Transcriptome-based discovery of genes and networks related to RSC3Q-mediated resistance to Soybean mosaic virus in soybean
Yuan Yuan A B , Yongqing Yang A , Jinlong Yin A , Yingchao Shen A , Bowen Li A , LiLiqun Wang A and Haijian Zhi A C
A C
A National Center for Soybean Improvement, National Key Laboratory for Crop Genetics and Germplasm Enhancement, Key Laboratory of Biology and Genetic Improvement of Soybean, Ministry of Agriculture, Nanjing Agricultural University, Weigang 1, Nanjing 210095, People’s Republic of China.
B Horticultural Research Institute, Shanghai Academy of Agricultural Sciences, Shanghai 201106, People’s Republic of China.
C Corresponding author. Email: zhj@njau.edu.cn
Crop and Pasture Science 71(12) 987-995 https://doi.org/10.1071/CP20253
Submitted: 18 July 2020 Accepted: 14 October 2020 Published: 7 December 2020
Journal Compilation © CSIRO 2020 Open Access CC BY-NC-ND
Abstract
Soybean mosaic virus (SMV) is a worldwide disease of soybean (Glycine max (L.) Merr.) that can cause serious reduction in yield and seed quality. Soybean cv. Qihuang-1 is an important source of resistance to SMV in China, carrying a resistance gene (RSC3Q) against SMV strain SC3. In order to discover genes and networks regulated by RSC3Q-mediated resistance in Qihuang-1, we analysed transcriptome data of a pair of near-isogenic lines, R (RSC3Q) and S (rSC3Q), from the cross Qihuang-1 × Nannong 1138-2 (rSC3Q), after SC3 inoculation. Many differentially expressed genes (DEGs) were identified in the R and S lines at 6, 20 and 48 h post-inoculation. Based on pathway-enrichment analysis of DEGs, three genes encoding calmodulin-like protein (Glyma03g28650, Glyma19g31395 and Glyma11g33790) with downregulated expression in the S line were identified in the plant–pathogen interaction pathway at 6 h post-inoculation. Analyses by quantitative real-time PCR were performed to verify that these three genes were not beneficial for SMV infection. Our results also revealed a complex plant-hormone signal network in RSC3Q-mediated resistance during the early stage of SMV infection. Expression of jasmonic acid repressor genes (TIFY/JAZ) and abscisic acid-induced genes (PP2C3a) was upregulated in the R line but not the S line. More DEGs related to indole-3-acetic acid were found in the R line than the S line, and no salicylic acid-related DEGs were identified. These results suggest that suppression of jasmonic acid or promotion of abscisic acid is important for RSC3Q-mediated resistance against SC3, and that salicylic acid may not act as a main regulator of RSC3Q-mediated resistance during early stages of SC3 infection. Growth and development were greatly affected through RSC3Q-mediated resistance responses after SC3 infection. Our understanding would be enhanced by identification of factors associated with RSC3Q that help to trigger the resistance response.
Keywords: CML, Glycine max, NILs, plant-hormone signal-transduction pathway, Soybean mosaic virus.
Introduction
Soybean mosaic virus (SMV) is a member of the largest genus of known plant viruses, Potyvirus (Adams et al. 2005). SMV is a worldwide disease of soybean (Glycine max (L.) Merr.) and can cause serious reduction in yield and seed quality. Based on their differential responses on resistant and susceptible soybean lines, SMV isolates have been grouped into seven strains (G1–G7) in the United States (Cho and Goodman 1979; Cho and Goodman1982), five strains (A–E) in Japan (Takahashi et al. 1980; Nakano 1982), and 22 strains in China (Li et al. 2010; Wang et al. 2018). Soybean has a two-layered innate immune system for combatting SMV: pathogen-associated molecular pattern-triggered immunity (PTI), and effector-triggered immunity (ETI) (Jones and Dangl 2006). ETI is induced when a strain-specific avirulent (Avr) protein from the pathogen associates directly or indirectly with a cognate plant resistance (R) protein (Jones and Dangl 2006). Currently, soybean resistance genes to SMV, Rsv1, Rsv3, Rsv4 and Rsv5, are identified as single dominant SMV resistance gene (R) loci in the USA (Yu et al. 1994; Hayes et al. 2000; Jeong et al. 2002; Klepadlo et al. 2017). Resistance to the strains from China is derived from the single dominant SMV Rsc loci, and these have been mapped to chromosomes 2, 13, 14, and 6 in the respective cultivars Kefeng-1, Qihuang-1, Dabaima and RN-9 (Ma et al. 2011; Wang et al. 2011; Zheng et al. 2014; Rui et al. 2017). To date, only the Rsv4 gene has been cloned (Ishibashi et al. 2019). Some downstream signalling components and pathways of Rsv-mediated resistance have been reported; however, there are no reports about Rsc-mediated resistance (Fu et al. 2009; Zhang et al. 2012; Wang et al. 2014).
Plant-hormone signalling pathways also play a crucial role in the process of the plant defence response, involving hormones such as salicylic acid (SA), jasmonic acid (JA) and abscisic acid (ABA). SA, a phenolic compound, is usually required for triggering innate immune responses (i.e. PTI and ETI). Localised resistance responses of plants, activation of programmed cell death, systemic acquired resistance (SAR) and small interfering RNA (siRNA) antiviral machinery are associated with SA and/or SA derivatives (Alamillo et al. 2006; Hunter et al. 2013; Baebler et al. 2014; Shigenaga and Cristiana 2016). JA is a lipid-derived signalling molecule. In the JA signal-transduction process, JASMONATE ZIM-DOMAIN (JAZ) proteins, as a subfamily of TIFY, are key regulators in JA hormonal response (Vanholme et al. 2007). Upon stress perception, plants accumulate the bioactive JA-Ile molecule ((+)-7-iso-JA-Ile), which induces the interaction between F-box CORONATINE INSENSITIVE 1 (COI1) and JAZ (Fonseca et al. 2009; Sheard et al. 2010). The JA-Ile-mediated COI1–JAZ interaction leads to the ubiquitination and subsequent degradation of the JAZ proteins. Several transcription factors including the JA master regulator MYC2 are liberated, which in turn induce JA-specific cellular reprogramming, such as defence responses (Chini et al. 2007; Kazan and Manners 2013). JA reportedly has positive and negative effects on defence against viruses (Alazem et al. 2018); at the early stage of virus infection, JA seems to support plant defence, but it decreases plant resistance at a later stage when it is induced or applied (Pacheco et al. 2012; Garcia-Marcos et al. 2013). ABA is a sesquiterpene compound produced by the cleavage of γ-carotene. When ABA receptors PYR/PYL/RCAR bind accumulated ABA, dephosphorylation of SnRK2s (SNF1-related protein kinase 2 family) mediated by protein phosphatase 2C (PP2C) is prevented. The active SnRK2 kinases phosphorylate and activate downstream transcription factors, and then induce the transcription of ABA‐responsive genes for developmental process and adaptive stress responses (Hauser et al. 2011; Alazem et al. 2018). During plant defence, the role of ABA depends on the stage of virus infection. ABA can resist viruses by mediating stomata closure or increasing callose deposition on plasmodesmata to restrict movement of viruses at early stages of infection. However, ABA can suppress hypersensitive response to decrease the production of reactive oxygen species (ROS) and SA, and weak distal SAR and siRNA systems (Alazem et al. 2018).
Qihuang-1 is an important resistance source for SMV in China. It carries a resistance gene (RSC3Q) to SMV strain SC3, a prevalent SMV strain in Huang, Huai and Chang Jiang Valleys in China (Wang et al. 2003). However, the mechanism underlying SMV resistance in Qihuang-1 is not clear. Next-generation sequencing has become the first choice for researching SMV resistance mechanisms, with a reduction in expense and time needed in sequencing. In this study, we applied transcriptome analysis of a pair of near-isogenic lines (NILs) from the cross Qihuang-1 (resistant to SC3) × Nannong 1138-2 (susceptible to SC3) in order to explore the transcript-accumulation patterns after SMV infection. This enabled us to discover genes and networks in Qihuang-1 that are regulated by RSC3Q-mediated resistance.
Materials and methods
Plant materials and inoculation
Near-isogenic lines (R and S lines) that have similar genetic background were derived from the cross Qihuang-1 × Nannong 1138-2 in a previous study (Zheng et al. 2014). The R line carries a resistance gene to SC3 (RSC3Q), like Qihuang-1, whereas S line carries a susceptible gene (rSC3Q), like Nannong 1138–2.
Qihuang-1, Nannong 1138-2, the R line and the S line were planted in a mixture of soil, perlite and vermiculite (volume ratio 3 : 1 : 1). Plants were grown in a greenhouse with temperatures of 25°C (day) and 20°C (night). Plants were inoculated with the SMV strain SC3 or phosphatic buffer solution (control) when the unifoliolate leaves were fully unfolded (V1 stage). The inoculation method was according to the previous study of Li et al. (2017).
Soybean RNA-Seq data
The RNA-Seq data from the NILs (R line and S line) were obtained previously (Li et al. 2017; Yuan et al. 2020). In brief, leaves were collected as samples from the two lines independently at 0, 6, 20 and 48 h post-inoculation (hpi) with SC3. After extracting total RNA from the samples, libraries were generated from eight samples with Ultra (New England Biolabs, Ipswich, MA, USA), following the manufacturer’s instructions, and samples were sequenced on a HiSeq 2000 (Illumina, San Diego, CA, USA). After removing impure data from the raw sequence (using FASTX-Toolkit version 0.0.13; Gordon and Hannon 2010), clean reads were obtained. The clean reads were aligned to the Williams 82 soybean mRNA reference and genome Glyma.Wm82.a1.v1 (Schmutz et al. 2010) by using SOAP2 version 2.21 with default settings (Li et al. 2009). Differentially expressed genes (DEGs) (|log2(fold-change)| ≥2 and P ≤ 0.01) were obtained by comparison of SMV-infected samples (at 6, 20 and 48 hpi) with 0 hpi in the R line and the S line, using Cufflinks v1.1.0 software (Trapnell et al. 2010). All of the original RNA-sequencing data have been submitted to the Sequence Read Archive database (SRA accession no. PRJNA668549, www.ncbi.nlm.nih.gov/sra; BioSample accession no. SAMN16414956, www.ncbi.nlm.nih.gov/biosample).
Soybean pathway enrichment analysis of the DEGs was performed by using EXPath 2.0 (Zheng et al. 2017). A heatmap graph was drawn by Origin 2019b (www.originlab.com/2019b) based on log2(fold-change) values.
RNA extraction and gene expression analysis by qPCR
Extraction of RNA from leaf tissues of soybean plants and quantitative real-time polymerase chain reaction (qPCR) were performed according to the previous study (Zheng et al. 2014; Luan et al. 2016). Each gene was tested with at least three biological replicates and the experiment was repeated at least twice. Gene IDs and primer sequences are provided in Supplementary Material Table S1 (available at the journal’s website).
Results
Virus content in R and S lines after SMV inoculation
In terms of SMV strain SC3 content in R and S lines after SC3 inoculation (Fig. 1a), there was a continuous decrease in the R line after 48 hpi, and virus could hardly be detected at 5 days post-infection (dpi). The SMV level in S line showed a trend of fluctuation. A small amount of virus was detected in inoculated leaves in the S line, but an abundance of virus in uninoculated distal leaves (the first trifoliate leaves at 7 dpi) (Fig. 1a). This suggested that the S line without RSC3Q also had some resistance to SC3; however, this resistance did not stop the movement of the virus. Some virus moved along the veins and stems to the distal leaves and replicated, which was detected at 7 dpi in the S line (Fig. 1a).
RNA sequencing assembly and assessment of sequencing quality
The eight samples from the R and S lines at four time-points subjected to Illumina sequencing generated 43–67 million clean reads (Table S2), 79.72–92.57% of which were mapped to the soybean reference genome. Around 40 000 genes were found to be expressed in each sample (Table S2).
The reliability of transcriptome analysis was tested via qPCR analysis on eight randomly selected genes. The results showed that seven of the genes had similar expression trends in the transcriptome and qPCR analysis, which indicated that transcriptome data were reliable (Fig. S1).
Identification of DEGs in response to SMV infection
In total, 779, 189 and 84 DEGs were obtained at three time-points (6, 20, and 48 hpi, respectively) in the R line, and 390, 162 and 65 DEGs were discovered at the same three time-points in the S line (Fig. 1b–d). The number of DEGs was greater in the R line than the S line at each time-point. Comparable numbers of DEGs with upregulated and downregulated expression were apparent; in the R line, more were upregulated than downregulated at three time-points, but in the S line, more were downregulated (Fig. 1b–d). Most of the DEGs were mobilised in response to SC3 infection before 20 hpi in both lines, which was likely an important aspect of soybean defence.
Pathway enrichment analyses
Pathway enrichment analyses by EXPath 2.0 showed that, whether in the R or S line, DEGs involved in metabolic-related pathways represented a large proportion at the three time-points (6, 20 and 48 hpi; Fig. S2). Among the different pathways, plant–pathogen interaction and plant-hormone signal-transduction pathways play important roles in plant defence.
Genes encoding calmodulin-like (CML) protein involved in the response to SMV in soybean
Three CML genes (Glyma03g28650, Glyma19g31395 and Glyma11g33790) with downregulated expression were found in the plant–pathogen interaction pathway in the S line only. CMLs are the members of the Ca2+ sensors, which interact with Ca2+ and regulate the function of diverse target proteins by direct binding or through phosphorylation (Aldon et al. 2018). CMLs are reported to be important regulators of plant defence against pathogens (Xu et al. 2017; Zhu et al. 2017; Lu et al. 2018). Therefore, we speculated that the three identified CML genes in the S line at 6 hpi probably positively regulated the early resistance response of soybean to SC3. Further, qPCR analysis was performed on the parent plants of R and S lines (Qihuang-1 and Nannong 1138-2). As shown in Fig. 2, expression of the three CML genes increased in Qihuang-1 but decreased significantly in Nannong 1138-2 at 6 hpi, indicating that inhibition of expression of the three CML genes was very likely to promote SMV infection.
Plant-hormone effects on soybean defence against SMV
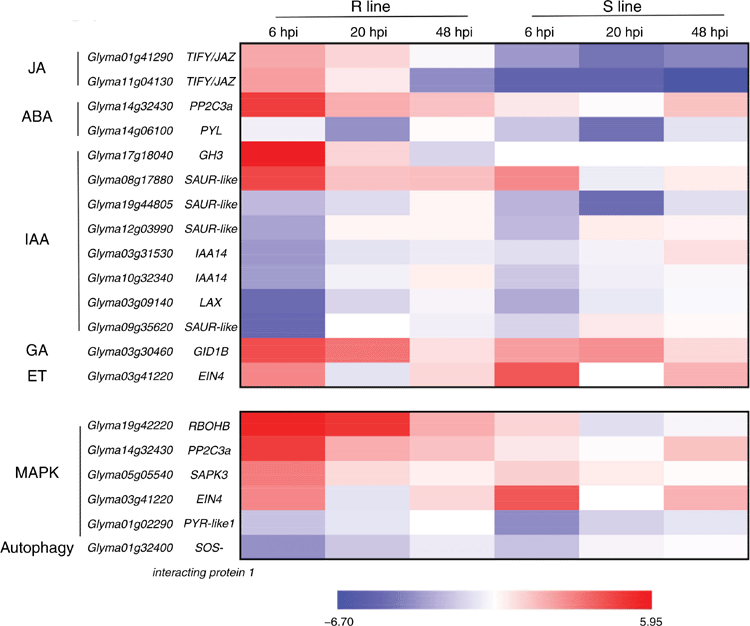
According to the pathway enrichment analyses, 14 DEGs were found in the plant-hormone signal-transduction pathway (Fig. 3). Glyma01g41290 and Glyma11g04130 encoding TIFY/JAZ proteins had downregulated expression in the S line, and upregulated in R line (Fig. 3), indicating that induction of Glyma01g41290 and Glyma11g04130 expression is advantageous for soybean resistance during early stages of SC3 infection. TIFY/JAZ proteins are a key negative regulator in JA hormonal response (Fonseca et al. 2009). This means that JA probably plays a role in response to SC3 infection, and repression of JA probably enhances soybean resistance to SC3 during early stages of SC3 infection.
Two DEGs (Glyma14g06100 and Glyma14g32430) were related to the ABA-signalling pathway. Glyma14g06100 encodes a PYR1-like protein (PYL), which is an ABA receptor and positively responds to ABA regulation (Hauser et al. 2011). Its expression was significantly downregulated in the S line. Glyma14g32430, as an ABA-induced PP2C3a (encoding a subset of the Type 2C protein phosphatase), is reported to be a key positive regulator of Rsv3-mediated extreme resistance against SMV strain G5H in soybean (Seo et al. 2014). In our study, PP2C3a had significantly upregulated expression at 6 hpi in the R line, but it had almost had no expression in the S line (Fig. 3). These results suggest that ABA was involved in plant response after SC3 infection and probably promoted RSC3-mediated resistance.
The auxin (IAA)-related DEGs with different expression patterns were mostly in the plant-hormone signal-transduction pathway (Fig. 3). This shows that the expression of DEGs was greatly affected by SMV infection at early time-points in the R line, but not in the S line. We speculate that growth and development were greatly affected through RSC3Q-mediated resistance responses during the early stages of SC3 infection. There was one DEG in each of the ethylene (ET) and gibberellin (GA) signalling pathways, encoding ET receptor EIN4 (ETHYLENE INSENSITIVE4) and GA receptor GID1B (GIBBERELLIN INSENSTIVE DWARF1), respectively (Fig. 3). This indicated that ET and GA participated in response to SC3 inoculation.
Other defence pathways possibly effecting soybean against SMV
Mitogen-activated protein kinase (MAPK) signalling pathway plays a critical role in plant immunity. Five DEGs were identified in the MAPK signalling pathway at 6 hpi in the R and S lines (Fig. 3). We speculated that these DEGs were probably regulated by RSC3Q-mediated resistance. Three of them (Glyma05g05540, Glyma14g32430 (PP2C3a) and Glyma19g42220) had significantly upregulated expression at 6 hpi in the R line, but they had almost no expression in the S line (Fig. 3). We speculate that these three DEGs maybe more important in RSC3Q-mediated resistance. Autophagy responses have also been reported in plant defences (Alazem et al. 2018). Only one DEG was involved in the autophagy pathway (Glyma01g32400), which had downregulated expression in both R and S lines.
Discussion
Qihuang-1 (RSC3Q) is an important resistance source for SMV strain SC3 in China, but the published literature includes little information on RSC3Q-regulated resistance mechanisms or pathways. Our findings provide some RSCQ-mediated, resistance-related genes and a general understanding of transcriptional activation of defence-related genes. Immediately downstream of the initial elicitor–receptor recognition, the activation of ion fluxes (including Ca2+) and the production of H2O2 are the initial responses detected in plant cells, which occur before transcriptional activation of defence-related genes (Ramos et al. 2008). In response to the stimuli, cytosolic free Ca2+ rises and binds to a plethora of sensors, including CML, which in turn activate subsequent reactions of plant immunity (Aldon et al. 2018). In our study, three CML genes (Glyma03g28650, Glyma19g31395 and Glyma11g33790) were enriched in the plant–pathogen interaction pathway in the S line only. Their expression was downregulated in the S line and the susceptible parent Nannong 1138-2 at 6 and 24 hpi, but was upregulated in resistant parent Qihuang-1 (Fig. 2). We consider that these three CML genes are beneficial for RSC3Q-mediated resistance. The functions of CML genes are diverse. CML8 and CML9 can promote Arabidopsis resistance against the phytopathogenic bacteria Pseudomonas syringae pv. tomato (strain DC3000) (Zhu et al. 2017). Disease-resistance pathways involving CML8 and CML9 are different. CML8 plays a role in SA-dependent processes, probably by modulating the effect of bacterial effectors. CML9 contributes to PTI (via a flagellin-dependent pathway) but also to SA-dependent processes (Zhu et al. 2017). CML9 as a negative regulator is involved in drought and salt stress. Arabidopsis CML42 and CML37 are also involved in plant defence and drought response (Vadassery et al. 2012a, 2012b; Scholz et al. 2014). Therefore, the functions of the three CML genes in this study can be investigated jointly from the aspects of plant defence and abiotic stress in subsequent experiments. These three genes might have different functions.
Some DEGs identified had involvement in plant-hormone signal transductions (Fig. 3). Plants defend against pathogen attack by modulating plant-hormone signalling pathways, such as those involving SA, JA, ET and ABA (Robert-Seilaniantz et al. 2011; Pieterse et al. 2012; Vos et al. 2013).
The role of JA in plant defence against viruses is controversial (Alazem and Lin 2015). JA has been known for its positive roles in a few compatible interactions; for example, the silencing of JA receptor gene COI1 can enhance virulence of Potato virus X and Potato virus Y in Nicotiana benthamiana (Garcia-Marcos et al. 2013). However, for incompatible interactions between Tobacco mosaic virus (TMV) and N. benthamiana, JA had negative roles. Oka et al. (2013) found that N-mediated resistance to TMV was enhanced in NtCOI1-RNAi line. There was an incompatible interaction between the R line and SC3, and a compatible interaction between the S line and SC3. Two DEGs (Glyma01g41290 and Glyma11g04130) encoding TIFY/JAZ, a negative regulator in JA pathway, had significantly downregulated expression in the R line and upregulated in S line following SC3 infection (Fig. 3). Therefore, we speculate that JA probably had a positive role in RSC3Q-mediated resistance during the early stages of infection, which was similar to previously reported findings as mentioned above (Oka et al. 2013; Alazem et al. 2018).
Xun et al. (2019) found that ABA might be related to R-mediated resistance to SMV in soybean. Overexpression of a candidate R gene against SMV in soybean increased ABA accumulation. In the present study, we found that ABA-related genes PYL (Glyma14g06100) and PP2C3a (Glyma14g32430) were differentially expressed. As shown in Fig. 3, PP2C3a was rapidly induced in the R line at 6 hpi, but not in the S line. PP2C3a, encoding a Type-2C protein phosphatase, was predicted to participate not only in the ABA signalling pathway, but also in the MAPK signalling pathway (Fig. 3). When plants are invaded by pathogens, callose deposition increases on plasmodesmata and restricts cell-to-cell movement of viruses (Nakashima et al. 2003). In this process, the MAPK signalling pathway plays a positive regulatory role. Inhibition of the MAPK signalling pathway will decrease callose deposition and reduce resistance of plants (Xu et al. 2019). It has been verified that PP2C3a is specifically involved in Rsv3-mediated extreme resistance in an ABA-dependent manner against SMV (strain G5H) in soybean. It controls the rapid accumulation of callose at the points of G5H infection to stop virus spread (Seo et al. 2014; Alazem et al. 2019). All of the above evidence indicates that ABA plays an important role in R-mediated resistance against SMV and that PP2C3a might be a key gene connecting the MAPK and ABA signalling pathways. The role of PP2C3a in two pathways needs further verification.
In plant defence against viruses, SA signalling constitutes the major defensive pathway, and is tightly connected to the majority of R genes (Alazem and Lin 2015). Although SMV multiplication was reported to be inhibited by SA treatment (Zhao et al. 2018), no SA-related DEGs were detected in our study. Perhaps SA did not act as a main regulator of RSC3Q-mediated resistance during the early stages of SC3 infection.
In addition to PP2C3a, Glyma19g42220 (encoding respiratory burst oxidase homologue B, RBOHB) was significantly upregulated at 6 and 20 hpi in the MAPK signalling pathway in the R line. RBOH is also called NADPH oxidase (NOX), a key enzyme of ROS generation, and playing vital roles in various biological processes including plant immunity. The important role of NOX/RBOH in plant immunity has been well reported, especially in Arabidopsis, tobacco and rice (Hu et al. 2020). However, there is little research into the NOX/RBOH–MAPK pathway in plant immunity. In N. benthamiana, NbRBOHB is an important player in both the PTI ROS burst and the ETI ROS burst. It is noteworthy that MAPK is responsible for the ETI ROS burst by transactivation of NbRbohB, but not for the PTI ROS burst (Yoshioka et al.2016). We assume that Glyma19 g42220 (RBOHB) has a similar function to NbRBOHB, which participates in the ETI ROS burst (the R-mediated resistance) through the MAPK cascade-signalling pathway.
In summary, CML, JA, ABA and MAPK, but not SA, are involved in the plant defence response during the early stages of SC3 infection. Suppression of JA or induction of ABA probably increased RSC3Q-mediated SMV resistance. Soybean resistance against SMV was mediated by complex gene families at different loci, and the resistance was specified via both SMV strain and soybean variety. Our understanding of RSC3Q-mediated resistance against SC3 would be enhanced by identification of factors associated with RSC3Q that help to trigger the resistance response.
Conflicts of interest
We declare no conflict of interest.
Author contributions
HZ, Y Yuan and Y Yang conceived and designed the experiments; Y Yuan wrote the article and performed major parts of the experiments; JL modified the language and provided technical support for RNA-Seq; LW performed parts of RNA-Seq experiment; YS and BL performed parts of plants cultivation and qRT experiment; HZ re-edited the article.
Acknowledgements
We thank Keshun Yu and Limei Wang for the help of this manuscript. This work was supported by the Fund of Transgenic Breeding for Soybean Resistance to Soybean mosaic virus (2016ZX08004-004), the Fundamental Research Funds for the Central Universities (KYT201801) and Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT_17R55), the National Natural Science Foundation of China (31571690), the National Soybean Industrial Technology System of China (CARS-004), Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP), and the National Key R&D Program of China (2017YFD0101501).
References
Adams MJ, Antoniw JF, Fauquet CM (2005) Molecular criteria for genus and species discrimination within the family Potyviridae. Archives of Virology 150, 459–479.| Molecular criteria for genus and species discrimination within the family Potyviridae.Crossref | GoogleScholarGoogle Scholar | 15592889PubMed |
Alamillo JM, Saénz P, García JA (2006) Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. The Plant Journal 48, 217–227.
| Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco.Crossref | GoogleScholarGoogle Scholar | 17018032PubMed |
Alazem M, Lin NS (2015) Roles of plant hormones in the regulation of host–virus interactions. Molecular Plant Pathology 16, 529–540.
| Roles of plant hormones in the regulation of host–virus interactions.Crossref | GoogleScholarGoogle Scholar | 25220680PubMed |
Alazem M, Tseng KC, Chang WC, Seo JK, Kim KH (2018) Elements involved in the Rsv3-mediated extreme resistance against an avirulent strain of soybean mosaic virus. Viruses 10, 581
| Elements involved in the Rsv3-mediated extreme resistance against an avirulent strain of soybean mosaic virus.Crossref | GoogleScholarGoogle Scholar |
Alazem M, Widyasari K, Kim KH (2019) An avirulent strain of soybean mosaic virus reverses the defensive effect of abscisic acid in a susceptible soybean cultivar. Viruses 11, 879
| An avirulent strain of soybean mosaic virus reverses the defensive effect of abscisic acid in a susceptible soybean cultivar.Crossref | GoogleScholarGoogle Scholar |
Aldon D, Mbengue M, Mazars C, Galaud JP (2018) Calcium signalling in plant biotic interactions. International Journal of Molecular Sciences 19, 665
| Calcium signalling in plant biotic interactions.Crossref | GoogleScholarGoogle Scholar |
Baebler Š, Witek K, Petek M, Stare K, Tušek-Žnidarič M, Pompe-Novak M, Renaut J, Szajko K, Strzelczyk-Żyta D, Marczewski W, Morgiewicz K, Gruden K, Hennig J (2014) Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against Potato virus Y infection in potato. Journal of Experimental Botany 65, 1095–1109.
| Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against Potato virus Y infection in potato.Crossref | GoogleScholarGoogle Scholar | 24420577PubMed |
Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garciacasado G, Lopezvidriero L, Lozano FM, Ponce MR, Micol JL, Solano R (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671.
| The JAZ family of repressors is the missing link in jasmonate signalling.Crossref | GoogleScholarGoogle Scholar | 17637675PubMed |
Cho EK, Goodman RM (1979) Strains of soybean mosaic virus: classification based on virulence in resistant soybean cultivars. Phytopathology 69, 467–470.
| Strains of soybean mosaic virus: classification based on virulence in resistant soybean cultivars.Crossref | GoogleScholarGoogle Scholar |
Cho EK, Goodman RM (1982) Evaluation of resistance in soybeans to soybean mosaic virus strains. Crop Science 22, 1133–1136.
| Evaluation of resistance in soybeans to soybean mosaic virus strains.Crossref | GoogleScholarGoogle Scholar |
Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R (2009) (+)-7-iso-Jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nature Chemical Biology 5, 344–350.
| (+)-7-iso-Jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate.Crossref | GoogleScholarGoogle Scholar | 19349968PubMed |
Fu DQ, Ghabrial S, Kachroo A (2009) GmRAR1 and GmSGT1 are required for basal, R gene-mediated and systemic acquired resistance in soybean. Molecular Plant–Microbe Interactions 22, 86–95.
| GmRAR1 and GmSGT1 are required for basal, R gene-mediated and systemic acquired resistance in soybean.Crossref | GoogleScholarGoogle Scholar | 19061405PubMed |
Garcia-Marcos A, Pacheco R, Manzano A, Aguilar E, Tenllado F (2013) Oxylipin biosynthesis genes positively regulate programmed cell death during compatible infections with the synergistic pair Potato virus X–Potato virus Y and Tomato spotted wilt virus. Journal of Virology 87, 5769–5783.
| Oxylipin biosynthesis genes positively regulate programmed cell death during compatible infections with the synergistic pair Potato virus X–Potato virus Y and Tomato spotted wilt virus.Crossref | GoogleScholarGoogle Scholar | 23487466PubMed |
Gordon A, Hannon GJ (2010) FASTX-toolkit. FASTQ/A short-reads preprocessing tools. Hannon Laboratory, CSHL, NY, USA. Available at: http://hannonlab.cshl.edu/fastx_toolkit/[Verified 4 November 2020]
Hauser F, Waadt R, Schroeder JI (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Current Biology 21, R346–R355.
| Evolution of abscisic acid synthesis and signaling mechanisms.Crossref | GoogleScholarGoogle Scholar | 21549957PubMed |
Hayes AJ, Ma G, Buss GR, Saghai Maroof MA (2000) Molecular marker mapping of Rsv4, a gene conferring resistance to all known strains of soybean mosaic virus. Crop Science 40, 1434–1437.
| Molecular marker mapping of Rsv4, a gene conferring resistance to all known strains of soybean mosaic virus.Crossref | GoogleScholarGoogle Scholar |
Hu CH, Wang PQ, Zhang PP, Nie XM, Li BB, Tai L, Liu WT, Li WQ, Chen KM (2020) NADPH oxidases: the vital performers and center hubs during plant growth and signaling. Cells 9, 437
| NADPH oxidases: the vital performers and center hubs during plant growth and signaling.Crossref | GoogleScholarGoogle Scholar |
Hunter LJR, Westwood JH, Heath G, Macaulay K, Smith AG, MacFarlane SA, Palukaitis P, Carr JP (2013) Regulation of RNA-dependent RNA polymerase 1 and isochorismate synthase gene expression in Arabidopsis. PLoS One 8, e66530
| Regulation of RNA-dependent RNA polymerase 1 and isochorismate synthase gene expression in Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
Ishibashi K, Saruta M, Shimizu T, Shu M, Anai T, Komatsu K, Yamada N, Katayose Y, Ishikawa M, Ishimoto M, Kaga A (2019) Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nature Communications 10, 4033
| Soybean antiviral immunity conferred by dsRNase targets the viral replication complex.Crossref | GoogleScholarGoogle Scholar | 31562302PubMed |
Jeong SC, Kristipati S, Hayes AJ, Maughanb PJ, Noffsingerc SL, Gunduza I, Bussa GR, Saghai Maroof MA (2002) Genetic and sequence analysis of markers tightly linked to the Soybean mosaic virus resistance gene, Rsv3. Crop Science 42, 265–270.
Jones JD, Dangl JL (2006) The plant immune system Nature 444, 323–329.
Kazan K, Manners JM (2013) MYC2: the master in action. Molecular Plant 6, 686–703.
| MYC2: the master in action.Crossref | GoogleScholarGoogle Scholar | 23142764PubMed |
Klepadlo M, Chen P, Shi A, Maughanb PJ, Noffsingerc SL, Gunduza I, Bussa GR, Saghai Maroof MA (2017) Single nucleotide polymorphism markers for rapid detection of the Rsv4 locus for soybean mosaic virus resistance in diverse germplasm. Molecular Breeding 37, 10
| Single nucleotide polymorphism markers for rapid detection of the Rsv4 locus for soybean mosaic virus resistance in diverse germplasm.Crossref | GoogleScholarGoogle Scholar |
Li C, Karthikeyan A, Yuan Y, Yin J, Ren R, Yang Y, Zhi H (2017) Identification of candidate genes for resistance to Soybean mosaic virus strain SC3 by using fine mapping and transcriptome analyses. Crop and Pasture Science 68, 156–166.
| Identification of candidate genes for resistance to Soybean mosaic virus strain SC3 by using fine mapping and transcriptome analyses.Crossref | GoogleScholarGoogle Scholar |
Li K, Yang QH, Zhi HJ, Gai JY (2010) Identification and distribution of Soybean mosaic virus strains in southern China. Plant Disease 94, 351–357.
| Identification and distribution of Soybean mosaic virus strains in southern China.Crossref | GoogleScholarGoogle Scholar | 30754253PubMed |
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967.
| SOAP2: an improved ultrafast tool for short read alignment.Crossref | GoogleScholarGoogle Scholar | 19497933PubMed |
Lu Y, Truman W, Liu X, Bethke G, Zhou M, Myers CL, Katagiri F, Glazebrook J (2018) Different modes of negative regulation of plant immunity by calmodulin-related genes. Plant Physiology 176, 3046–3061.
| Different modes of negative regulation of plant immunity by calmodulin-related genes.Crossref | GoogleScholarGoogle Scholar | 29449432PubMed |
Luan H, Shine MB, Cui X, Chen X, Ma N, Kachroo P, Zhi H, Kachroo A (2016) The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiology 172, 221–234.
| The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis.Crossref | GoogleScholarGoogle Scholar | 27356973PubMed |
Ma Y, Wang DG, Li HC, Zheng GJ, Yang YQ, Li HW, Zhi HJ (2011) Fine mapping of the RSC14Q locus for resistance to soybean mosaic virus. Euphytica 181, 127–135.
| Fine mapping of the RSC14Q locus for resistance to soybean mosaic virus.Crossref | GoogleScholarGoogle Scholar |
Nakano M (1982) Further study on strains of soybean mosaic virus in Kyushu. Proceedings of the Association for Plant Protection of Kyushu 28, 24–25.
| Further study on strains of soybean mosaic virus in Kyushu.Crossref | GoogleScholarGoogle Scholar |
Nakashima J, Laosinchai W, Cui X, Brown RM (2003) New insight into the mechanism of cellulose and callose biosynthesis: proteases may regulate callose biosynthesis upon wounding. Cellulose 10, 369–389.
| New insight into the mechanism of cellulose and callose biosynthesis: proteases may regulate callose biosynthesis upon wounding.Crossref | GoogleScholarGoogle Scholar |
Oka K, Kobayashi M, Mitsuhara I, Seo S (2013) Jasmonic acid negatively regulates resistance to Tobacco mosaic virus in tobacco. Plant and Cell Physiology 54, 1999–2010.
| Jasmonic acid negatively regulates resistance to Tobacco mosaic virus in tobacco.Crossref | GoogleScholarGoogle Scholar | 24071744PubMed |
Pacheco R, Garcia Marcos A, Manzano A, De Lacoba MG, Camanes G, Garcia Agustin P, Diazruiz JR, Tenllado F (2012) Comparative analysis of transcriptomic and hormonal responses to compatible and incompatible plant–virus interactions that lead to cell death. Molecular Plant-Microbe Interactions 25, 709–723.
| Comparative analysis of transcriptomic and hormonal responses to compatible and incompatible plant–virus interactions that lead to cell death.Crossref | GoogleScholarGoogle Scholar | 22273391PubMed |
Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology 28, 489–521.
| Hormonal modulation of plant immunity.Crossref | GoogleScholarGoogle Scholar |
Ramos AC, Façanha AR, Feijó JA (2008) Proton (H+) flux signature for the presymbiotic development of the arbuscular mycorrhizal fungi. New Phytologist 178, 177–188.
| Proton (H+) flux signature for the presymbiotic development of the arbuscular mycorrhizal fungi.Crossref | GoogleScholarGoogle Scholar | 18208473PubMed |
Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just JASMONATE–SALICYLATE antagonism. Annual Review of Phytopathology 49, 317–343.
| Hormone crosstalk in plant disease and defense: more than just JASMONATE–SALICYLATE antagonism.Crossref | GoogleScholarGoogle Scholar | 21663438PubMed |
Rui R, Liu S, Karthikeyan A, Wang T, Niu H, Yin J, Yang Y, Wang L, Yang Q, Zhi H, Li K (2017) Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to Soybean mosaic virus. Theoretical and Applied Genetics 130, 2395–2410.
| Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to Soybean mosaic virus.Crossref | GoogleScholarGoogle Scholar | 28825113PubMed |
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht JE, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the paleopolyploid soybean. Nature 463, 178–183.
| Genome sequence of the paleopolyploid soybean.Crossref | GoogleScholarGoogle Scholar | 20075913PubMed |
Scholz SS, Vadassery J, Heyer M, Reichelt M, Bender KW, Snedden WA, Boland W, Mith€ofer A (2014) Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory. Molecular Plant 7, 1712–26.
| Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory.Crossref | GoogleScholarGoogle Scholar | 25267731PubMed |
Seo JK, Kwon SJ, Cho WK, Choi HS, Kim KH (2014) Type 2C protein phosphatase is a key regulator of antiviral extreme resistance limiting virus spread. Scientific Report S4, 5905 https://doi.org/10.1038/srep05905
Sheard LB, Tan X, Mao H, Withers J, Bennissan G, Hinds TR, Kobayashi Y, Hsu F, Sharon M, Browse J, He SY, Rizo J, Howe GA, Zheng N (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405.
| Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor.Crossref | GoogleScholarGoogle Scholar | 20927106PubMed |
Shigenaga AM, Cristiana TA (2016) No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Seminars in Cell & Developmental Biology 56, 174–189.
| No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens.Crossref | GoogleScholarGoogle Scholar |
Takahashi K, Tanaka T, Iida W, Tsuda Y (1980) Studies on virus diseases and causal viruses of soybean in Japan. Bulletin of the Tohoku National Agricultural Experiment Station 62, 1–130.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515.
| Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation.Crossref | GoogleScholarGoogle Scholar | 20436464PubMed |
Vadassery J, Reichelt M, Hause B, Gershenzon J, Boland W, Mithofer A (2012a) CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiology 159, 1159–1175.
| CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 22570470PubMed |
Vadassery J, Scholz SS, Mith€ofer A (2012b) Multiple calmodulin-like proteins in Arabidopsis are induced by insect-derived (Spodoptera littoralis) oral secretion. Plant Signaling & Behavior 7, 1277–1280.
| Multiple calmodulin-like proteins in Arabidopsis are induced by insect-derived (Spodoptera littoralis) oral secretion.Crossref | GoogleScholarGoogle Scholar |
Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G (2007) The tify family previously known as ZIM. Trends in Plant Science 12, 239–244.
| The tify family previously known as ZIM.Crossref | GoogleScholarGoogle Scholar | 17499004PubMed |
Vos IA, Pieterse CMJ, Van Wees SCM (2013) Costs and benefits of hormone-regulated plant defences. Plant Pathology 62, 43–55.
| Costs and benefits of hormone-regulated plant defences.Crossref | GoogleScholarGoogle Scholar |
Wang D, Li K, Zhi H (2018) Progresses of resistance on soybean mosaic virus in soybean. Zhongguo Nong Ye Ke Xue 51, 3040–3059.
Wang D, Ma Y, Yang Y, Liu N, Li C, Song Y, Zhi H (2011) Fine mapping and analyses of RSC8 resistance candidate genes to soybean mosaic virus in soybean. Theoretical and Applied Genetics 122, 555–565.
| Fine mapping and analyses of RSC8 resistance candidate genes to soybean mosaic virus in soybean.Crossref | GoogleScholarGoogle Scholar | 20981404PubMed |
Wang J, Shine MB, Gao QM, Navarre D, Jiang W, Liu C, Chen Q, Hu G, Kachroo A (2014) Enhanced disease susceptibility1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiology 165, 1269–1284.
| Enhanced disease susceptibility1 mediates pathogen resistance and virulence function of a bacterial effector in soybean.Crossref | GoogleScholarGoogle Scholar | 24872380PubMed |
Wang X, Gai J, Pu Z (2003) Classification and distribution of strain groups of soybean mosaic virus in middle and lower Huang-Huai and Changjiang valleys. Dadou Kexue 22, 102–107.
Xu B, Cheval C, Laohavisit A, Hocking B, Chiasson D, Olsson TSG, Shirasu K, Faulkner C, Gilliham M (2017) A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytologist 215, 77–84.
| A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses.Crossref | GoogleScholarGoogle Scholar | 28513846PubMed |
Xu LW, Wu XZ, Jia ML, Li TJ, Wen F (2019) Research advances on MAPK cascade and their roles in plant disease resistance. Acta Laser Biology Sinica 6, 488–495.
Xun H, Yang X, He H, Wang M, Guo P, Wang Y, Pang J, Dong Y, Feng X, Wang S, Liu B (2019) Over-expression of GmKR3, a TIR–NBS–LRR type R gene, confers resistance to multiple viruses in soybean. Plant Molecular Biology 99, 95–111.
| Over-expression of GmKR3, a TIR–NBS–LRR type R gene, confers resistance to multiple viruses in soybean.Crossref | GoogleScholarGoogle Scholar | 30535849PubMed |
Yoshioka H, Adachi H, Nakano T, Miyagawa N, Asai S, Ishihama N, Yoshioka M (2016) Hierarchical regulation of NADPH oxidase by protein kinases in plant immunity. Physiological and Molecular Plant Pathology 95, 20–26.
| Hierarchical regulation of NADPH oxidase by protein kinases in plant immunity.Crossref | GoogleScholarGoogle Scholar |
Yu YG, Maroof MAS, Buss GR, Maughan PJ, Tolin SA (1994) RFLP and microsatellite mapping of a gene for soybean mosaic virus resistance. Phytopathology 84, 60–64.
| RFLP and microsatellite mapping of a gene for soybean mosaic virus resistance.Crossref | GoogleScholarGoogle Scholar |
Yuan Y, Yang Y, Shen Y, Yu K, Wang L, Ren R, Yin J, Zhi H (2020) Mapping and functional analysis of candidate genes involved in resistance to soybean (Glycine max) mosaic virus strain SC3. Plant Breeding 139, 618–625.
| Mapping and functional analysis of candidate genes involved in resistance to soybean (Glycine max) mosaic virus strain SC3.Crossref | GoogleScholarGoogle Scholar |
Zhang C, Grosics S, Whitham SA, Hill JH (2012) The requirement of multiple defense genes in soybean Rsv1-mediated extreme resistance to Soybean mosaic virus. Molecular Plant–Microbe Interactions 25, 1307–1313.
| The requirement of multiple defense genes in soybean Rsv1-mediated extreme resistance to Soybean mosaic virus.Crossref | GoogleScholarGoogle Scholar | 22712511PubMed |
Zhao Q, Li H, Sun H, Li A, Liu S, Yu R, Cui X, Zhang D, Wuriyanghan H (2018) Salicylic acid and broad spectrum of NBS-LRR family genes are involved in SMV–soybean interactions. Plant Physiology and Biochemistry 123, 132–140.
| Salicylic acid and broad spectrum of NBS-LRR family genes are involved in SMV–soybean interactions.Crossref | GoogleScholarGoogle Scholar | 29232653PubMed |
Zheng GJ, Yang YQ, Ma Y, Ma Y, Yang X, Chen S, Ren R, Wang D, Yang Z, Jian H (2014) Fine mapping and candidate gene analysis of resistance gene RSC3Q to Soybean mosaic virus in Qihuang 1. Journal of Integrative Agriculture 13, 2608–2615.
| Fine mapping and candidate gene analysis of resistance gene RSC3Q to Soybean mosaic virus in Qihuang 1.Crossref | GoogleScholarGoogle Scholar |
Zheng HQ, Wu NY, Chow CN, Tseng KC, Chien CH, Hung YC, Li GZ, Chang WC (2017) EXPath tool: a system for comprehensively analyzing regulatory pathways and coexpression networks from high-throughput transcriptome data. DNA Research 24, 371–375.
| EXPath tool: a system for comprehensively analyzing regulatory pathways and coexpression networks from high-throughput transcriptome data.Crossref | GoogleScholarGoogle Scholar | 28338930PubMed |
Zhu X, Perez M, Aldon D, Galaud JP (2017) Respective contribution of CML8 and CML9, two arabidopsis calmodulin-like proteins, to plant stress responses. Plant Signaling & Behavior 12, e1322246
| Respective contribution of CML8 and CML9, two arabidopsis calmodulin-like proteins, to plant stress responses.Crossref | GoogleScholarGoogle Scholar |