Successful creation of seedless (sterile) leucaena germplasm developed from interspecific hybridisation for use as forage
D. Real A * , C. Revell A , Y. Han A B , C. Li B , M. Castello A and C. D. Bailey C
A * , C. Revell A , Y. Han A B , C. Li B , M. Castello A and C. D. Bailey C
A Department of Primary Industries and Regional Development, South Perth, WA 6151, Australia.
B Western Crop Genetics Alliance, College of Science, Health, Engineering and Education, Murdoch University, Murdoch, WA 6150, Australia.
C Department of Biology, New Mexico State University, Las Cruces, NM 88003, USA.
Crop & Pasture Science - https://doi.org/10.1071/CP22281
Submitted: 12 August 2022 Accepted: 20 October 2022 Published online: 1 December 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: The legume shrub/tree leucaena (Leucaena leucocephala (Lam.) de Wit.) is highly regarded as a cattle fodder, although it is considered an environmental weed in many parts of Australia.
Aims: We investigated the feasibility of developing a forage variety of triploid leucaena through interspecific hybridisation that is sterile (without seeds) as a strategy to mitigate its weed risk.
Methods: A conventional breeding strategy was used to create triploid hybrids from hand-crosses between tetraploid and diploid species of leucaena. Molecular markers were developed to confirm successful crosses and flow cytometry was used to confirm the ploidy level. The plants are being evaluated for flowering behaviour, seed and herbage production across two environmentally diverse sites at Carnarvon and Kununurra in northern Western Australia.
Key results: During 2018/19 and 2019/20, 2260 crosses between 45 different combinations of diploid females by tetraploid males and their reciprocals have created over 3000 putative triploid hybrid plants. This is the first time that triploids have successfully been created in Australia and 10 new parental combinations were created as a world-first.
Conclusions: A cohort of sterile triploid plants has been identified and selections will be made based on their edible biomass productivity, insect tolerance, capacity for regrowth after cutting, nutritive value and plant chemistry (including anti-methanogenic responses). A sterile leucaena variety will have application not only in Australian grazing systems but also in other tropical regions of the world.
Implications: A cost-effective technology for mass vegetative propagation of leucaena will be required for commercialisation of a sterile variety.
Keywords: breeding, flow cytometry, flowering, legume, molecular markers, seed production, sterility, triploids.
Introduction
The genus Leucaena contains 24 species of leguminous shrubs/trees native to the north of South America, Central America and southern North America. The genus is genetically diverse, containing five allotetraploid and 19 diploid species (Hughes 1998; Govindarajulu et al. 2011a, 2011b; Brewbaker 2016). While several species of leucaena have been used historically as minor foods in south-central Mexico (Hughes et al. 1995), Leucaena leucocephala (Lam.) de Wit. has been broadly adopted for use across the tropics, providing valuable forage (Takahashi and Ripperton 1949; Beutel et al. 2018), wood pulp (Khanna et al. 2019) and bioenergy (Haruthaithanasan et al. 2010; Tanavat et al. 2011). However, these introductions have come at a considerable cost to native ecosystems, with L. leucocephala becoming a notoriously invasive weed (Dadant 1953; Petty 1969; Campbell et al. 2019; Idol 2019). In Australia, a leucaena/grass pasture industry has been extensively utilised for decades in Queensland for cattle production (Buck et al. 2019; Shelton et al. 2021), but leucaena use in other states is constrained by its environmental weed status (Revell et al. 2019).
The production of sterile (seedless) lines of leucaena has the potential to alleviate the invasive characteristics of these plants while retaining the benefits of this highly regarded forage (Real et al. 2019). One key strategy to breed sterile leucaena plants includes the generation of triploids through the hybridisation of diploid and tetraploid species or lines (Real et al. 2019). This creates the potential for meiotic induced sterility in the resulting first filial generation (F1) hybrid. Some historical plant breeding examples of sterile triploids include watermelons (Lozanov 1968), citrus (Nishiura 1965), bananas (Raman et al. 1971), guava (Ito and Nakasone 1968) and cucumber (Galcenko 1961).
Through large-scale, laborious crossing studies using different species of leucaena, Sorensson and Brewbaker (1994) evaluated the compatibility of interspecific crosses between tetraploid and diploid species and demonstrated that triploids could be created by hand-crossing. This breeding strategy was successfully applied to create sterile triploids in Hawaii (Brewbaker 2013, 2016) and India (Khanna et al. 2019), with a focus on timber production. A comprehensive list of historically produced leucaena triploids and their sterility level is presented in Table 1.

|
The development of these triploid lines, primarily selected for forestry related uses, led us to consider the production of similar sterile triploids that would be useful as forage in Australia. Meat & Livestock Australia (MLA) co-funded a project with the Department of Primary Industries and Regional Development (DPIRD) and The University of Queensland (UQ) to create sterile leucaena for Australia (McMillan et al. 2019; Real et al. 2019). The generation and downstream use of sterile forage leucaena triploids in Australia requires the development of local crossing populations, as triploids created elsewhere in the past are either not alive or their vegetative material cannot be easily imported. The subsequent selection of sterile populations also needs to be amenable to mass clonal propagation for regional distribution to graziers. Therefore, the objective of this research work was to conduct interspecific crosses between tetraploid and diploid species of leucaena with excellent forage attributes and to evaluate the resulting seed grown plants for their production of seed. We hypothesised that (1) triploids could be created for the first time in Australia, (2) some combinations of parental material would be easier to hybridise, (3) tetraploids used as the female parent would have a higher crossing success rate than the reciprocals as reported by Sorensson and Brewbaker (1994) and (4) the sterility of interspecific hybrid triploids would be confirmed under field conditions.
Materials and methods
Plant materials
A review of the relevant literature (Hughes 1998; Real et al. 2019) describing characteristics related to high forage potential led to the selection of two tetraploid and nine diploid species for this Western Australia (WA) centred breeding program (Table 2).

|
A collection of 224 accessions of leucaena species to commence the breeding program was assembled by the Department of Primary Industries and Regional Development (DPIRD) from five different sources: Australian Pasture Genebank (APG), United States Department of Agriculture (USDA), International Center for Tropical Agriculture (CIAT), University of Queensland (UQ) and collection of seeds from naturalised leucaena stands in the north of WA (Table 3).

|
Interspecific crosses
Interspecific crosses between the diploid and tetraploid parents were conducted in an insect-proof glasshouse in Perth from March 2018 to February 2019 to create ‘Generation 1 hybrids’ and from November 2019 to May 2020 to create ‘Generation 2 hybrids’. For both programs, the crosses involved diploid species as female or male parents and tetraploid species as male or female parents.
The plants utilised to create Generation 1 hybrids were not selected for any particular agronomic attribute, rather they simply represented the high forage quality tetraploid and diploid species that were flowering at the same time. The purpose of these first crosses was to test the crossing techniques reported by Sorensson (1988); Sorensson and Sun (1990) and Dalzell et al. (2013) and to make our own adjustments/improvements based on many years of experience hand-crossing other legumes species from the genera Lotus, Trifolium, Medicago, Bituminaria, Lotononis, Ornithopus, Lupinus, Cicer, Brassica and Lens (Dalla Rizza et al. 2003; Real et al. 2004, 2006, 2007, 2014; Real and Altier 2005; Nelson et al. 2006; Clements et al. 2008; Kalve and Tadege 2017).
A total of 825 crosses were made to produce the Generation 1 cohort. The tetraploid species utilised were L. leucocephala (with two subspecies: leucocephala and glabrata) and Leucaena diversifolia. L. leucocephala ssp. glabrata is the main commercial species with demonstrated long-term forage attributes (Brewbaker 2016). L. diversifolia is another productive species, with slightly lower forage quality than L. leucocephala (Hughes 1998), which also has a history of forage use (Jones et al. 1998; Sotelo 2017). The diploid parents utilised were Leucaena collinsii, Leucaena retusa, Leucaena shannonii, Leucaena trichandra, Leucaena macrophylla, Leucaena pulverulenta and Leucaena zacapana.
To produce the Generation 2 cohort, a total of 1435 crosses were made, involving parental accessions that were selected based on their performance at two WA field sites located at Carnarvon and Kununurra representing the target soils and climate (Revell et al. 2019), and sown in May 2018. Hereafter we refer to these parental accessions as the ‘selected’ stock. Selected parent plants were either cloned from the field sites or plants from the same accession of original individuals (Real et al. 2019). Selected plants from six tetraploids (L. leucocephala ssp. leucocephala and glabrata, L. diversifolia, L. diversifolia × L. leucocephala, L. leucocephala ssp. glabrata × L. pallida, and L. pallida × L. leucocephala x L. diversifolia) and eight diploids (L. collinsii, L. zacapana, L. retusa, L. shannonii, L. macrophylla, L. greggii, L. lanceolata, and L. pulverulenta) were utilised as parents.
Growth of hybrid seedlings
A total of 183 seedlings were grown from 13 Generation 1 hybrid parental combinations (tetraploids × diploid) and 2819 seedlings from 44 Generation 2 hybrid parental combinations. Seeds were hand-scarified by cutting a very small edge of each seed to allow water to enter the seed. Seeds were placed in Petri dishes lined with filter paper, watered with distilled water and placed in a growth cabinet at 25°C with 24 h light. Seedlings were transplanted to 64 cell seedling trays filled with commercial premium potting mix when roots were about 0.5–1 cm long. Seedlings were raised in a naturally lit glasshouse without temperature control at DPIRD, Perth. Seedlings were inoculated by watering plants with a slurry mixture of Leucaena CB3060/Desmanthus CB3126 peat-based inoculum strains (New Edge Microbials) at the 2–3 leaf stage.
Molecular markers and KASP genotyping
Gene variants among leucaena species were detected by mapping transcriptome sequences to the draft genome from L. trichandra (Abair et al. 2019). Homozygous single-nucleotide polymorphisms (SNPs) were identified from the in-house database and the 57 most common variants in WA crosses were further selected to develop a minimum marker pool for genotype differentiation. Among them, 39 Kompetitive Allele Specific (KASP) polymerase chain reaction (PCR) markers (Supplementary Table S1) were designed based on the L. trichandra reference sequence using the Geneious Prime software (https://www.geneious.com/prime/).
Genomic DNA was extracted from the roots of all parental accessions using the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson 1980), or from the young leaves of all F1 seedlings using the AquaGenomic kit (MoBiTec, Germany) following the manufacturer’s instructions. KASP reactions consisted of 2.5 μL master mix (LGC Biosearch Tech™), 0.5 μL MilliQ water, 1 μL DNA (25 ng), and 1 μL primer mix. KASP assays were performed with the Applied Biosystems ViiA 7 system. The thermal cycling protocol was 30°C for 59 s, a hot start Taq activation step at 94°C for 15 min, 10 cycles of touchdown PCR at 94°C for 20 s, annealing at 60°C to 55°C (decreasing 0.6°C per cycle) for 40 s, and 29 cycles of standard amplification steps consisting of 94°C for 20 s, 55°C for 40 s, and the sample reading stage at 30°C for 59 s. Subsequently, the re-read protocol followed starting with the amplification step at 94°C for 20 s, and 57°C for 60 s for three cycles. The protocol is for amplifying weak PCR targets for strong signals.
The markers were first tested using a set of DNA consisting of an artificial DNA mix (50%:50%) of the two parental accessions. Successfully detected polymorphic markers were then used to screen individuals of each cross with two technical replicates. The results were analysed using the QuantStudio™ Real-Time PCR Software.
Ploidy level
Flow cytometry was utilised to confirm the ploidy level of the different species and specific parents selected from regional field evaluation. Genome size estimates for some leucaena species have been conducted previously using a fluorescence-activated cell sorting (FACS) flow cytometer (Govindarajulu et al. 2011a). Selected plants from the Generation 2 cohort were analysed using a Sysmex CyFlow single tube ploidy analyser. For each plant, a leaf sample of 1 cm2 was chopped into 400 μL nucleic extraction buffer with 200 μL 10% polyvinylpyrrolidone (H. Gamage, University of Queensland, 2020/21, pers. comm.), incubated for 1 min, filtered through a 40 μM filter and stained with 1600 μL staining buffer containing the DAPI stain (4′,6-diamidino-2-phenylindole; Galbraith et al. 1983; Doležel et al. 2007). The machine was standardised in each run using red blood cells from trout and the diploid and tetraploid parents as controls. A total of 46 diploid parents, 71 tetraploid parents and 659 F1 seedlings were analysed by flow cytometry.
Field locations
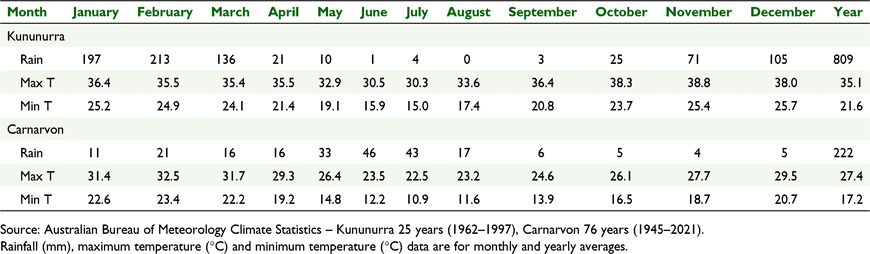
Two regional field sites were selected in WA (Revell et al. 2019) for the evaluation of the range of leucaena species sown in May 2018, Generation 1 hybrids sown in May 2020 and Generation 2 hybrids sown in May 2021, on land owned by the WA State Government where there is freedom to evaluate species that are otherwise not approved for use on pastoral lease (or require a diversification permit). The most northerly site was situated in Kununurra (15.65°S, 128.72°E, Elev. 35 m) on a grey-black self-mulching clay soil (Vertosol) associated with the alluvial plains of the Ord River. The southern site was situated in Carnarvon (24.86°S, 113.73°E, Elev. 12 m) on a red-brown sandy clay loam soil (Kandosol) associated with the alluvial plains of the Gascoyne River. Kununurra is a seasonally dry tropical environment, with a 5–6-month wet season and high annual rainfall. Carnarvon is a low rainfall site on the transition between subtropical and temperate environments. Mean monthly maximum and minimum temperatures are consistently higher in Kununurra than in Carnarvon (Table 4). All sites have the capability for irrigation that allowed sowing in the dry season. Kununurra is flood irrigated in raised beds and Carnarvon uses a pressurised dripper system. The Kununurra site was irrigated only during the dry season, while the Carnarvon site was irrigated all year around.
|
Both field sites were top-dressed with a basal all-purpose fertiliser before sowing and thereafter had no further nutrient applications. Seedlings were generally between 10 and 25 cm tall at the time of transplanting. At Kununurra the arrangement of transplanted seedlings in 2018 and 2020 was spaced plants on a 1.8 m (row centres) × 3 m grid. In 2021 the arrangement was spaced plants on a 1.8 m (row centres) × 2.5 m grid. An unsown buffer row was left every third bed. At Carnarvon, the arrangement of transplanted seedlings in 2018 and 2020 was spaced plants on a 3 m (row centres) × 3 m grid. In 2021 the arrangement was spaced plants on a 3 m (row centres) × 2.2 m grid. Each row across both sites contained between 33 and 56 spaced plants. Irrigation commenced immediately after seedlings were transplanted.
Field assessments
Flowering and pod setting were evaluated as visual scores from 0 to 5 in half unit increments. Zero was assigned when there were no flowers or no pods, and five was assigned when the plant was in full flower or full pod set. At Carnarvon, Generation 1 hybrids transplanted in May 2020 were assessed on the 28 January 2021, 15 March 2021, 6 May 2021, 29 July 2021, 21 October 2021 and 9 June 2022. Generation 2 hybrids transplanted in May 2021 were assessed on the 10 June 2022. At Kununurra, Generation 1 hybrids transplanted in May 2020 were assessed on the 11 May 2021, 18 November 2021 and 15 June 2022. Generation 2 hybrids transplanted in May 2021 were assessed on the 15 June 2022.
Results
Generation 1 hybrids (2018/2019)
The number of successful crosses in Generation 1 for each parental combination that produced viable seeds are presented in Table 5. An overall success rate of 19% was achieved from the 825 crosses. Crosses with diploid parents as females achieved an average 16% success rate, with the cross between L. pulverulenta and L. leucocephala ssp. glabrata being the most successful at 64%.

|
From the 160 crosses harvested, a subset of 183 hybrid seeds representing the 13 different parental combinations were grown (Table 6). Seventy-two were transplanted at Carnarvon on 19 June 2020 and 111 were transplanted at Kununurra on 23 June 2020. In addition to the 183 hybrid seedlings, 50 and 46 control tetraploid plants were transplanted at the same time next to the hybrids at Carnarvon and Kununurra, respectively.

|
Generation 2 hybrids (2019/2020)
The number of successful crosses in Generation 2 that produced viable seed are presented in Table 7 for crosses where diploid species were used as female parents and Table 8 where tetraploid species were used as female parents. Using diploids as female parents, overall crossing success was 20% (Table 7). The highest number of crosses was made with L. pulverulenta and this species (as a female parent) also achieved a high crossing success rate of 53%. Using tetraploids as female parents, overall crossing success was increased to 34% (Table 8). An average level of 55–56% crossing success was achieved with L. leucocephala ssp. glabrata and ssp. leucocephala (as a female parent). One of the most successful combinations was between L. leucocephala ssp. leucocephala and L. zacapana with a success rate of 82%.
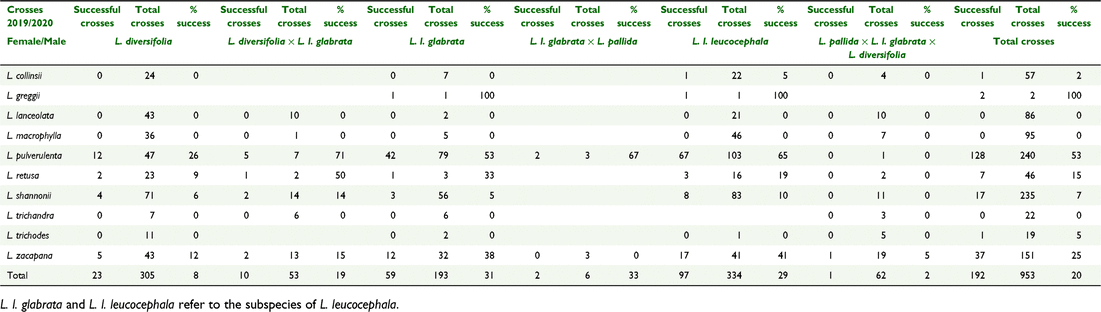
|

|
From the 358 crosses harvested, a subset of 2819 seeds representing 44 combinations (Table 9) were grown, with 1410 transplanted at Carnarvon on 6 May 2021 and 1312 transplanted at Kununurra on 10 May 2021. The commercial cultivar Wondergraze of L. leucocephala ssp. glabrata was transplanted at the same time as a high frequency control spread regularly across both field sites, with 26 and 24 Wondergraze seedlings transplanted at Carnarvon and Kununurra, respectively. Ninety-seven hybrid plants representing all combinations were kept at the South Perth glasshouse for close monitoring and observation.

|
Confirmation of hybrid offspring using KASP markers
All F1 individuals were tested with KASP markers to confirm the successful hybridisation and parental status. About 82% and 46% of the F1s were confirmed to be hybrids from their planned cross in Generation 1 and 2, respectively. The most robust makers were 6, 9, 13 and 17, which were preferred in genotyping of most combinations (Supplementary Table S1). Notably, the percentage of confirmed hybrids for Generation 1 was much higher than that for Generation 2. Generation 2 had 2173 individuals sharing the species L. leucocephala (ssp. leucocephala or glabrata) or a hybrid with L. leucocephala as one parent.
Confirmation of triploid status by flow cytometry
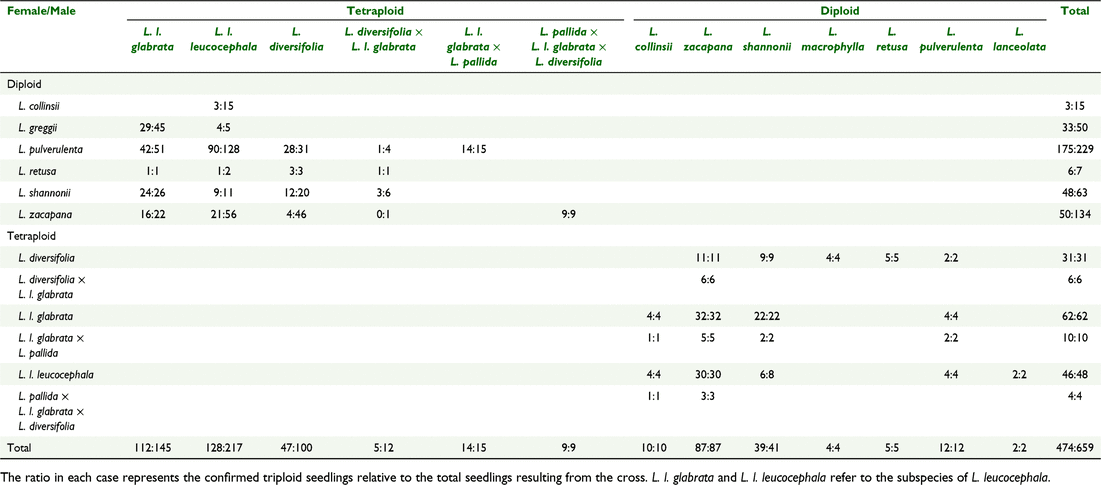
Generation 2 hybrids were evaluated by flow cytometry to assess diploid, triploid, or tetraploid status using genome size as the proxy (Fig. 1). All 46 diploid parents evaluated were confirmed as diploids and all 71 tetraploid parents evaluated were confirmed as tetraploids. From the 659 interspecific hybrids evaluated, 474 (72%) were confirmed as triploids (Table 10). The highest proportion of triploids (almost 100%) were achieved when a tetraploid species was used as the female parent. The diploids L. collinsii and L. zacapana used as the female parent produced the least successful crosses.
|
|
|
Time required for control tetraploid plants to flower
The percentage of 1- and 2-year old control tetraploid plants sown in May 2018 of L. leucocephala ssp. leucocephala and glabrata flowering in Kununurra and Carnarvon is presented in Table 11. Flowering occurred most rapidly at the Carnarvon site, with most plants flowering after 18 months and almost 100% of plants flowering by 30 months. Flowering was slowest at the Kununurra site with 13% of plants yet to flower after 30 months.

|
Preliminary sterility information of triploids
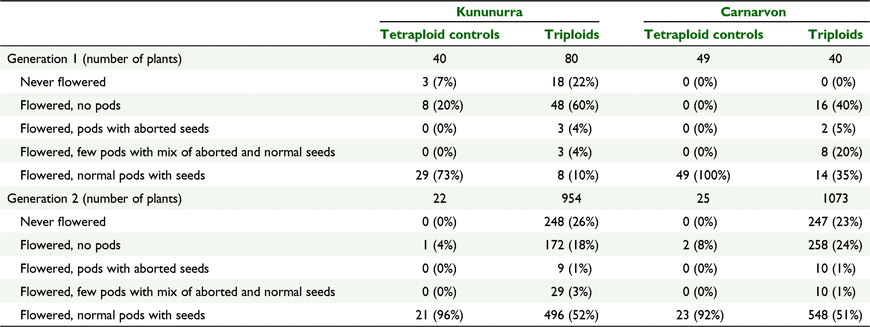
Flowering, fruiting, and viable seed status for Generation 1 and 2 hybrids characterised at Kununurra up to the 15 June 2022 and at Carnarvon up to the 10 June 2022 are presented in Tables 12. All tetraploid controls planted with Generation 1 hybrids at Carnarvon flowered and produced seed. Amongst the triploid cohort, 35% (14 plants) flowered and set seed while 40% (16 plants) flowered but produced no pods. At Kununurra, a higher proportion of plants had not yet flowered at the time of assessment (22%; 18 plants), but 60% (48 plants) of the triploid cohort had flowered and produced no pods. For the Generation 2 hybrids at both sites, 92% or greater of the tetraploid controls flowered and set seed, while for the triploid cohort, 51–52% flowered and set seed. At Carnarvon 24% (258 plants) of the triploid cohort flowered but produced no pods compared with 18% (172 plants) at Kununurra. For both generations, there was a small proportion (generally less than 5%, with the exception of Carnarvon-Generation 1) of the triploid cohort that flowered producing either few pods or pods with aborted seed. Collectively, across both generations and sites there were 494 single plants that flowered but did not produce pods (430 plants from the parent crosses in Generation 2). Up to 26% of the Generation 2 hybrid cohorts had not flowered at the time of assessment (13 months after transplanting).
Tables 13 presents the flowering behaviour of the Generation 2 cohort at the Kununurra and Carnarvon sites according to the ploidy of the female parent in the cross. Over 70% of the triploid cohort with tetraploid female parents flowered and produced seed, compared with only 25–27% with diploid female parents. Amongst the plants that flowered but have not produced pods, the highest proportion came from crosses with diploid female parents (22–33% cf. 13–15% with tetraploid female parents).

|
Discussion
The goal of this study was to investigate the feasibility of generating sterile lines of leucaena suitable for grazing across northern Australia. Through traditional crossing methods, we set out to hybridise diploid and tetraploid Leucaena spp. parental lines to generate putative sterile triploids.
The first hypothesis that triploids could be created for the first time in Australia was supported. Through a combination of (1) KASP genotyping to confirm hybrid seed resulting from the specific parental cross, and (2) flow cytometric-based genome size estimates to confirm that hybrid plants are triploids, we confirmed successful interspecific hybridisation and the creation of a large number of sterile triploid plants. From the list of triploids created in the past reported in Table 1 (Sorensson and Brewbaker 1994; Brewbaker 2016; Khanna et al. 2019) with L. leucocephala ssp. glabrata or L. diversifolia as one of the parent plants, we succeeded in re-creating nearly all of them in Australia for the first time, confirmed by ploidy level by flow cytometry (Table 10). The exceptions were the L. diversifolia × L. collinsii cross that was not evaluated in the subset tested with flow cytometry and for the crosses with diploid species that we did not include in our breeding program due to their lower forage quality such as Leucaena esculenta (Hughes 1998). In addition, we created 10 new world-first parental combinations, also confirmed by ploidy level (Table 10). These novel triploids were created with tetraploid hybrids of (a) L. diversifolia × L. leucocephala ssp. glabrata with diploids L. pulverulenta, L. retusa, L. shannonii and L. zacapana; (b) L. leucocephala ssp. glabrata × L. pallida with diploids L. collinsii, L. pulverulenta, L. shannonii and L. zacapana and (c) L. pallida × L. leucocephala ssp. glabrata × L. diversifolia with diploids L. collinsii and L. zacapana.
The second hypothesis that some combination of parental material would be easier to hybridise was supported. We found considerable variability in the success rate of specific crosses for both Generation 1 and 2. Considering the successful crosses that produced viable seed, highly productive diploid female plants included L. greggii, L. pulverulenta, L. retusa, L. shannonii, and L. zacapana while productive tetraploid males included the two subspecies of L. leucocephala. Similarly, all tested tetraploid females were highly productive and productive diploid males included L. collinsii, L. pulverulenta, L. shannonii, and L. zacapana. This is consistent with the work of Sorensson and Brewbaker (1994) who found highly compatible combinations between L. leucocephala or L. diversifolia with the diploid species L. pulverulenta. This high compatibility is also expected as both L. leucocephala and L. diversifolia are allotetraploids and L. pulverulenta is the ancestral maternal line for both species (Govindarajulu et al. 2011a).
The third hypothesis that tetraploids used as the female parent would have a larger crossing success rate was supported. Tetraploids used as females produced an average success rate of 34% vs 20% for diploid female parents. However, some of these successful crosses might be through accidental selfing due to the fact that tetraploids are self-compatible (Sorensson and Brewbaker 1994; Hughes 1998) and, therefore, easier to accidentally self while hand-crossing.
The fourth hypothesis that sterility of the interspecific hybrid would be confirmed under field conditions was supported in the experience to date, but further observation of the hybrids is required. Leucaena is a long-lived perennial plant species that can remain productive for over 30 years (Shelton et al. 2021). The time required to flower for the first time and the best environmental conditions conducive for prolific flowering are not well understood. Leucaena flowers throughout the year when moisture and temperature are adequate, although it has been reported that trees may not flower in the first year (Cook 2007). The reason for the more rapid flowering of plants at the Carnarvon site compared with Kununurra is unclear. It is counter intuitive as Kununurra is a more tropical environment with hotter and wetter summers (Table 4). Carnarvon is nine degrees further south in latitude, so slightly shorter daylengths (longer nights) maybe influential. Carnarvon is therefore an important site for the evaluation of flowering and sterility, allowing plants to express their true flowering and seed production behaviour in a much shorter time frame. The large numbers of plants that flowered without producing pods (64 from 120 in Generation 1 and 430 from 2027 in Generation 2 is very encouraging for long-term sterility, as flowering had taken place for periods of 6–24 months and the plants had been visited by honeybees constantly with a suite of pollen from nearby plants of different leucaena species. However, there are still 495 triploids that did not flower after 13 months for Generation 2, so we expect to have many more individuals in the target category that flower but do not produce pods. Generation 2 needs to be evaluated for at least another year to confirm the sterility of the triploids.
Molecular markers represent a robust and versatile means to characterise genetic diversity within and among plant species and populations. The advent of sequencing technology has greatly promoted the mapping of plant genomes and facilitated genome-wide and sequence-based marker development. Here, KASP markers transformed from SNPs among leucaena species effectively differentiated the F1 parentage in crosses. The method offers a high throughput approach that outperforms some other genetic markers such as sequence-related amplified polymorphism that requires intensive, large-scale and time-consuming PCR, gel electrophoresis and scoring (Li and Quiros 2001). However, the marker cross-species transferability was relatively low within the genus Leucaena since only four markers out of the total 39 showed good amplification in other species. The conservativeness of markers is dependent on plant evolutionary stability (Baird et al. 2010) and genetic heterozygosity. Diploid leucaena species have a self-incompatibility and out-crossing habit so the plants may have higher heterozygosity and therefore, sequence variations are more abundant and common across plant individuals. The low amplification of markers from some accessions indicates that the primers could not bind onto the DNA template for successful PCR. This may be due to the fact that the primers were designed based on the L. trichandra genome and none of the two subspecies of L. leucocephala were included in the variant database, thus the markers were not specific to successfully diagnose the L. leucocephala genetic background.
Conclusions
The final number of sterile (non-seeding) triploids for evaluation is yet to be determined but could be as high as 494 plants. Selections within this group will be made based on their edible biomass productivity, insect tolerance, capacity for regrowth after cutting and assessments of nutritive value (feed quality) and plant chemistry (including anti-methanogenic responses). The combination of KASP markers and flow cytometry techniques developed in this program will continue to be used to confirm the triploid status of these selections. As sterile lines with high forage potential are developed, parallel studies will be required to characterise how easily clonal plants of these lines can be produced. A cost-effective technology for mass vegetative propagation of Leucaena will be required for commercialisation of a sterile variety. Such a product will have application not only in Australian grazing systems but also in other tropical regions of the world where leucaena presents a significant weed risk.
Supplementary material
Supplementary material is available online.
Data availability
Raw data are available upon request to Daniel Real.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This research was funded by Meat & Livestock Australia Donor Company, grant number P.PSH.0884 and the Department of Primary Industries and Regional Development, WA, Australia.
Acknowledgements
The authors thank Meat & Livestock Australia (MLA) and the Department of Primary Industries and Regional Development (DPIRD) for funding the ‘Sterile leucaena project’. We also thank Mengistu Yadete, Daalacha Roba and Geoff Moore (DPIRD, South Perth), Kaylie Simpfendorfer, Helena O’Dwyer, Mark Warmington and Clare Atkins (DPIRD, Kununurra) and Anastasia Van Blommestein, James Barr and Graeme Sinclair (DPIRD, Carnarvon) for providing field and glasshouse support at various times for the project. The Leucaena trichandra genome and broader transcriptome data used to develop the KASP genotyping derived from work supported by the US National Science Foundation grant 1238731 (CDB)
References
Abair A, Hughes CE, Bailey CD (2019) The evolutionary history of Leucaena: recent research, new genomic resources and future directions. Tropical Grasslands-Forrajes Tropicales 7, 65–73.| The evolutionary history of Leucaena: recent research, new genomic resources and future directions.Crossref | GoogleScholarGoogle Scholar |
Baird RE, Wadl PA, Allen T, McNeill D, Wang X, Moulton JK, Rinehart TA, Abbas HK, Shier T, Trigiano RN (2010) Variability of United States isolates of Macrophomina phaseolina based on simple sequence repeats and cross genus transferability to related genera within botryosphaeriaceae. Mycopathologia 170, 169–180.
| Variability of United States isolates of Macrophomina phaseolina based on simple sequence repeats and cross genus transferability to related genera within botryosphaeriaceae.Crossref | GoogleScholarGoogle Scholar |
Beutel TS, Corbet DH, Hoffmann MB, Buck SR, Kienzle M (2018) Quantifying leucaena cultivation extent on grazing land. The Rangeland Journal 40, 31–38.
| Quantifying leucaena cultivation extent on grazing land.Crossref | GoogleScholarGoogle Scholar |
Brewbaker JL (2013) ‘KX4-Hawaii’, seedless interspecific hybrid leucaena. HortScience 48, 390–391.
| ‘KX4-Hawaii’, seedless interspecific hybrid leucaena.Crossref | GoogleScholarGoogle Scholar |
Brewbaker JL (2016) Breeding Leucaena: tropical multipurpose leguminous tree. In ‘Plant breeding reviews. Vol. 40’. (Ed. J Janick) pp. 43–123. (Wiley-Blackwell, Inc.)
Buck S, Rolfe J, Lemin C, English B (2019) Adoption, profitability and future of leucaena feeding systems in Australia. Tropical Grasslands-Forrajes Tropicales 7, 303–314.
| Adoption, profitability and future of leucaena feeding systems in Australia.Crossref | GoogleScholarGoogle Scholar |
Campbell S, Vogler W, Brazier D, Vitelli J, Brooks S (2019) Weed leucaena and its significance, implications and control. Tropical Grasslands-Forrajes Tropicales 7, 280–289.
| Weed leucaena and its significance, implications and control.Crossref | GoogleScholarGoogle Scholar |
Clements J, Prilyuk L, Quealy J, Francis G (2008) Interspecific crossing among the new world lupin species for Lupinus mutabilis crop improvement. In ‘Lupins for health and wealth. Proceedings of the 12th international lupin conference’. (Eds JA Palta, JB Berger) pp. 324–327. (International Lupin Association: Canterbury, New Zealand)
Cook B (2007) Leucaena – pastures Australia factsheet. Available at https://keys.lucidcentral.org/keys/v3/pastures/Html/Leucaena.htm#:∼:text=Leucaena%20flowers%20throughout%20the%20year,seed%20set%20in%20dry%20years [Accessed 30 July 2022]
Dadant R (1953) The chemical control of weeds. Revue Agricole de la Nouvelle Caledonie 4, 31–32.
Dalla Rizza M, Real D, Quesenberry K (2003) Determinacion molecular del sistema reproductivo en Lotononis bainesii y repercusion taxonomica. In ‘Presentación oral y Poster en 2das Jornadas de Bioquímica y Biología molecular’. (Sociedad de bioquimica y Biologia molecular (SByBM), IIBCE: Uruguay)
Dalzell SA, Robertson LM, Lambrides CJ, Brewbaker JL, Greenway D, Dieters M, Max Shelton H (2013) Selection of psyllid-resistant forage varieties from an inter-specific breeding program of Leucaena leucocephala with L. pallida. Tropical Grasslands-Forrajes Tropicales 1, 66–68.
| Selection of psyllid-resistant forage varieties from an inter-specific breeding program of Leucaena leucocephala with L. pallida.Crossref | GoogleScholarGoogle Scholar |
Doležel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols 2, 2233–2244.
| Estimation of nuclear DNA content in plants using flow cytometry.Crossref | GoogleScholarGoogle Scholar |
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049–1051.
| Rapid flow cytometric analysis of the cell cycle in intact plant tissues.Crossref | GoogleScholarGoogle Scholar |
Galcenko N (1961) Cucumber hybrids not requiring bee pollination. Kartofel’ i Ovoshchi 6, 54–56.
Govindarajulu R, Hughes CE, Alexander PJ, Bailey CD (2011a) The complex evolutionary dynamics of ancient and recent polyploidy in Leucaena (Leguminosae; Mimosoideae). American Journal of Botany 98, 2064–2076.
| The complex evolutionary dynamics of ancient and recent polyploidy in Leucaena (Leguminosae; Mimosoideae).Crossref | GoogleScholarGoogle Scholar |
Govindarajulu R, Hughes CE, Bailey CD (2011b) Phylogenetic and population genetic analyses of diploid Leucaena (Leguminosae; Mimosoideae) reveal cryptic species diversity and patterns of divergent allopatric speciation. American Journal of Botany 98, 2049–2063.
| Phylogenetic and population genetic analyses of diploid Leucaena (Leguminosae; Mimosoideae) reveal cryptic species diversity and patterns of divergent allopatric speciation.Crossref | GoogleScholarGoogle Scholar |
Haruthaithanasan M, Haruthaithanasan K, Thanawat A, Phromlert S, Baysangchan A (2010) Potential of Leucaena leucocephala, Eucalyptus camaldulensis, Acacia mangium and Acacia spp. (mangium × auriculaeformis) as plantation crops for energy. In ‘Proceedings of the 48th Kasetsart University annual conference’. (Kasetsart University: Thailand)
Hughes CE (1998) ‘Leucaena. A genetic resource handbook.’ (Oxford University Press: Oxford)
Hughes CE, Sorensson CT, Bray RA, Brewbaker JL (1995) Leucaena germplasm collections, genetic conservation and seed increase. In ‘Leucaena – opportunities and limitations. Proceedings of a workshop’. (Eds HM Shelton, CM Piggin, JL Brewbaker) pp. 66–74. (ACIAR: Bogor, Indonesia)
Idol T (2019) A short review of leucaena as an invasive species in Hawaii. Tropical Grasslands-Forrajes Tropicales 7, 290–294.
| A short review of leucaena as an invasive species in Hawaii.Crossref | GoogleScholarGoogle Scholar |
Ito PJ, Nakasone HY (1968) Compatibility and the inheritance of a seedling character in guava (Psidium guajava). Proceedings Tropical Region, American Society for Horticultural Science 12, 216–221.
Jones RJ, Galgal KK, Castillo AC, Palmer B, Deocareza A, Bolam M (1998) Animal production from five species of Leucaena. In ‘Leucaena – adaptation, quality and farming systems. ACIAR Proceedings 86.’ (Eds HM Shelton, RC Gutteridge, BF Mullen, RA Bray) pp. 247–252. (ACIAR: Hanoi, Vietnam)
Kalve S, Tadege M (2017) A comprehensive technique for artificial hybridization in Chickpea (Cicer arietinum). Plant Methods 13, 52
| A comprehensive technique for artificial hybridization in Chickpea (Cicer arietinum).Crossref | GoogleScholarGoogle Scholar |
Khanna NK, Shukla OP, Gogate MG, Narkhede SL (2019) Leucaena for paper industry in Gujarat, India: case study. Tropical Grasslands-Forrajes Tropicales 7, 200–209.
| Leucaena for paper industry in Gujarat, India: case study.Crossref | GoogleScholarGoogle Scholar |
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theoretical and Applied Genetics 103, 455–461.
| Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica.Crossref | GoogleScholarGoogle Scholar |
Lozanov P (1968) The use of polyploidy in breeding water melons. Gradinarstvo 10, 19–21.
McMillan HE, Liu G, Shelton HM, Dalzell SA, Godwin ID, Gamage H, Sharman C, Lambrides CJ (2019) Sterile leucaena becomes a reality? Tropical Grasslands-Forrajes Tropicales 7, 74–79.
| Sterile leucaena becomes a reality?Crossref | GoogleScholarGoogle Scholar |
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8, 4321–4326.
| Rapid isolation of high molecular weight plant DNA.Crossref | GoogleScholarGoogle Scholar |
Nelson MN, Castello M, Thomson LJ, Cousin A, Yan G, Cowling WA (2006) Microspore culture from interspecific hybrids of Brassica napus and B. carinata produces fertile progeny. In ‘Breeding for success: diversity in action’. (Ed. CF Mercer) (Australiasian Plant Breeding Conference)
Nishiura M (1965) Natural mutation and its utilization in the selection of citrus fruits. In ‘Gamma Field Symposia. Number 4. The use of spontaneous mutations in plant breeding’. 1965, pp. 27–42.
Petty DR (1969) Weed problems of Papua and New Guinea. In ‘Weed control basic to agriculture development. Proceedings 1st Asian-Pacific weed control interchange’. Hawaii. pp. 136–137.
Raman VS, Alikhan WM, Manimekalai G, Bhakthavathsalu CM (1971) A study of the cytomorphology of some banana hybrids. Madras Agricultural Journal 58, 55–62.
Real D, Altier N (2005) Breeding for disease resistance, forage, and seed production in Lotononis bainesii Baker. New Zealand Journal of Agricultural Research 48, 93–100.
| Breeding for disease resistance, forage, and seed production in Lotononis bainesii Baker.Crossref | GoogleScholarGoogle Scholar |
Real D, Dalla Rizza M, Quesenberry KH, Echenique M (2004) Reproductive and molecular evidence for allogamy in Lotononis bainesii Baker. Crop Science 44, 394–400.
Real D, Warden J, Nelson M, Sandral GA, Turner N, Acuna T, Johnson R (Eds) (2006) Preliminary studies of inheritance of autogamy in Lotus corniculatus L. In ‘Groundbreaking Stuff: proceedings of the 13th Australian agronomy conference’. (The Regional Institute Ltd: Perth, Western Australia)
Real D, Dalla Rizza M, Reyno R, Quesenberry KH (2007) Breeding system of the aerial flowers in an amphicarpic clover species: Trifolium polymorphum. Crop Science 47, 1401–1406.
| Breeding system of the aerial flowers in an amphicarpic clover species: Trifolium polymorphum.Crossref | GoogleScholarGoogle Scholar |
Real D, Oldham CM, Nelson MN, Croser J, Castello M, Verbyla A, Pradhan A, Van Burgel A, Méndez P, Correal E, Teakle NL, Revell CK, Ewing MA (2014) Evaluation and breeding of tedera for Mediterranean climates in southern Australia. Crop & Pasture Science 65, 1114–1131.
| Evaluation and breeding of tedera for Mediterranean climates in southern Australia.Crossref | GoogleScholarGoogle Scholar |
Real D, Han Y, Bailey CD, Vasan S, Li C, Castello M, Broughton S, Abair A, Crouch S, Revell C (2019) Strategies to breed sterile leucaena for Western Australia. Tropical Grasslands-Forrajes Tropicales 7, 80–86.
| Strategies to breed sterile leucaena for Western Australia.Crossref | GoogleScholarGoogle Scholar |
Revell C, Moore G, Real D, Crouch S (2019) Environmental adaptation of leucaena in Western Australia – challenges and opportunities. Tropical Grasslands-Forrajes Tropicales 7, 112–119.
| Environmental adaptation of leucaena in Western Australia – challenges and opportunities.Crossref | GoogleScholarGoogle Scholar |
Shelton M, Dalzell S, Tomkins N, Buck S (2021) ‘Leucaena. The productive and sustainable forage legume.’ (Meat & Livestock Australia Limited: Brisbane, Qld, Australia)
Sorensson CT (1988) Pollinating and emasculating techniques for Leucaena species. Leucaena Research Reports 9, 127–130.
Sorensson CT, Brewbaker JL (1994) Interspecific compatibility among 15 Leucaena species (Leguminosae: Mimosoideae) via artificial hybridizations. American Journal of Botany 81, 240–247.
| Interspecific compatibility among 15 Leucaena species (Leguminosae: Mimosoideae) via artificial hybridizations.Crossref | GoogleScholarGoogle Scholar |
Sorensson CT, Sun W (1990) Production of leucaena hybrid seed without emasculation. Leucaena Research Reports 11, 129–132.
Sotelo M (2017) Use of Leucaena on sustainable livestock systems in the tropics: evaluation of agronomy, productivity, green house gasses and soil parameters. In ‘Workshop on leucaena breeding and development’. (International Center for Tropical Agriculture, CIAT: Perth, WA)
Takahashi M, Ripperton JC (1949) ‘Koa Haole (Leucaena glauca). Its establishment, culture and utilisation as a forage crop.’ Hawaii Agricultural Experiment Station, Bulletin No. 100. p. 56. (University of Hawaii)
Tanavat E, Haruthaithanasan M, Puangchit L, Thaiutsa B, Haruthaithanasan K (2011) Nutrient storage in aboveground biomass and nutrient return in fast growing tree species planted for bio-energy. In ‘Proceedings of the 49th Kasetsart University annual conference’. 1–4 February 2011. pp. 616–623. (Kasetsart University: Bangkok, Thailand)