongly as VIP >1 were analysed further using either (a–c) two-way ANOVA or (d) one-way ANOVA. Untransformed means of statistically significant different genera (P < 0.05) are shown. (a) Black open bars, RG_nil (ryegrass without endophyte); black closed bars, RG_EP (ryegrass infected with Epichloë sp.); blue open bars, MF_nil (meadow fescue without endophyte); blue closed bars, MF_EP (meadow fescue infected with Epichloë uncinata U2); red open bars, TF_nil (tall fescue without endophyte); red closed bars, TF_EP (tall fescue infected with Epichloë coenophiala AR542). (b) Open bars, nil (grasses without endophyte); closed bars, grasses infected with Epichloë sp. endophyte. (c) Black bars, RG (ryegrass); blue bars, MF (meadow fescue); red bars, TF (tall fescue). (d) Open bars, RG_nil; closed bars, RG_WT (ryegrass infected with E. festucae var. lolii wild-type); downward striped bars, RG_AR1 (ryegrass infected with E. festucae var. lolii AR1); upward striped bars, RG_AR37 (ryegrass infected with Epichloë sp. LpTG-3 strain AR37); dotted bars, RG_NEA2 (ryegrass infected with E. festucae var. lolii NEA2). Abundancies were Box–Cox transformed prior to ANOVA; within each genus, treatment means with the same letter are not significantly different according to Tukey’s HSD at P = 0.05.
Discussion
There is increasing evidence that aboveground vegetation interacts with belowground microbial populations, with significant outcomes for cycling of both C and N in soils (Wardle et al. 2004; van der Heijden et al. 2008; Chen et al. 2022; Tomazelli et al. 2023). In this study, we used a random spatial arrangement of carefully chosen aboveground treatments (plant species/functional traits and N fertilisation level in Experiment 1; grass species and endophyte infection extended phenotype in Experiment 2) across an initially common soil mineralogy and community. Use of a common soil systematically avoids confounding effects of soil biophysical and biochemical characteristics per se on microbial communities (Ettema and Wardle 2002; Young and Crawford 2004). Using DNA-based high-throughput sequencing techniques, we are able to focus on those soil organisms that contribute to any significant differences as the soil community responds, while avoiding being overwhelmed by the substantial majority of organisms that might remain common.
Effects of plant species/traits and N level on soil fungal and bacterial communities
In our study, by far the strongest impacts on soil fungal communities were seen in Experiment 1. Most differences were associated with the distinction between grass and legumes, with relatively minor differences arising between grass cultivars (high vs low sugar), between legume species, or due to N fertilisation level of grasses. Overall, fungal communities were dominated by Asco- and Basidiomycota, with an almost even distribution of these two phyla in all soils except those planted with white clover, in which substantially fewer Basidiomycota were detected. The two dominant phyla in grass soils have also been observed in other studies (Borer et al. 2014; Prober et al. 2015; Yu et al. 2021). A sequencing study of the fungal ITS region in soils from grasslands along elevation gradients in the Western Swiss Alps found an even distribution of these two phyla (Pellissier et al. 2014). Another sequencing study of the fungal ITS region in soils from 25 temperate grassland sites of the global Nutrient Network experiment (Borer et al. 2014) found a strong dominance of Ascomycota in most soils, comprising up to 77% of fungi in tallgrass prairie soils from central USA. Ascomycota, Basidiomycota and Glomeromycota also reportedly dominate fungal communities in the typical steppe soils of northern China (Yu et al. 2021). However, unlike our study, none of those studies were performed on monocultures but rather on mixtures of grasses, and notably, where legumes were involved, a mixture of grasses and legumes was used, and so the effects of plant species and N input were confounded.
Within the Ascomycota, we found that Epicoccum sp., Fusarium sp., Ilyonectria sp. (syn. Cylindrocarpon) and Plectosphaerella sp. were substantially more abundant in legume than grass soils. Species of Epicoccum have been shown to form phenolic substances from non-aromatic compounds, which either accumulate as recalcitrant ‘humic acid’ polymers in soils (Filip et al. 1972, 1974) or act as antifungal and antibacterial compounds (Brown et al. 1987; Dzoyem et al. 2017). Fusarium sp. and Ilyonectria sp. have been described as plant pathogens (McGee and Kellock 1974) often associated with legumes (Brown et al. 1987; Zahid et al. 2001; Benitez et al. 2016). Plectosphaerella sp. has also been described as a plant pathogen (Domsch and Gams 1969). These fungi have been shown to have high pectinase activity (Domsch and Gams 1969) and their high abundance in legume soils might reflect the differences in grass versus dicot cell wall composition, whereby dicot primary cell walls contain up to 7-fold higher levels of pectin (Vogel 2008). In contrast to the above genera, Crucellisporium sp., Leohumicola sp., Chaetomium sp. and Phaeosphaeriopsis sp. were more abundant in grass soils than legume soils, and all of these have been described as endophytic fungi (Sánchez Márquez et al. 2010; Takashima et al. 2014), but very little is known about their metabolism. The abundance of Pestalotiopsis sp., Monographella sp. and Anthostomella sp. was also high in grass soils (data not shown); these genera contain members capable of decomposing lignin (Osono 2007).
We also found that the lignin-decomposing fungi in Basidiomycota were more abundant in grass soils. Within the Basidiomycota, several fungal classes (Mycenaceae, Ceratobasidiaceae, Polyporaceae) belonging to the Agaricomycetes were highly represented in soils under grasses compared with those under legumes. Mycenaceae and Polyporaceae are major fungal classes known to contain a large number of ligninolytic white-rot fungal genera (Osono 2007, 2010; Morgenstern et al. 2008; Floudas et al. 2012; Barrasa et al. 2014; Oliveira et al. 2020; Atiwesh et al. 2022). In this study, the most abundant genus, Mycena, was almost absent in soils under white clover and significantly lower in abundance in soils under bird’s-foot trefoil, whereas Polyporus sp. was most abundant in ryegrass soils under high N fertilisation and least abundant in white clover soils. Both of these genera are white-rot fungi able to degrade lignin, a plant cell-wall phenolic polymer of complex structure that is recalcitrant to decomposition by most organisms, and which forms a protective shield around cellulose (Osono 2007; Dungait et al. 2012). These ligninolytic fungi excrete a special set of enzymes: manganese, versatile peroxidases and laccases (Osono 2007; Morgenstern et al. 2008; Floudas et al. 2012). Most studies on white-rot fungi have focused on wood decay; only little is known about lignin degradation in grasses (Osono 2010; Suhara et al. 2012; Nuchdang et al. 2015; Shirkavand et al. 2016; Oliveira et al. 2020; Atiwesh et al. 2022). Cell walls of grasses differ significantly from those of dicots (such as legumes) in that they have a higher lignin content and especially high levels of ferulic and p-coumaric acid, which crosslink lignin to cellulose, thereby preventing access for microbes to cellulose and other cell wall carbohydrates (Carpita 1996; Grabber et al. 2004; Vogel 2008; Oliveira et al. 2020; Atiwesh et al. 2022). Our results showing a very high abundance of white-rot fungi in grass soils most probably reflect the differences between grasses and legumes, indicating that the chemical composition of plant cell walls that differs between grasses and legumes might be the main driver of shifts in these communities. Qu et al. (2016) also found that plant species composition was the key factor influencing the variability of microbial composition within soils.
Ceratobasidium sp. (syn. Rhizoctonia) and Sebacina sp. were also more abundant in grass soils than legume soils. Ceratobasidium sp. is a fungal pathogen, known to cause diseases in grasses (Toda et al. 1999). Sebacinales are mycorrhizal and endophytic fungi (Weiß et al. 2016), some of which (e.g. Sebacina vermifera) have been shown to enhance biomass production in switchgrass (Panicum virgatum L.) and other plants (Ghimire and Craven 2011; Weiß et al. 2016). The only Basidiomycota that was more abundant in legume soils was the Holtermannia sp. yeast. Yeasts utilise monomeric and dimeric carbohydrates and amino acids either excreted in root exudates or released during the decomposition of plant cell walls and proteins by other soil microbes (Bisaria and Ghose 1981; Tomme et al. 1995); our results might indicate that these types of sugars are more prevalent in soils with legume cover.
Only minor differences in fungal communities were associated with differences in cultivar within the grass species (L. perenne), level of N fertilisation or legume species (T. repens vs L. pedunculatus). Glomus sp., an arbuscular mycorrhizal fungus, was more abundant in HSG soil with low N fertilisation than in the legume soils, whereas there were no significant differences among the other grass and legume soils. Olpidium sp., a virus-transmitting member of class Chytridiomycetes (Rochon et al. 2004), was more abundant in RG soils than legume soils, whereas HSG soils did not differ from any of the soils. The only fungal genus which was affected by legume species was Phaeosphaeriopsis sp., which was lower in WCl soils than all other soils including LoP soils.
In this study, soil bacterial communities were dominated by Proteobacteria, Acidobacteria and Bacteroidetes, which is consistent with other studies on grassland soils (Nemergut et al. 2008; Fierer et al. 2009; Nacke et al. 2011; Yu et al. 2021). Bacterial communities, particularly the Rhizobium, Nitrospira, an unclassified Comamonadaceae, Pedobacter and Byssovorax spp., appeared to be affected by plant species and N levels as well. We found that the N-fixing Rhizobium sp. was generally less abundant in grass soils at low N than in legume soils. This was expected because the Rhizobium spp. can form symbiotic associations with legumes (Mylona et al. 1995). Rhizobium spp. can also live non-symbiotically in soils, where they assimilate simple sugars and organic and amino acids, and it has been shown that growth of rhizobia is stimulated by the presence of legumes and non-leguminous dicots, whereas grasses maintain smaller numbers of rhizobia (Tuzimura and Watanabe 1962). In this study, the nitrifying Nitrospira sp. and an unclassified genus of the Comamonadaceae family containing denitrifiers (Patureau et al. 1996; Khan et al. 2002) were also less abundant in grass soils at low N than in legume soils, consistent with a study showing that legume species enriched Nitrospira in the soil microbiome (Zhou et al. 2017). These findings indicate that N fixed by Rhizobium associated with legumes might not only promote plant growth but also provide soil with N-containing substrates that promote propagation of the Nitrospira and denitrifiers in the soils. Nitrospira are chemolithoautotrophic nitrite-oxidising bacteria, which catalyse the last step in nitrification (Murphy et al. 1985; Kowalchuk and Stephen 2001; Leininger et al. 2006), and denitrifiers are bacteria capable of performing denitrification as part of the N cycle (Knowles 1982). All of those play key roles in the biogeochemical N cycle and in the release of nitrous oxide, a potent greenhouse gas, into the atmosphere (Wrage et al. 2001). As such, the potential linkage between Rhizobium, Nitrospira and denitrifiers in the legume soils may require further study. Compared with the grass soils at low N in this study, a high abundance of Pedobacter sp. and a lower abundance of Byssovorax sp. were generally found in legume soils. Pedobacter sp. has been described as an endophytic bacteria preferentially associating with legumes (Dudeja and Giri 2014; Piechulla et al. 2017; Wemheuer et al. 2017), and Byssovorax sp. was shown to be a cellulose decomposer also able to use nitrate as a source of N (Reichenbach et al. 2006). Our results above imply that N fertilisation and biological N fixation have strong effects on bacterial communities, especially on nitrifying and denitrifying genera.
By far the most abundant bacterial genus affected by our treatments was Flavobacterium, which was more abundant in legume soils and much less abundant in grass soils at high N. Flavobacterium has been described as plant growth-promoting bacterial genus through the production of growth hormones (Hartmann et al. 2009; Kolton et al. 2016) and phosphate solubilisation from soils (Goldstein 1986). Interestingly, the Adhaeribacter, Luteolibacter and Rhodococcus were also found to be more abundant in legume soils than in grass soils. The Adhaeribacter and Luteolibacter have been shown to have acid and/or alkaline phosphatases (Yoon et al. 2008; Elderiny et al. 2017), and these might be able to release phosphorus (P) from insoluble-P sources in the soil and supply legumes, which are often P-limited due to nodulation (Uhde-Stone et al. 2003), with additional P. Rhodococcus has also been shown to solubilise P by excreting organic acids (Chen et al. 2006). These results suggest that plant species and N fertilisation appear to affect bacterial communities associating soil phosphate solubilisation.
In this study, the genera Flavobacterium, Cellvibrio, Novosphingobium and Armatimonas, and an unclassified Micromonosporaceae, were generally affected by plant species as well. Like the impacts on fungi described above, the main impacts of plant species on those genera appear to be associated with the capabilities of bacteria metabolising complex carbohydrates in plants. The genera Flavobacterium, Cellvibrio and Novosphingobium were all found to be more abundant in legume soils than in grass soils. The xyloglucan production by Flavobacterium in dicots is reportedly up to 20-fold higher than that in grass cell walls (Harris and Smith 2006; Vogel 2008). Cellvibrio has been shown to produce many enzymes involved in cell wall and storage carbohydrate degradation, especially pectins and galactomannans (DeBoy et al. 2008), which are more abundant in legume than in grass cell walls (Vogel 2008). Novosphingobium has been shown to produce enzymes that degrade aromatic compounds such as hydroxybenzoate, vanillate and protocatechuic acid (Ohta et al. 2015), all of which were detected in relatively high concentrations in legumes (Klejdus et al. 2008). By contrast, Armatimonas sp. and Micromonosporaceae (both less abundant in legume soils than grass soils) have been shown, respectively, to be oligo-heterotrophic bacteria, preferring nutrient-poor environments and complex carbohydrates over soluble sugars (Tamaki et al. 2011; Dunfield et al. 2012), or to be degraders of complex cell wall polysaccharides (Yeager et al. 2017). These results indicate again that the differences in cell wall composition of grasses vs legumes are major drivers of differences in soil bacteria communities.
Effects of plant extended phenotype on soil fungal and bacterial communities
Previous experimental reports showed that soils with endophyte-infected grasses had an altered composition of the associated fungal and bacterial communities (Casas et al. 2011; Wakelin et al. 2015; Rojas et al. 2016). In Experiment 2, fungal and bacterial soil communities also differed between soils with endophyte-infected and uninfected grasses. Soils with infected grasses showed low abundance of Ascomycota (here Epicoccum, Verticillium and Phoma) and high abundance of Glomeromycota (e.g. Glomus) fungi, findings that agree with previous experimental results (Rojas et al. 2016). As discussed, some Epicoccum species produce antifungal and antibacterial compounds (e.g. Brown et al. 1987; Dzoyem et al. 2017), members of Verticillium and Phoma are plant pathogens (Harris 1986; Klosterman et al. 2009), and Glomus spp. are beneficial fungi of plants. The reduced abundance of detrimental Ascomycota fungi in soils could be associated with the generally observed Epichloë-based inhibition of fungal phytopathogens (and potentially mycopathogens as well) in plants (Card et al. 2021; Kou et al. 2021). The high abundance of Glomeromycota fungi in soils would be associated with the alteration in the composition of root exudates usually observed in Epichloë-symbiotic plants, which promotes the growth and reproduction of members of this phylum (i.e. mycorrhizal fungi) (Vignale et al. 2018; Zhong et al. 2022). Epichloë infection appeared to influence community diversity and abundance of N-cycling bacteria as well (e.g. the ammonia oxidation, nitrite reduction and nitrous oxide reduction bacteria), a pattern that agrees with previous experimental results (Yang et al. 2021; Chen et al. 2022). Furthermore, soil with Epichloë-infected grasses exhibited reduced abundance of N-fixing Rhizobium. A similar outcome was documented in soils conditioned by endophyte-associated Lolium multiflorum Lam. plants, which reduced the nodulation of N-fixing bacteria in legumes (García-Parisi et al. 2017). Additional results in that study showed that despite the reduced abundance of this bacterial group, endophyte-conditioned soils increased plant growth and N acquisition when the availability of Rhizobium was low (García-Parisi et al. 2017).
Conclusions
Results associated with Experiment 1 showed that neither N fertilisation level, nor elevated levels of soluble sugars (in HSG litter) or condensed tannins (in the LoP treatment), had a substantial effect on soil fungal communities. The main driver of shifts in these communities appears to be the chemical composition of plant cell walls, which differs between grasses and legumes. In the case of soil bacterial communities, N fertilisation and biological N fixation affected the abundance of nitrifying and denitrifying genera, while plant species and N fertilisation seemed to affect the bacterial groups associating with the solubilisation of phosphate. Results associated with Experiment 2 showed that Epichloë changed the abundance of soil fungal and bacterial groups, including reduction of fungal phyto- and myco-pathogens, an increment of mycorrhizal fungi, and reduction of N-fixing bacteria. The changes in soil microbial communities could be explained in part by the altered root exudate composition exhibited by Epichloë-infected plants (Patchett and Newman 2021). Further investigations are warranted to evaluate whether the Epichloë-mediated changes in soil microbial communities affect plant responses to the environment and functionality/productivity of plant communities (Bastías et al. 2022; Bastías and Gundel 2023).
Data availability
The data that support this study will be shared upon reasonable request to the corresponding author.
Declaration of funding
This work was partially funded by New Zealand Ministry of Business, Innovation and Employment (Contract ID C10X0903).
Author contributions
Conceptualisation: SR and AJP; methodology: SR, JR and QL; investigation: SR, AJP, JR and QL; writing: SR, AJP, QL and DAB.
Acknowledgements
We thank Professor Grant Edwards and Dr Racheal Bryant (Lincoln University, NZ) for setting up and maintaining the field trials and for their continued support; Drs David Hume and Wei Zhang (both at AgResearch Ltd) and the Endophyte Innovation management team for reviewing the earlier version of the manuscript. DAB acknowledges the research support provided by the Strategic Science Investment Fund (SSIF) from the New Zealand Ministry of Business, Innovation and Employment.
References
Atiwesh G, Parrish CC, Banoub J, Le T-AT (2022) Lignin degradation by microorganisms: a review. Biotechnology Progress 38(2), e3226.
| Crossref | Google Scholar | PubMed |
Barrasa JM, Blanco MN, Esteve-Raventós F, Altés A, Checa J, Martinez AT, et al. (2014) Wood and humus decay strategies by white-rot basidiomycetes correlate with two different dye decolorization and enzyme secretion patterns on agar plates. Fungal Genetics and Biology 72(11), 106-114.
| Crossref | Google Scholar |
Bastías DA, Gundel PE (2023) Plant stress responses compromise mutualisms with Epichloë endophytes. Journal of Experimental Botany 74(1), 19-23.
| Crossref | Google Scholar | PubMed |
Bastías DA, Balestrini R, Pollmann S, Gundel PE (2022) Environmental interference of plant−microbe interactions. Plant, Cell & Environment 45(12), 3387-3398.
| Crossref | Google Scholar | PubMed |
Benitez M-S, Taheri WI, Lehman RM (2016) Selection of fungi by candidate cover crops. Applied Soil Ecology 103(7), 72-82.
| Crossref | Google Scholar |
Bezemer TM, Lawson CS, Hedlund K, Edwards AR, Brook AJ, Igual JM, et al. (2006) Plant species and functional group effects on abiotic and microbial soil properties and plant-soil feedback responses in two grasslands. Journal of Ecology 94(5), 893-904.
| Crossref | Google Scholar |
Bisaria VS, Ghose TK (1981) Biodegradation of cellulosic materials: substrates, microorganisms, enzymes and products. Enzyme and Microbial Technology 3(2), 90-104.
| Crossref | Google Scholar |
Borer ET, Harpole WS, Adler PB, Lind EM, Orrock JL, Seabloom EW, et al. (2014) Finding generality in ecology: a model for globally distributed experiments. Methods in Ecology and Evolution 5(1), 65-73.
| Crossref | Google Scholar |
Brisson N, Gate P, Gouache D, Charmet G, Oury F-X, Huard F (2010) Why are wheat yields stagnating in Europe? A comprehensive data analysis for France. Field Crops Research 119(1), 201-212.
| Crossref | Google Scholar |
Brown AE, Finlay R, Ward JS (1987) Antifungal compounds produced by Epicoccum purpurascens against soil-borne plant pathogenic fungi. Soil Biology and Biochemistry 19(6), 657-664.
| Crossref | Google Scholar |
Cai Y, Sun Y (2011) ESPRIT-Tree: hierarchical clustering analysis of millions of 16S rRNA pyrosequences in quasilinear computational time. Nucleic Acids Research 39(14), e95.
| Crossref | Google Scholar | PubMed |
Card SD, Bastías DA, Caradus JR (2021) Antagonism to plant pathogens by Epichloë fungal endophytes – a review. Plants 10(10), 1997.
| Crossref | Google Scholar | PubMed |
Carpita NC (1996) Structure and biogenesis of the cell walls of grasses. Annual Review of Plant Physiology and Plant Molecular Biology 47, 445-476.
| Crossref | Google Scholar | PubMed |
Casas C, Omacini M, Montecchia MS, Correa OS (2011) Soil microbial community responses to the fungal endophyte Neotyphodium in Italian ryegrass. Plant and Soil 340, 347-355.
| Crossref | Google Scholar |
Chen YP, Rekha PD, Arun AB, Shen FT, Lai W-A, Young CC (2006) Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Applied Soil Ecology 34(1), 33-41.
| Crossref | Google Scholar |
Chen Z, White JF, Malik K, Chen H, Jin Y, Yao X, Wei X, Li C, Nan Z (2022) Soil nutrient dynamics relate to Epichloë endophyte mutualism and nitrogen turnover in a low nitrogen environment. Soil Biology and Biochemistry 174, 108832.
| Crossref | Google Scholar |
Cripps MG, Edwards GR, McKenzie SL (2013) Grass species and their fungal symbionts affect subsequent forage growth. Basic and Applied Ecology 14(3), 225-234.
| Crossref | Google Scholar |
De Deyn GB, Cornelissen JHC, Bardgett RD (2008) Plant functional traits and soil carbon sequestration in contrasting biomes. Ecology Letters 11(5), 516-531.
| Crossref | Google Scholar | PubMed |
DeBoy RT, Mongodin EF, Fouts DE, Tailford LE, Khouri H, Emerson JB, et al. (2008) Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicas. Journal of Bacteriology 190(15), 5455-5463.
| Crossref | Google Scholar | PubMed |
Domsch KH, Gams W (1969) Variability and potential of a soil fungus population to decompose pectin, xylan and carboxymethyl-cellulose. Soil Biology and Biochemistry 1(1), 29-36.
| Crossref | Google Scholar |
Dudeja SS, Giri R (2014) Beneficial properties, colonization, establishment and molecular diversity of endophytic bacteria in legumes and non legumes. African Journal of Microbiology Research 8(15), 1562-1572.
| Crossref | Google Scholar |
Dunfield PF, Tamas I, Lee KC, Morgan XC, McDonald IR, Stott MB (2012) Electing a candidate: a speculative history of the bacterial phylum OP10. Environmental Microbiology 14(12), 3069-3080.
| Crossref | Google Scholar | PubMed |
Dungait JAJ, Hopkins DW, Gregory AS, Whitmore AP (2012) Soil organic matter turnover is governed by accessibility not recalcitrance. Global Change Biology 18(6), 1781-1796.
| Crossref | Google Scholar |
Dzoyem JP, Melong R, Tsamo AT, Maffo T, Kapche DGWF, Ngadjui BT, et al. (2017) Cytotoxicity, antioxidant and antibacterial activity of four compounds produced by an endophytic fungus Epicoccum nigrum associated with Entada abyssinica. Revista Brasileira de Farmacognosia 27(2), 251-253.
| Crossref | Google Scholar |
Eady C (2021) The impact of alkaloid-producing Epichloë endophyte on forage ryegrass breeding: a New Zealand perspective. Toxins 13(2), 158.
| Crossref | Google Scholar | PubMed |
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16), 2194-2200.
| Crossref | Google Scholar | PubMed |
Elderiny N, Lee J-J, Lee Y-H, Park S-J, Lee S-Y, Park S, et al. (2017) Adhaeribacter terrae sp. nov., a novel bacterium isolated from soil. International Journal of Systematic and Evolutionary Microbiology 67(8), 2922-2927.
| Crossref | Google Scholar | PubMed |
Ettema CH, Wardle DA (2002) Spatial soil ecology. Trends in Ecology & Evolution 17(4), 177-183.
| Crossref | Google Scholar |
Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC (2009) Global patterns in belowground communities. Ecology Letters 12(11), 1238-1249.
| Crossref | Google Scholar | PubMed |
Filip Z, Haider K, Martin JP (1972) Influence of clay minerals on the formation of humic substances by Epicoccum nigrum and Stachybotrys chartarum. Soil Biology and Biochemistry 4(2), 147-154.
| Crossref | Google Scholar |
Filip Z, Haider K, Beutelspacher H, Martin JP (1974) Comparisons of IR-spectra from melanins of microscopic soil fungi, humic acids and model phenol polymers. Geoderma 11(1), 37-52.
| Crossref | Google Scholar |
Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, et al. (2012) The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336(6089), 1715-1719.
| Crossref | Google Scholar | PubMed |
García-Parisi PA, Lattanzi FA, Grimoldi AA, Druille M, Omacini M (2017) Three symbionts involved in interspecific plant-soil feedback: epichloid endophytes and mycorrhizal fungi affect the performance of rhizobia-legume symbiosis. Plant and Soil 412, 151-162.
| Crossref | Google Scholar |
Ghahramani A, Howden SM, del Prado A, Thomas DT, Moore AD, Ji B, Ates S (2019) Climate change impact, adaptation, and mitigation in temperate grazing systems: a review. Sustainability 11(24), 7224.
| Crossref | Google Scholar |
Ghimire SR, Craven KD (2011) Enhancement of switchgrass (Panicum virgatum L.) biomass production under drought conditions by the ectomycorrhizal fungus Sebacina vermifera. Applied and Environmental Microbiology 77(19), 7063-7067.
| Crossref | Google Scholar | PubMed |
Goldstein AH (1986) Bacterial solubilization of mineral phosphates: historical perspective and future prospects. American Journal of Alternative Agriculture 1(2), 51-57.
| Crossref | Google Scholar |
Grabber JH, Ralph J, Lapierre C, Barrière Y (2004) Genetic and molecular basis of grass cell-wall degradability. I. Lignin-cell wall matrix interactions. Comptes Rendus Biologies 327(5), 455-465.
| Crossref | Google Scholar | PubMed |
Hancock KR, Collette V, Fraser K, Greig M, Xue H, Richardson K, et al. (2012) Expression of the R2R3-MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiology 159(3), 1204-1220.
| Crossref | Google Scholar | PubMed |
Harris JR (1986) The association of Phoma sclerotioides with root diseases of cereals, legumes and weeds. Australasian Plant Pathology 15, 14-17.
| Crossref | Google Scholar |
Harris PJ, Smith BG (2006) Plant cell walls and cell-wall polysaccharides: structures, properties and uses in food products. International Journal of Food Science and Technology 41(s2), 129-143.
| Crossref | Google Scholar |
Hartmann M, Lee S, Hallam SJ, Mohn WW (2009) Bacterial, archaeal and eukaryal community structures throughout soil horizons of harvested and naturally disturbed forest stands. Environmental Microbiology 11(12), 3045-3062.
| Crossref | Google Scholar | PubMed |
Jones MB, Donnelly A (2004) Carbon sequestration in temperate grassland ecosystems and the influence of management, climate and elevated CO2. New Phytologist 164(3), 423-439.
| Crossref | Google Scholar |
Kelliher FM, Parfitt RL, van Koten C, Schipper LA, Rys G (2013) Use of shallow samples to estimate the total carbon storage in pastoral soils. New Zealand Journal of Agricultural Research 56(1), 86-90.
| Crossref | Google Scholar |
Khan ST, Horiba Y, Yamamoto M, Hiraishi A (2002) Members of the family Comamonadaceae as primary poly(3-hydroxybutyrate-co-3-hydroxyvalerate)-degrading denitrifiers in activated sludge as revealed by a polyphasic approach. Applied and Environmental Microbiology 68(7), 3206-3214.
| Crossref | Google Scholar | PubMed |
Klejdus B, Vacek J, Lojková L, Benešová L, Kubáñ V (2008) Ultrahigh-pressure liquid chromatography of isoflavones and phenolic acids on different stationary phases. Journal of Chromatography A 1195(1–2), 52-59.
| Crossref | Google Scholar | PubMed |
Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV (2009) Diversity, pathogenicity, and management of Verticillium species. Annual Review of Phytopathology 47, 39-62.
| Crossref | Google Scholar | PubMed |
Knowles R (1982) Denitrification. Microbiological Reviews 46(1), 43-70.
| Crossref | Google Scholar | PubMed |
Kolton M, Erlacher A, Berg G, Cytryn E (2016) The Flavobacterium genus in the plant Holobiont: ecological, physiological, and applicative insights. In ‘Microbial models: from environmental to industrial sustainability. Vol. 1. Microorganisms for sustainability’. (Ed. S Castro-Sowinski) pp. 189–207. (Springer: Singapore) doi:10.1007/978-981-10-2555-6_9
Kou M-Z, Bastías DA, Christensen MJ, Zhong R, Nan Z-B, Zhang X-X (2021) The plant salicylic acid signalling pathway regulates the infection of a biotrophic pathogen in grasses associated with an Epichloë endophyte. Journal of Fungi 7(8), 633.
| Crossref | Google Scholar | PubMed |
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annual Review of Microbiology 55, 485-529.
| Crossref | Google Scholar | PubMed |
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, et al. (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442(7104), 806-809.
| Crossref | Google Scholar | PubMed |
Mahmud K, Lee K, Hill NS, Mergoum A, Missaoui A (2021) Influence of tall fescue Epichloë endophytes on rhizosphere soil microbiome. Microorganisms 9(9), 1843.
| Crossref | Google Scholar |
Martin KJ, Rygiewicz PT (2005) Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiology 5, 28.
| Crossref | Google Scholar | PubMed |
McGee DC, Kellock AW (1974) Fusarium avenaceum, a seed-borne pathogen of subterranean clover roots. Australian Journal of Agricultural Research 25, 549-557.
| Crossref | Google Scholar |
McSherry ME, Ritchie ME (2013) Effects of grazing on grassland soil carbon: a global review. Global Change Biology 19(5), 1347-1357.
| Crossref | Google Scholar | PubMed |
Morgenstern I, Klopman S, Hibbett DS (2008) Molecular evolution and diversity of lignin degrading heme peroxidases in the Agaricomycetes. Journal of Molecular Evolution 66(3), 243-257.
| Crossref | Google Scholar | PubMed |
Mudge PL, Kelliher FM, Knight TL, O’Connell D, Fraser S, Schipper LA (2017) Irrigating grazed pasture decreases soil carbon and nitrogen stocks. Global Change Biology 23(2), 945-954.
| Crossref | Google Scholar | PubMed |
Murphy WM, Gotlieb AR, Dugdale DT (1985) The effects of Fusarium wilt and weed control on survival of birdsfoot trefoil. Canadian Journal of Plant Science 65(2), 329-334.
| Crossref | Google Scholar |
Mylona P, Pawlowski K, Bisseling T (1995) Symbiotic nitrogen fixation. The Plant Cell 7(7), 869-885.
| Crossref | Google Scholar | PubMed |
Nacke H, Thürmer A, Wollherr A, Will C, Hodac L, Herold N, et al. (2011) Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE 6(2), e17000.
| Crossref | Google Scholar | PubMed |
Nemergut DR, Townsend AR, Sattin SR, Freeman KR, Fierer N, Neff JC, et al. (2008) The effects of chronic nitrogen fertilization on alpine tundra soil microbial communities: implications for carbon and nitrogen cycling. Environmental Microbiology 10(11), 3093-3105.
| Crossref | Google Scholar | PubMed |
Nuchdang S, Vatanyoopaisarn S, Phalakornkule C (2015) Effectiveness of fungal treatment by Coprinopsis cinerea and Polyporus tricholoma on degradation and methane yields of lignocellulosic grass. International Biodeterioration & Biodegradation 104, 38-45.
| Crossref | Google Scholar |
Oenema O, Wrage N, Velthof GL, van Groenigen JW, Dolfing J, Kuikman PJ (2005) Trends in global nitrous oxide emissions from animal production systems. Nutrient Cycling in Agroecosystems 72, 51-65.
| Crossref | Google Scholar |
Ohta Y, Nishi S, Kobayashi K, Tsubouchi T, Iida K, Tanizaki A, et al. (2015) Draft genome sequence of Novosphingobium sp. strain MBES04, isolated from sunken wood from Suruga Bay, Japan. Genome Announcements 3(1), e01373–14.
| Crossref | Google Scholar | PubMed |
Oliveira DM, Mota TR, Grandis A, de Morais GR, de Lucas RC, Polizeli MLTM, et al. (2020) Lignin plays a key role in determining biomass recalcitrance in forage grasses. Renewable Energy 147(1), 2206-2217.
| Crossref | Google Scholar |
Osono T (2007) Ecology of ligninolytic fungi associated with leaf litter decomposition. Ecological Research 22(6), 955-974.
| Crossref | Google Scholar |
Osono T (2010) Decomposition of grass leaves by ligninolytic litter-decomposing fungi. Grassland Science 56(1), 31-36.
| Crossref | Google Scholar |
Parsons AJ, Rowarth JS, Rasmussen S (2011) High sugar grasses. In ‘Plant sciences reviews 2011’. (Ed. D Hemming) pp.187–195. (CABI Reviews: Wallingford, UK) doi:10.1079/PAVSNNR20116046
Parsons AJ, Thornley JHM, Newton PCD, Rasmussen S, Rowarth JS (2013) Soil carbon dynamics: the effects of nitrogen input, intake demand and off-take by animals. Science of The Total Environment 465, 205-215.
| Crossref | Google Scholar | PubMed |
Parsons AJ, Thornley JHM, Rasmussen S, Rowarth JS (2016) Some clarification of the impacts of grassland intensification on food production, nitrogen release, greenhouse gas emissions and carbon sequestration: using the example of New Zealand. CAB Reviews 11(54), 1-19.
| Crossref | Google Scholar |
Patchett A, Newman JA (2021) Comparison of plant metabolites in root exudates of Lolium perenne infected with different strains of the fungal endophyte Epichloë festucae var. lolii. Journal of Fungi 7(2), 148.
| Crossref | Google Scholar | PubMed |
Patureau D, Bernet N, Moletta R (1996) Study of the denitrifying enzymatic system of Comamonas sp. strain SGLY2 under various aeration conditions with a particular view on nitrate and nitrite reductases. Current Microbiology 32, 25-32.
| Crossref | Google Scholar |
Pellissier L, Niculta-Hirzel H, Dubuis A, Pagni M, Guex N, Ndiribe C, et al. (2014) Soil fungal communities of grasslands are environmentally structured at a regional scale in the Alps. Molecular Ecology 23, 4274-4290.
| Crossref | Google Scholar | PubMed |
Piechulla B, Lemfack MC, Kai M (2017) Effects of discrete bioactive microbial volatiles on plants and fungi. Plant, Cell & Environment 40, 2042-2067.
| Crossref | Google Scholar | PubMed |
Prober SM, Leff JW, Bates ST, Borer ET, Firn J, Harpole WS, et al. (2015) Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecology Letters 18(1), 85-95.
| Crossref | Google Scholar | PubMed |
Qu T-B, Du W-C, Yuan X, Yang Z-M, Liu D-B, Wang D-L, Yu L-J (2016) Impacts of grazing intensity and plant community composition on soil bacterial community diversity in a steppe grassland. PLoS ONE 11(7), e0159680.
| Crossref | Google Scholar |
Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ (2011) Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12(1), 38.
| Crossref | Google Scholar |
Reichenbach H, Lang E, Schumann P, Spröer C (2006) Byssovorax cruenta gen. nov., sp. nov., nom. Rev., a cellulose-degrading myxobacterium: rediscovery of ‘Myxococcus cruentus’ Thaxter 1897. International Journal of Systematic and Evolutionary Microbiology 56(10), 2357-2363.
| Crossref | Google Scholar |
Reisinger A, Ledgard S (2013) Impact of greenhouse gas metrics on the quantification of agricultural emissions and farm-scale mitigation strategies: a New Zealand case study. Environmental Research Letters 8, 025019.
| Crossref | Google Scholar |
Rochon DA, Kakani K, Robbins M, Reade R (2004) Molecular aspects of plant virus transmission by Olpidium and plasmodiophorid vectors. Annual Review of Phytopathology 42, 211-241.
| Crossref | Google Scholar | PubMed |
Rojas X, Guo J, Leff JW, McNear DH, Jr., Fierer N, McCulley RL (2016) Infection with a shoot-specific fungal endophyte (Epichloë) alters tall fescue soil microbial communities. Microbial Ecology 72(1), 197-206.
| Crossref | Google Scholar | PubMed |
Schardl CL, Florea S, Pan J, Nagabhyru P, Bec S, Calie PJ (2013) The epichloae: alkaloid diversity and roles in symbiosis with grasses. Current Opinion in Plant Biology 16(4), 480-488.
| Crossref | Google Scholar | PubMed |
Schipper LA, Parfitt RL, Ross C, Baisden WT, Claydon JJ, Fraser S (2010) Gains and losses in C and N stocks of New Zealand pasture soils depend on land use. Agriculture, Ecosystems & Environment 139(4), 611-617.
| Crossref | Google Scholar |
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology 75(23), 7537-7541.
| Crossref | Google Scholar | PubMed |
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences 109(16), 6241-6246.
| Crossref | Google Scholar |
Shirkavand E, Baroutian S, Gapes DJ, Young BR (2016) Combination of fungal and physicochemical processes for lignocellulosic biomass pretreatment – a review. Renewable and Sustainable Energy Reviews 54, 217-234.
| Crossref | Google Scholar |
Soussana J-F, Lemaire G (2014) Coupling carbon and nitrogen cycles for environmentally sustainable intensification of grasslands and crop-livestock systems. Agriculture, Ecosystems & Environment 190, 9-17.
| Crossref | Google Scholar |
Suhara H, Kodama S, Kamei I, Maekawa N, Meguro S (2012) Screening of selective lignin-degrading basidiomycetes and biological pretreatment for enzymatic hydrolysis of bamboo culms. International Biodeterioration & Biodegradation 75, 176-180.
| Crossref | Google Scholar |
Sánchez Márquez S, Bills GF, Dominguez Acuña L, Zabalgogeazcoa I (2010) Endophytic mycobiota of leaves and roots of the grass Holcus lanatus. Fungal Diversity 41(1), 115-123.
| Crossref | Google Scholar |
Takashima Y, Narisawa K, Hidayat I, Rahayu G (2014) First report on fungal symbionts of Lycopodiaceae root from Mount Gede Pangrango National Park Indonesia. Journal of Developments in Sustainable Agriculture 9, 81-88.
| Crossref | Google Scholar |
Tamaki H, Tanaka Y, Matsuzawa H, Muramatsu M, Meng X-Y, Hanada S, et al. (2011) Armatimonas rosea gen. nov., sp. nov., of a novel bacterial phylum, Armatimonadetes phyl. nov., formally called the candidate phylum OP10. International Journal of Systematic and Evolutionary Microbiology 61(6), 1442-1447.
| Crossref | Google Scholar |
Terrill TH, Rowan AM, Douglas GB, Barry TN (1992) Determination of extractable and bound condensed tannin concentrations in forage plants, protein concentrate meals and cereal grains. Journal of the Science of Food and Agriculture 58(3), 321-329.
| Crossref | Google Scholar |
Toda T, Hyakumachi M, Suga H, Kageyama K, Tanaka A, Tani T (1999) Differentiation of Rhizoctonia AG-D isolates from turfgrass into subgroups I and II based on rDNA and RAPD analyses. European Journal of Plant Pathology 105, 835-846.
| Crossref | Google Scholar |
Tomazelli D, Klauberg-Filho O, Mendes SDC, Baldissera TC, Garagorry FC, Tsai SM, Pinto CE, Mendes LW, Goss-Souza D (2023) Pasture management intensification shifts the soil microbiome composition and ecosystem functions. Agriculture, Ecosystems & Environment 346(11), 108355.
| Crossref | Google Scholar |
Tomme P, Driver DP, Amandoron EA, Miller RC, Jr, Antony R, Warren J, et al. (1995) Comparison of a fungal (family I) and bacterial (family II) cellulose-binding domain. Journal of Bacteriology 177(15), 4356-4363.
| Crossref | Google Scholar | PubMed |
Tuzimura K, Watanabe I (1962) The effect of rhizosphere of various plants on the growth of Rhizobium. Soil Science and Plant Nutrition 8(4), 13-17.
| Crossref | Google Scholar |
Uhde-Stone C, Zinn KE, Ramirez-Yañez M, Li A, Vance CP, Allan DL (2003) Nylon filter arrays reveal differential gene expression in proteoid roots of white lupin in response to phosphorus deficiency. Plant Physiology 131(3), 1064-1079.
| Crossref | Google Scholar | PubMed |
van der Heijden MGA, Bardgett RD, van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecology Letters 11(3), 296-310.
| Crossref | Google Scholar | PubMed |
van der Wal A, Geydan TD, Kuyper TW, de Boer W (2013) A thready affair: linking fungal diversity and community dynamics to terrestrial decomposition processes. FEMS Microbiology Reviews 37(4), 477-494.
| Crossref | Google Scholar | PubMed |
Vignale MV, Iannone LJ, Scervino JM, Novas MV (2018) Epichloë exudates promote in vitro and in vivo arbuscular mycorrhizal fungi development and plant growth. Plant and Soil 422(1-2), 267-281.
| Crossref | Google Scholar |
Vogel J (2008) Unique aspects of the grass cell wall. Current Opinion in Plant Biology 11(3), 301-307.
| Crossref | Google Scholar | PubMed |
Wakelin S, Harrison S, Mander C, Dignam B, Rasmussen S, Monk S, Fraser K, O’Callaghan M (2015) Impacts of endophyte infection of ryegrass on rhizosphere metabolome and microbial community. Crop & Pasture Science 66(10), 1049-1057.
| Crossref | Google Scholar |
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 73(16), 5261-5267.
| Crossref | Google Scholar | PubMed |
Wardle DA, Bardgett RD, Klironomos JN, Setälä H, van der Putten WH, Wall DH (2004) Ecological linkages between aboveground and belowground biota. Science 304(5677), 1629-1633.
| Crossref | Google Scholar | PubMed |
Weiß M, Waller F, Zuccaro A, Selosse M-A (2016) Sebacinales – one thousand and one interactions with land plants. New Phytologist 211(1), 20-40.
| Crossref | Google Scholar | PubMed |
Wemheuer F, Kaiser K, Karlovsky P, Daniel R, Vidal S, Wemheuer B (2017) Bacterial endophyte communities of three agricultural important grass species differ in their response towards management regimes. Scientific Reports 7, 40914.
| Crossref | Google Scholar | PubMed |
Wrage N, Velthof GL, van Beusichem ML, Oenema O (2001) Role of nitrifier denitrification in the production of nitrous oxide. Soil Biology and Biochemistry 33(12-13), 1723-1732.
| Crossref | Google Scholar |
Yang Z, Jin Y, Hou F, Bowatte S (2021) Soil microbial and chemical responses to foliar Epichloë fungal infection in Lolium perenne, Hordeum brevisubulatum and Achnatherum inebrians. Fungal Ecology 53, 101091.
| Crossref | Google Scholar |
Yeager CM, Gallegos-Graves LV, Dunbar J, Hesse CN, Daligault H, Kuske CR (2017) Polysaccharide degradation capability of Actinomycetales soil isolates from a semiarid grassland of the Colorado Plateau. Applied and Environmental Microbiology 83(6), e03020–16.
| Crossref | Google Scholar | PubMed |
Yoon J, Matsuo Y, Adachi K, Nozawa M, Matsuda S, Kasai H, et al. (2008) Description of Persicirhabdus sediminis gen. nov., sp. nov., Roseibacillus ishigakijimensis gen. nov., sp. nov., Roseibacillus ponti sp. nov., Roseibacillus persicicus sp. nov., Luteolibacter pohnpeiensis gen. nov., sp. nov. and Luteolibacter algae sp. nov., six marine members of the phylum ‘Verrucomicrobia’, and emended descriptions of the class Verrucomicrobiae, the order Verrucomicrobiales and the family Verrucomicrobiaceae. International Journal of Systematic and Evolutionary Microbiology 58(4), 998-1007.
| Crossref | Google Scholar |
Young IM, Crawford JW (2004) Interactions and self-organization in the soil-microbe complex. Science 304(5677), 1634-1637.
| Crossref | Google Scholar | PubMed |
Yu Y, Zheng L, Zhou Y, Sang W, Zhao J, Liu L, et al. (2021) Changes in soil microbial community structure and function following degradation in a temperate grassland. Journal of Plant Ecology 14(3), 384-397.
| Crossref | Google Scholar |
Zahid MI, Gurr GM, Nikandrow A, Fulkerson WJ, Nicol HI (2001) Pathogenicity of root and stolon-colonising fungi to white clover. Australian Journal of Experimental Agriculture 41(6), 763-771.
| Crossref | Google Scholar |
Zak DR, Pregitzer KS, Burton AJ, Edwards IP, Kellner H (2011) Microbial responses to a changing environment: implications for the future functioning of terrestrial ecosystems. Fungal Ecology 4, 386-395.
| Crossref | Google Scholar |
Zhong R, Zhang L, Zhang X (2022) Allelopathic effects of foliar Epichloë endophytes on belowground arbuscular mycorrhizal fungi: a meta-analysis. Agriculture 12(11), 1768.
| Crossref | Google Scholar |
Zhou Y, Zhu H, Fu S, Yao Q (2017) Variation in soil microbial community structure associated with different legume species is greater than that associated with different grass species. Frontiers in Microbiology 8, 1007.
| Crossref | Google Scholar |


