Predictive characterisation identifies global sources of acyanogenic germplasm of a key forage species
Rosa María García Sánchez A B F , Mauricio Parra-Quijano C , Stephanie Greene D and José María Iriondo E
A B F , Mauricio Parra-Quijano C , Stephanie Greene D and José María Iriondo E
A Escuela Internacional de Doctorado, Universidad Rey Juan Carlos, Calle Tulipán s/n, 28933 Móstoles, Madrid, Spain.
B Centro Nacional de Recursos Fitogenéticos, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria, Autovía A-2 Km 36, 28800 Alcalá de Henares, Madrid, Spain.
C Facultad de Ciencias Agrarias, Universidad Nacional de Colombia, Sede Bogotá, Ciudad Universitaria, A.A. 14490, Bogotá D.C., Colombia.
D National Laboratory for Genetic Resources Preservation, USDA ARS, Fort Collins, CO, USA.
E Área de Biodiversidad y Conservación, Universidad Rey Juan Carlos, Calle Tulipán s/n, 28933 Móstoles, Madrid, Spain.
F Corresponding author. Email: rosamaria.garcia@inia.es
Crop and Pasture Science 70(6) 546-554 https://doi.org/10.1071/CP18346
Submitted: 19 July 2018 Accepted: 23 April 2019 Published: 19 June 2019
Journal Compilation © CSIRO 2019 Open Access CC BY-NC-ND
Abstract
Forage breeding is essential for animal production, and its effectiveness depends on available genetic diversity. However, breeding is challenged when there is limited evaluation of genebank accessions. Predictive characterisation based on ecogeographic information is a promising approach to address the urgent need to expedite evaluation of target traits in existing collections of forage genetic resources. Using white clover (Trifolium repens L.) as an example, we applied predictive characterisation to model the expression of cyanogenesis, an important process related to the generation of anti-quality compounds. Data on genebank accessions and other population occurrences were divided into two subsets, one including accessions that had been evaluated for this trait, and the other with those that had not. The occurrence sites of the records with the best geo-referencing quality were characterised ecogeographically. The cyanogenesis trait was predicted using the calibration method, in which some selected ecogeographic variables were used as independent variables. Thus, we identified 470 populations with high probability of being acyanogenic. A small sample of populations (18 accessions) was evaluated to ratify the usefulness of this approach. Seventeen of the evaluated accessions showed a complete acyanogenic response and one showed 95% acyanogenic plants. Our study also expanded the areas previously rated as highly acyanogenic. In conclusion, our results contribute in a predictive way and with minimum cost to increase the knowledge of wild populations and genebank accessions in relation to a target trait. This facilitation in the generation of evaluation data may encourage greater investment in forage plant breeding and boost germplasm utilisation.
Introduction
In 2016, human food requirements and eating habits necessitated the transport of over 13 Mt of forage products to feed livestock worldwide (www.fao.org/faostat/en/#data/TP). Because of increasing global demand for meat, an essential goal of forage breeders is the improved performance of animals consuming forage. Main forage-breeding targets include maximising yield of forage that has optimum nutritional value, is free of anti-quality compounds, has high persistence in the field, and can be produced in an environmentally friendly manner (Brummer et al. 2009).
The perennial life form of many forage species requires that evaluations extend over multiple years to assess certain traits adequately. This, along with the many different species that can be cultivated as forage crops, has contributed to the slower pace of genetic gain in forage crops and to smaller breeding efforts than in grain crops (Brummer et al. 2009). Additionally, only a small amount of genetic diversity present in natural populations is preserved in gene banks, and just a minority of those accessions has been evaluated for target traits sought by plant breeders, which further impedes the progress of forage-plant breeding. Therefore, there is an urgent need to expedite the evaluation of target traits in existing forage genetic resources.
Predictive characterisation is a recently developed concept that uses ecogeographic data from collection sites and phenotypes to search for specific traits in a larger set of germplasm (Thormann et al. 2016). Complementing the Focussed Identification of Germplasm Strategy (FIGS, Mackay and Street 2004), this concept is applied not only to landrace seed accessions conserved in gene banks but also to accessions and wild populations of crop wild relatives (Thormann et al. 2016). The search for interesting traits can be carried out by either the ecogeographic filtering method or the calibration method. The ecogeographic filtering method selects occurrence records present in environments that are likely to impose selection pressure for the adaptive trait investigated. In the calibration method, partial evaluation data for the target trait are required to identify an association between ecogeographic variables and the target trait. The calibrated predictive model based on association is then applied to non-evaluated populations. Both methods result in a subset of populations that have a higher probability than random subsets of possessing the target trait. The usefulness of these methods was demonstrated in several studies; for example, El Bouhssini et al. (2011) identified sources of resistance to Russian wheat aphid (Diuraphis noxia) in bread wheat (Triticum aestivum L.) by applying the ecogeographic filtering method, and Bari et al. (2012, 2014) predicted resistance to stem rust (Puccinia graminis f. sp. tritici) and stripe rust (Puccinia striiformis f. sp. tritici) in accessions of wheat landraces by using the calibration method.
We selected white clover (Trifolium repens L.) for a case study to gain further insight into the potential of this approach for increasing the effectiveness of trait evaluation in forage breeding. White clover is a common perennial legume found in fields, lawns and pastures of temperate regions worldwide (Gibson and Cope 1985). It is adapted to a wide range of climatic, edaphic and management conditions (Lane et al. 2000) and, thus, has become naturalised on every continent to which it was introduced with European settlement (Gibson and Hollowell 1966). It is an insect-pollinated and obligate outcrossing species, with vegetative propagation occurring by stolons (Olsen and Ungerer 2008). It is the most widely grown temperate forage legume (Frame and Newbould 1986) and the most common legume in pastures grazed by cattle and sheep (Laidlaw and Teuber 2001). It is generally considered a high-quality forage (Ulyatt 1981) with good persistence under grazing (Caradus et al. 1995). Estimated consumption of white clover globally ranges between 9000 and 10 000 Mt (Mather et al. 1996), and it is ranked the third most used legume pasture species after lucerne (Medicago sativa L.), at 160 000 Mt, and red clover (Trifolium pratense L.), at 15 000 Mt (Mather et al. 1996).
As with changes to breeding targets in other forage crops, the focus in clover species has shifted from attempting to achieve a nutritionally balanced sward to considering the effects of genetic variation on animal performance and the environment, i.e. the impact of forage diets on meat and milk quality, and the contribution of forage to direct and indirect diffusion of nitrogen and phosphorus pollution (Abberton and Marshall 2005). Cyanogenesis, a process that generates anti-quality compounds in some Trifolium species, is therefore a matter of concern in some countries because of its negative effects on large herbivores; as such, it is given consideration in several breeding programs (Crush and Caradus 1995).
The cyanoglucosides present in some cultivars of white clover form free hydrogen cyanide (HCN) when ingested by ruminants (Caradus et al. 1995). Although there are no recorded instances of mortality due to cyanide toxicity in livestock (Caradus et al. 1995), indirect effects on metabolism of iodine (Butler et al. 1957; Greer et al. 1966), selenium (Gutzwiller 1993) and sulfur (Caradus et al. 1995) in animals may have serious consequences for nutrient availability. Because of this, there is a growing interest in acyanogenic germplasm.
Clues on the distribution of the frequency of cyanogenic clover have emerged through the publication of ecological studies of white clover. Some studies have shown an inverse relationship between the frequency of cyanogenic plants and altitude (Till 1987; Till-Bottraud et al. 1988; Pederson et al. 1996; Richards and Fletcher 2002; Oliveira et al. 2013), not only in the native Eurasian species range but also in the non-native range, where populations have become naturalised (reviewed by Olsen and Ungerer 2008). A strong selective pressure maintaining the cyanogenesis polymorphism seems to be responsible for the rapid evolution of cyanogenic clines (Olsen and Ungerer 2008). The frequency of cyanogenic plants has also been positively correlated with temperature and negatively correlated with precipitation (López-Díaz et al. 2009). However, Richards and Fletcher (2002) suggested that summer drought, together with winter cold, might select against cyanogenic phenotypes. Grazing by large herbivores does not favour cyanogenesis, whereas invertebrate herbivory (i.e. molluscs and insects) may favour cyanogenesis (Dirzo and Harper 1982a, 1982b; Pederson and Brink 1998; Richards and Fletcher 2002; Saucy et al. 1999; Viette et al. 2000).
The selective factors favouring cyanogenic phenotypes are fairly well understood; however, the factors that favour acyanogenic plants in colder climates are not (Olsen and Ungerer 2008). Two main hypotheses have been proposed. The first focuses on abiotic stress and proposes that cyanogenic plants may suffer decreased freezing tolerance because cell rupture from freezing could lead to auto-toxicity and tissue death following HCN release (Daday 1958, 1965; Hughes 1991; Olsen and Ungerer 2008). The second hypothesis focuses on biotic interactions and suggests that in cooler climates, where generalist herbivores are less abundant, plants investing in growth instead of cyanogenesis might be more competitive (Kakes 1987).
Although previous studies have shown an association of cyanogenesis with different single abiotic factors, until now no efforts have been made to model and predict the presence or absence of the cyanogenic trait in Trifolium germplasm based on these factors.
Our main objective in this study was to develop a procedure that would enable us to find wild populations and genebank accessions of white clover that are more likely to be acyanogenic. We hypothesised that the association of evaluation data for cyanogenesis with the environmental conditions of the locations where the evaluated accessions were collected would help us to identify new sources of acyanogenic germplasm (among accessions and wild populations) at minimum cost. By applying the calibration predictive characterisation method, we aimed to answer the following questions of: (i) which ecogeographic variables influence cyanogenesis based on available evaluation data in the USDA white clover collection; (ii) which model best explains the relationship between ecogeographic variables and the expression of the trait; and (iii) which non-evaluated populations of white clover are more likely to have the acyanogenic trait.
Materials and methods
Species datasets
Occurrence information, including geographical coordinates, was compiled for white clover. Data on accessions conserved in gene banks were obtained from the following databases: the European Cooperative Program for Plant Genetic Resources (www.ecpgr.cgiar.org); the United States Department of Agriculture, National Plant Germplasm System’s GRIN (Germplasm Resources Information Network) Global Project (www.ars-grin.gov/npgs/); and the CGIAR System-Wide Information Network for Genetic Resources (SINGER) or its successor Genesys Plant Genetic Resources (www.genesys-pgr.org). Additional data on accessions conserved in gene banks were provided by AgResearch (www.agresearch.co.nz) and the Australian Plant Genetic Resources Information System (www.agric.wa.gov.au/). Evaluation data for cyanogenesis were obtained from the USDA white clover collection. These data had been collected in 1992 and 1993 on 543 accessions by testing two or three leaves from each of 20 plants grown in the greenhouse at Mississippi State, MS, USA. Cyanogenesis was determined by the picric acid test (Hogg and Ahlgren 1942) and ranged between 0 (completely acyanogenic accessions) and 100 (completely cyanogenic accessions). Data on population occurrences from sources other than gene banks were obtained from the Global Biodiversity Information Facility (GBIF, https://www.gbif.org/en/). Species names were standarised by using GRIN Taxonomy (Wiersema and Remsen 2018).
Data on white clover populations without geographic coordinates were removed. The dataset with coordinates was divided into two subsets: one including only the accessions that had been evaluated for cyanogenesis (‘evaluated set’), and the other accessions that had not been evaluated (‘non-evaluated set’). Evaluated accessions recorded as breeding materials or improved cultivars were discarded. Therefore, only wild, weedy and landrace accessions from the evaluated set were considered in the subsequent analysis. Because white clover is an outcrossing species, we considered localities spaced <3000 m apart to be from the same population (Visscher and Seeley 1982). Therefore, in this study, handling spatial duplicates in both evaluated and non-evaluated sets consisted of selecting only one representative locality in each 3000-m-diameter circular area.
Records from the evaluated and non-evaluated sets were subjected to a geo-referencing quality evaluation, using the GEOQUAL procedure from the CAPFITOGEN toolkit (Parra-Quijano et al. 2015). This procedure detects coordinates with a high level of uncertainty due to low accuracy and geographical inconsistencies. Following García et al. (2017), records with geo-referencing quality values below TOTALQUAL = 80 were discarded for subsequent analyses.
Expert knowledge survey for ecogeographic variables related to cyanogenesis
A questionnaire was sent to 10 researchers involved in white clover breeding. Taking into account available scientific literature on this subject and additional consideration of other potential factors, researchers were asked to select from the following list the ecogeographic variables of most influence on the expression of cyanogenesis: annual mean temperature, annual precipitation, elevation, latitude–longitude, slope, aspect, sun radiation, soil pH, soil texture, content of organic carbon in the soil, salinity–sodicity and soil drainage. Considering the range of expression of cyanogenesis data gathered, the researchers were also asked to select a threshold value to discern accessions with desirable low levels of cyanogenesis. For this purpose, they were informed about the method used to quantify the content of cyanogenic compounds and the minimum and maximum values observed in the USDA Trifolium collection.
Ecogeographic characterisation
The ecogeographic variables selected by at least 75% of the breeders answering the questionnaire were used to characterise the occurrence sites of the evaluated and non-evaluated sets.
The values of the variables selected for ecogeographic characterisation were extracted from global raster layers with 5 arc-minute resolution (~10 km at the Equator) for all of the white clover geo-referenced genebank accessions and occurrence records. The bioclimatic, geophysical and edaphic global raster layers were obtained from Worldclim (Hijmans et al. 2005), SRTM Digital Elevation Models (Jarvis et al. 2008) and Harmonized World Soil Database (FAO/IIASA/ISSCAS/JRC 2012), respectively. Redundancy of the variables was tested through bivariate correlation analysis. When a pair of variables had Pearson correlation coefficients > |0.50| and P-value <0.05, one of the variables was randomly discarded. Both the ecogeographic characterisation and the bivariate correlation analysis were carried out by using SelecVar from the CAPFITOGEN toolkit.
Calibration method
The cyanogenesis trait was predicted by the calibration method of the predictive characterisation approach. To this end, the strictest threshold proposed by experts was used in the binarisation of the original levels of expression of cyanogenesis, so that desirable low levels of expression were assigned a value of 1, and undesirable levels a value of 0. The binarised variable was used as the dependent variable, and the selected ecogeographic variables were used as explanatory variables in the construction of several alternative models.
The evaluated set was partitioned into two subsets: one set containing 75% of the data points, which was used to calibrate all models (training data); and a second set containing 25% of the data points, which was used to evaluate the model (test data). Eight modelling techniques implemented in the biomod2 R package (Thuiller et al. 2014) were used in this analysis: artificial neural networks, classification tree analysis, flexible discriminant analysis, generalised additive model, generalised boosted model, generalised linear model (GLM), multivariate adaptive regression splines, and random forest. The biomod2 package was originally conceived for species distribution modelling, but in this study it was used to model the presence–absence of a trait in a population. Biomod2 allowed us to perform multiple modelling algorithms efficiently, to work with ecogeographic variables (in a GIS layer format), and to project the results on maps.
Using the function BIOMOD_Modelling in the biomod2 package, 100 models were run for each of the eight techniques. In each model, the distribution of the data points for the training and test sets was randomly assigned; however, the original proportion of the number of 1s and 0s of the entire evaluated set was maintained.
The predictive power of each model was evaluated with the true skill statistic (TSS). Considering that the receiver operating characteristic curve cannot be constructed for presence–absence predictions (Allouche et al. 2006), and that the kappa statistic is the most widely used measure for the performance of models generating presence–absence predictions but is affected by prevalence (Allouche et al. 2006), we decided to use TSS. TSS takes into account both omission and commission errors, and success, as a result of random guessing. It ranges from –1 to +1, where +1 indicates perfect agreement and values of zero or less indicate a performance no better than random. However, in contrast to kappa, TSS is not affected by prevalence (Allouche et al. 2006). The average TSS for 100 runs was calculated for each of the eight techniques.
The function variables_importance in biomod2, which assigns to each variable a value from 0 (no influence on the model) to 1, allowed us to identify the most influential variables on the model.
Identification of populations of potential interest
The run with the highest TSS value from the modelling approach with the highest average TSS value was used to predict the levels of cyanogenesis in the non-evaluated set with the function BIOMOD_Projection (biomod2 package, Thuiller et al. 2014). The predictions for the populations of the non-evaluated set were obtained in the form of their probability (0–1000) of having low levels of cyanogenesis according to the selected model. The populations were then ranked according to their probability of having low levels of cyanogenesis. Thus, a priority set of populations of white clover that may have desirable low levels of cyanogenic plants was identified.
The projection of the selected model on the area of study with the function BIOMOD_Projection allowed us to identify the areas where environmental conditions would favour the occurrence of the acyanogenic trait and, thus, may suggest high priority for further collecting activities.
Evaluation of the predicted acyanogenic set
Because the populations that are of greatest interest for the breeders are those that are acyanogenic, only accessions ranked highest for probability of having low levels of cyanogenesis were considered for evaluation. Thus, 18 accessions from the first 40 USDA accessions ranked highest for probability of having low levels of cyanogenesis were evaluated to determine the percentage of plants that were cyanogenic, following the same method applied in the initial evaluation of the USDA accessions (Pederson et al. 1996). To guarantee the comparability of the evaluation of this subset with the previous evaluation tests, five completely cyanogenic and five completely acyanogenic accessions from the original evaluated set were included in the trial. Seeds of each accession were scarified with sandpaper and germinated on water agar. Forty seedlings per accession were transplanted into flats with a growth medium of peat moss and vermiculite (1 : 1, v/v). Rhizobium subterraneum inoculum was watered into the growth medium following planting of the seedlings. The plants were grown in a greenhouse for 22–25 weeks. Leaves of weight 0.15 g were harvested from 20 plants per accession, and the presence or absence of hydrocyanic acid was determined by the picric acid test (Hogg and Ahlgren 1942). For each cyanogenic plant, the intensity of the cyanogenic reaction was scored on a 1–4 scale (HCN intensity score) where 1 is no reaction and 4 is most intense reaction.
Results
Processing of species datasets
After the elimination of the duplicated and low geo-referencing quality records, 3182 occurrence records remained in the study (Table 1). The evaluated set corresponded to 5% of the study dataset.

|
Accessions used in the study were distributed all over the world, but came mainly from Europe (80%). By contrast, occurrence data from natural populations obtained from GBIF mainly belonged to other continents (79%) (Fig. 1).
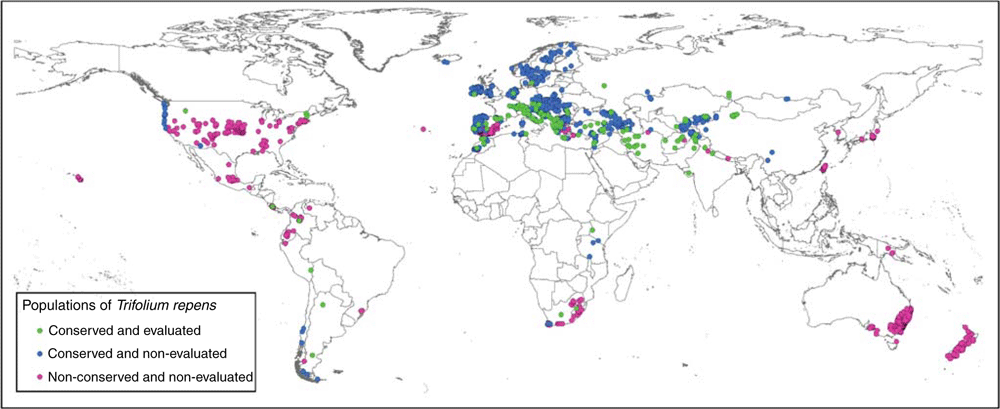
|
Selection of ecogeographic variables
Based on the responses of the expert survey, we identified five potentially influential ecogeographical variables: sun radiation, annual mean temperature, annual precipitation, elevation and soil pH. Because we did not have a global cover layer with sun radiation data when the layers were compiled, we utilised global layers for slope, northness and latitude as a proxy for sun radiation.
From the 3182 population records selected for the study, complete ecogeographic data were obtained for 3072 records (150 populations from the evaluated set and 2922 populations from the non-evaluated set). The records without complete ecogeographical data correspond to populations in urban environments, where edaphic information is not available.
Calibration method
The threshold proposed by the breeders to discern desirable and undesirable levels of cyanogenesis was 0% cyanogenesis. The GLM method yielded the best fit, with the highest TSS value of a run and the highest ‘lower and upper’ TSS values (Table 2).
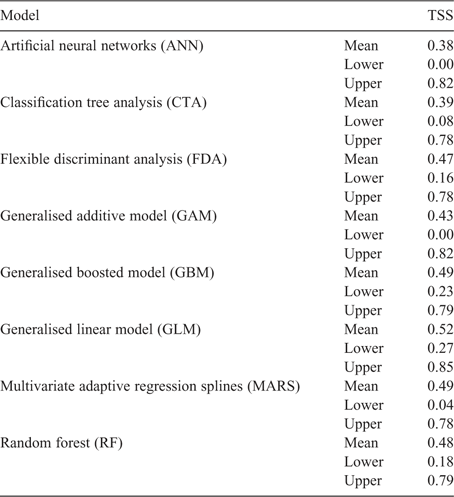
|
The most influential variable on the model was annual mean temperature (0.93), followed by annual precipitation (0.43) and altitude (0.13). The other four ecogeographic variables did not have any influence on the model.
Identification of populations of potential interest
The projection of the run of the GLM technique with the highest TSS value (0.85) on the non-evaluated set identified 470 populations with higher probability of being acyanogenic (Fig. 2). The Southern Cone and areas above the Tropic of Cancer in America, as well as central and northern Eurasia, were identified as areas with high probability of occurrence of acyanogenesis (Fig. 3).

|
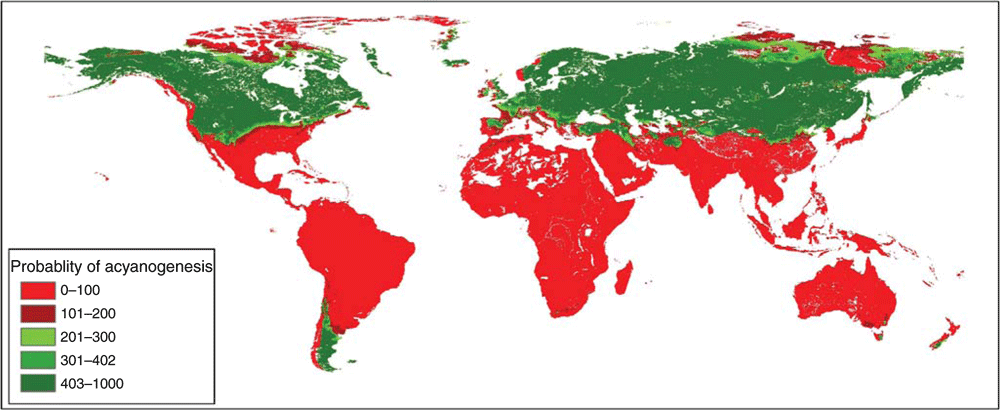
|
Validation of prediction results
Only a portion of the USDA top-rank predicted acyanogenic accessions could be evaluated owing to limited seed availability. These accessions were located between positions 3 and 351 in the ranking, and their predicted probabilities of being acyanogenic ranged between 930 and 626 (Table 3). Of the 18 evaluated accessions, 17 were completely acyanogenic, whereas the other accession, which had the lowest probability of being acyanogenic of the evaluated sample, had 95% of acyanogenic plants (Table 3). The five cyanogenic and five acyanogenic accessions that were included in the present trial as a control had the same results as in previous testing.
Discussion
As far as we know, this is the first study to use predictive characterisation to identify potential sources of desirable germplasm in a forage species. This approach allowed us to model the expression of cyanogenesis in white clover and to predict which populations might have desired levels of this trait, based on the ecogeographic variables involved in the expression of the trait. The evaluation carried out in this study confirmed the reliability of the prediction, given that the 18 accessions selected by predictive characterisation and later evaluated were completely or almost completely acyanogenic. A larger random sample of non-evaluated populations could have been used to conduct a more complete validation of the model, but this was not possible owing to limited seed availability.
The variables of greatest importance in the selected model were congruent with those highlighted by the scientific literature as influencing cyanogenesis. Thus, populations predicted as acyanogenic were associated with temperate areas with low annual mean temperatures, high altitude and high annual precipitation, which agreed with previous studies on the distribution of cyanogenic clover (Pederson et al. 1996; Richards and Fletcher 2002; López-Díaz et al. 2009; Oliveira et al. 2013).
The model selected to predict the cyanogenic response in the non-evaluated set achieved a TSS value classified as high according to Hodd et al. (2014). Our methodological approach departed from previous FIGS approaches in two steps of the prediction process. First, the evaluated set was split into a training set and a test set (Endresen 2010; Endresen et al. 2011; Bari et al. 2012, 2014). But because the proportion of acyanogenic populations was <25% of the evaluated set, we decided to keep the original proportions of acyanogenic and cyanogenic accessions of the entire evaluated set in the random distribution of data points for the training and the test sets. Second, we used TSS instead of kappa and area under the curve of the receiver operating characteristics to measure for the performance of models and to compare them. TSS has been recommended in order to avoid the effect of prevalence (Allouche et al. 2006), and we consider this option the best way to assess the performance of the models. Finally, for adequate performance of the calibration method, it is vital to make selective use of high-quality georeferenced records. We discarded 13% of the initial number of white clover geo-referenced accessions and occurrence records during the pre-processing stage to minimise the probability of generating erroneous results in the analysis. Other authors have assessed the importance of the quality of geo-referencing in spatial analysis and concluded that geographic inaccuracy affects diversity patterns more than taxonomic uncertainty (Maldonado et al. 2015) and that models run with data subject to random location errors showed less accuracy in many species (Graham et al. 2008).
Our study expanded the areas previously rated as highly acyanogenic (Pederson et al. 1996). The previous exploration of the distribution of the dataset of our species allowed us to detect an important spatial bias in the non-evaluated set. This set, mainly composed of population occurrences from sources other than gene banks, contained only two populations with geographic coordinates from the area comprising the United Kingdom, Germany, Denmark, Sweden and Norway, despite there being ≥25 000 genebank accession records of white clover from this area. The identification of populations of potential interest in the study is biased by the lack of high-quality georeferenced data of the records from these countries. Despite this, new countries should be added to the list of those most likely to have acyanogenic white clover: Sweden, Bulgaria, Poland and the Netherlands (44% of the 470 populations predicted as acyanogenic according to the selected model occur in these countries).
Despite having been introduced in some continents (Gibson and Hollowell 1966) relatively recently, those populations introduced by European settlers seem to have had time to adapt to the new environmental conditions. This is an essential issue to be able to establish relations or patterns between the environmental conditions of the site and the presence or absence of the target trait (Mackay and Street 2004). A short time since the introduction in a new place could not guarantee the plants have adapted to the new environmental conditions.
The predicted expression levels of cyanogenesis obtained in this study can be used as a criterion to assign collecting priorities for white clover. In the same way that García et al. (2017) identified populations with potential tolerance to drought and salinity within the ecogeographic gaps of the Spanish Aegilops collections, populations predicted as acyanogenic could be considered as a higher priority for collecting among other candidate populations. Prediction of phenotypes that can have specific use in plant breeding may enhance the use of genetic resource diversity, especially when characterisation and evaluation data held by gene banks is incomplete or lacking.
Conclusions
By using a predictive characterisation approach, we modelled the expression of cyanogenesis in white clover and predicted which accessions and wild populations would have desired levels of this trait. Our results contribute to increasing, in a predictive way and with a minimum cost, the knowledge of wild populations and genebank accessions in relation to a target trait. This improvement in the available evaluation data may encourage greater investment in forage plant breeding. Other opportunities for the application of this methodology include its use in the assessment of cyanogenesis and prioritised collecting of wild relatives of white clover. Furthermore, the predictive characterisation of other interesting traits of white clover, such as autumn or spring recovery, winter hardiness, stand survival and flower production, could be explored.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank Dr Beat Boller, Dr Zulfi Jahufer, Dr Kenneth H. Quesenberry and Dr Tomas Vymyslicky, for their generous contribution by responding the questionnaire about cyanogenesis. We are also very grateful to Dr Tomás Ruiz de Argüeso and Dr David Durán (Center for Biotechnology and Plant Genomics, Madrid) for kindly providing the inoculum for the seedlings. We thank Marta García Díaz (Faculty of Biology, Complutense University, Madrid) for her help in planning the laboratory tests. We are grateful to Marcos Méndez (Biodiversity and Conservation Area, Rey Juan Carlos University, Madrid) for comments that greatly improved the manuscript. This work was funded by the Horizon 2020 Framework Programme of the European Union under grant agreement number: 774271(Farmer’s Pride project), the Ministry of Economy and Competitiveness of Spain (grant CGLC2016-77377-R), and the Madrid Regional Government (grant REMEDINAL-3). The cost of the publication was funded by project EUC2014-51611 ‘Strengthening of the European Projects Office of Rey Juan Carlos University’ of the Ministry of Economy and Competitiveness of Spain.
References
Abberton MT, Marshall AH (2005) Progress in breeding perennial clovers for temperate agriculture. The Journal of Agricultural Science 143, 117–135.| Progress in breeding perennial clovers for temperate agriculture.Crossref | GoogleScholarGoogle Scholar |
Allouche O, Tsoar A, Kadmon R (2006) Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). Journal of Applied Ecology 43, 1223–1232.
| Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS).Crossref | GoogleScholarGoogle Scholar |
Bari A, Street K, Mackay M, Endresen DTF, De Pauw E, Amri A (2012) Focused identification of germplasm strategy (FIGS) detects wheat stem rust resistance linked to environmental variables. Genetic Resources and Crop Evolution 59, 1465–1481.
| Focused identification of germplasm strategy (FIGS) detects wheat stem rust resistance linked to environmental variables.Crossref | GoogleScholarGoogle Scholar |
Bari A, Amri A, Street K, Mackay M, De Pauw E, Sanders R, Nazari K, Humeid B, Konopka J, Alo F (2014) Predicting resistance to stripe (yellow) rust (Puccinia striiformis) in wheat genetic resources using focused identification of germplasm strategy. The Journal of Agricultural Science 152, 906–916.
| Predicting resistance to stripe (yellow) rust (Puccinia striiformis) in wheat genetic resources using focused identification of germplasm strategy.Crossref | GoogleScholarGoogle Scholar |
Brummer EC, Bouton JH, Casler MD, McCaslin MH, Waldron BL (2009) Grasses and legumes: genetics and plant breeding. In ‘Grassland quietness and strength for a new American agriculture’. (Eds F Wedin, SL Fales) pp. 157–172. (ASA-CSSA-SSSA: Madison, WI, USA)
Butler GW, Flux DS, Peterson GB, Wright EW, Glenday AC, Jhonson JM (1957) Goitrogenic effect of white clover (Trifolium repens L.) II. New Zealand Journal of Science and Technology 38, 793–802.
Caradus JR, McNabb W, Woodfield DR, Waghorn GC, Keogh R (1995) Improving quality characteristics of white clover. In Proceedings 25th Agronomy Society Conference’. Lincoln, New Zealand. pp. 7–12. (Agronomy Society of New Zealand)
Crush JR, Caradus JR (1995) Cyanogenesis potential and iodine concentration in white clover (Trifolium repens L.) cultivars. New Zealand Journal of Agricultural Research 38, 309–316.
| Cyanogenesis potential and iodine concentration in white clover (Trifolium repens L.) cultivars.Crossref | GoogleScholarGoogle Scholar |
Daday H (1958) Gene frequencies in wild populations of Trifolium repens L. III. World distribution. Heredity 12, 169–184.
| Gene frequencies in wild populations of Trifolium repens L. III. World distribution.Crossref | GoogleScholarGoogle Scholar |
Daday H (1965) Gene frequencies in wild populations of Trifolium repens L. IV. Mechanisms of natural selection. Heredity 20, 355–365.
| Gene frequencies in wild populations of Trifolium repens L. IV. Mechanisms of natural selection.Crossref | GoogleScholarGoogle Scholar |
Dirzo R, Harper JL (1982a) Experimental studies on slug-plant interactions. III. Differences in the acceptability of individual plants of Trifolium repens to slugs and snails. Journal of Ecology 70, 101–117.
| Experimental studies on slug-plant interactions. III. Differences in the acceptability of individual plants of Trifolium repens to slugs and snails.Crossref | GoogleScholarGoogle Scholar |
Dirzo R, Harper JL (1982b) Experimental studies on slug-plant interactions. IV. The performance of cyanogenic and acyanogenic morphs of Trifolium repens in the field. Journal of Ecology 70, 119–138.
| Experimental studies on slug-plant interactions. IV. The performance of cyanogenic and acyanogenic morphs of Trifolium repens in the field.Crossref | GoogleScholarGoogle Scholar |
El Bouhssini M, Street K, Amri A, Mackay M, Ogbonnaya FC, Omran A, Abdalla O, Baum M, Dabbous A, Rihawi F (2011) Sources of resistance in bread wheat to Russian wheat aphid (Diuraphis noxia) in Syria identified using the Focused Identification of Germplasm Strategy (FIGS). Plant Breeding 130, 96–97.
| Sources of resistance in bread wheat to Russian wheat aphid (Diuraphis noxia) in Syria identified using the Focused Identification of Germplasm Strategy (FIGS).Crossref | GoogleScholarGoogle Scholar |
Endresen DTF (2010) Predictive association between trait data and ecogeographic data for Nordic barley landraces. Crop Science 50, 2418–2430.
| Predictive association between trait data and ecogeographic data for Nordic barley landraces.Crossref | GoogleScholarGoogle Scholar |
Endresen DTF, Street K, Mackay M, Bari A, De Pauw E (2011) Predictive association between biotic stress traits and ecogeographic data for wheat and barley landraces. Crop Science 51, 2036–2055.
| Predictive association between biotic stress traits and ecogeographic data for wheat and barley landraces.Crossref | GoogleScholarGoogle Scholar |
FAO/IIASA/ISSCAS/JRC (2012) Harmonized World Soil Database (version 1.2). Food and Agriculture Organization of the United Nations, Rome; International Institute for Applied Systems Analysis, Laxenburg, Austria. Available at: http://webarchive.iiasa.ac.at/Research/LUC/External-World-soil-database/HTML/ (accessed 1 April 2013)
Frame J, Newbould P (1986) Agronomy of white clover. Advances in Agronomy 40, 1–88.
| Agronomy of white clover.Crossref | GoogleScholarGoogle Scholar |
García RM, Parra-Quijano M, Iriondo JM (2017) Identification of ecogeographic gaps in the Spanish Aegilops collections with potential tolerance to drought and salinity. PeerJ 5, e3494
| Identification of ecogeographic gaps in the Spanish Aegilops collections with potential tolerance to drought and salinity.Crossref | GoogleScholarGoogle Scholar | 28761779PubMed |
Gibson PB, Cope WA (1985) White clover. In ‘Clover science and technology’. Agronomy Monograph 25. (Ed. NL Taylor) pp. 471–490. (ASA-CSSA-SSSA: Madison, WI, USA)
Gibson PB, Hollowell EA (1966) ‘White clover.’ US Department of Agriculture Handbook 314. (US Department of Agriculture: Washington, DC)
Graham CH, Elith J, Hijmans RJ, Guisan A, Peterson AT, Loisells BA (2008) The influence of spatial errors in species occurrence data used in distribution models. Journal of Applied Ecology 45, 239–247.
| The influence of spatial errors in species occurrence data used in distribution models.Crossref | GoogleScholarGoogle Scholar |
Greer MA, Stott AK, Milne KA (1966) Effect of thiocyanate, perchlorate and other anions on thyroidal iodine metabolism. Endocrinology 79, 237–247.
| Effect of thiocyanate, perchlorate and other anions on thyroidal iodine metabolism.Crossref | GoogleScholarGoogle Scholar | 4288428PubMed |
Gutzwiller A (1993) The effect of a diet containing cyanogenetic glycosides on the selenium status and the thyroid function of sheep. Animal Production 57, 415–419.
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25, 1965–1978.
| Very high resolution interpolated climate surfaces for global land areas.Crossref | GoogleScholarGoogle Scholar |
Hogg PG, Ahlgren HL (1942) A rapid method for determining hydrocyanic acid content of single plants of Sudan grass. Journal - American Society of Agronomy 34, 199–200.
| A rapid method for determining hydrocyanic acid content of single plants of Sudan grass.Crossref | GoogleScholarGoogle Scholar |
Hodd RL, Bourke D, Skeffington MS (2014) Projected range contractions of European protected oceanic montane plant communities: focus on climate change impacts is essential for their future conservation. PLoS One 9, e95147
| Projected range contractions of European protected oceanic montane plant communities: focus on climate change impacts is essential for their future conservation.Crossref | GoogleScholarGoogle Scholar | 24752011PubMed |
Hughes MA (1991) The cyanogenic polymorphism in Trifolium repens L. (white clover). Heredity 66, 105–115.
| The cyanogenic polymorphism in Trifolium repens L. (white clover).Crossref | GoogleScholarGoogle Scholar |
Jarvis A, Reuter HI, Nelson A, Guevara E (2008) Digital elevation models (DEM) of the Shuttle Radar Topography Mission (SRTM). Hole-filled SRTM for the Globe Version 4. CGIAR Consortium for Spatial Information. Available at: http://srtm.csi.cgiar.org/ (accessed 7 January 2016)
Kakes P (1987) On the polymorphism for cyanogenesis in natural populations of Trifolium repens L. in the Netherlands. I. Distribution of the genes Ac and Li. Acta Botanica Neerlandica 36, 59–69.
| On the polymorphism for cyanogenesis in natural populations of Trifolium repens L. in the Netherlands. I. Distribution of the genes Ac and Li.Crossref | GoogleScholarGoogle Scholar |
Laidlaw AS, Teuber N (2001) Temperate forage grass-legume mixtures: advances and perspectives. In ‘Grassland ecosystems: an outlook into the 21st Century. Proceedings XIX International Grassland Congress’. Sao Paulo, Brazil. pp. 85–92. (Brazilian Society of Animal Husbandry/International Grasslands Congress)
Lane LA, Ayres JF, Lovett JV (2000) The pastoral significance, adaptive characteristics, and grazing value of white clover (Trifolium repens L.) in dryland environments in Australia: a review. Australian Journal of Experimental Agriculture 40, 1033–1046.
| The pastoral significance, adaptive characteristics, and grazing value of white clover (Trifolium repens L.) in dryland environments in Australia: a review.Crossref | GoogleScholarGoogle Scholar |
López-Díaz JE, González-Arráez E, Oliveira JA (2009) Variabilidad cianogénica y agronómica en poblaciones naturales de trébol blanco recolectadas en la Cordillera Cantábrica. In ‘La multifuncionalidad de los pastos: producción ganadera sostenible y gestión de los ecosistemas’. (Eds RJ Reiné, O Barrantes, A Broca, C Ferrer) pp. 177–183. (Sociedad Española para el Estudio de los Pastos: Huesca, Spain)
Mackay MC, Street K (2004) Focused identification of germplasm strategy—FIGS. In ‘Cereals 2004. Proceedings 54th Australian Cereal Chemistry Conference and 11th Wheat Breeders’ Assembly’. (Eds CK Black, JF Panozzo, GJ Rebetzke) pp. 138–141. (Cereal Chemistry Division, Royal Australian Chemical Institute: Melbourne)
Maldonado C, Molina CI, Zizka A, Persson C, Taylor CM, Albán J, Chilquillo E, Ronsted N, Antonelli A (2015) Estimating species diversity and distribution in the era of Big Data: to what extent can we trust public databases? Global Ecology and Biogeography 24, 973–984.
| Estimating species diversity and distribution in the era of Big Data: to what extent can we trust public databases?Crossref | GoogleScholarGoogle Scholar | 27656106PubMed |
Mather RDJ, Melhuish DT, Herlihy M (1996) Trends in the global marketing of white clover cultivars. In ‘White clover: New Zealand’s competitive edge’. Grassland Research and Practice Series B, No. 6. (Ed. DR Woodfield) pp. 1–14. (New Zealand Grassland Association: Palmerston North, New Zealand)
Oliveira JA, López JE, Palencia P (2013) Agromorphological characterization, cyanogenesis and productivity of accessions of white clover (Trifolium repens L.) collected in Northern Spain. Czech Journal of Genetics and Plant Breeding 49, 24–35.
| Agromorphological characterization, cyanogenesis and productivity of accessions of white clover (Trifolium repens L.) collected in Northern Spain.Crossref | GoogleScholarGoogle Scholar |
Olsen KM, Ungerer MC (2008) Freezing tolerance and cyanogenesis in white clover (Trifolium repens L. Fabaceae). International Journal of Plant Sciences 169, 1141–1147.
| Freezing tolerance and cyanogenesis in white clover (Trifolium repens L. Fabaceae).Crossref | GoogleScholarGoogle Scholar |
Parra Quijano M, Torres E, Iriondo JM, López F (2015) ‘CAPFITOGEN tools. User manual version 2.0.’ (International Treaty on Plant Genetic Resources for Food and Agriculture: Rome)
Pederson GA, Brink GE (1998) Cyanogenesis effect on insect damage to seedling white clover in a Bermuda grass sod. Agronomy Journal 90, 208–210.
| Cyanogenesis effect on insect damage to seedling white clover in a Bermuda grass sod.Crossref | GoogleScholarGoogle Scholar |
Pederson GA, Fairbrother TE, Green SL (1996) Cyanogenesis and climatic relationship in US white clover germplasm collection and core subset. Crop Science 36, 427–433.
| Cyanogenesis and climatic relationship in US white clover germplasm collection and core subset.Crossref | GoogleScholarGoogle Scholar |
Richards AJ, Fletcher A (2002) The effects of altitude, aspect, grazing and time on the proportion of cyanogenics in neighbouring populations of Trifolium repens L. (white clover). Heredity 88, 432–436.
| The effects of altitude, aspect, grazing and time on the proportion of cyanogenics in neighbouring populations of Trifolium repens L. (white clover).Crossref | GoogleScholarGoogle Scholar | 12180084PubMed |
Saucy F, Studer J, Aerni V, Scheneiter B (1999) Preference for acyanogenic white clover (Trifolium repens) in the vole Arvicola terrestris. I. Experiments with two varieties. Journal of Chemical Ecology 25, 1441–1454.
| Preference for acyanogenic white clover (Trifolium repens) in the vole Arvicola terrestris. I. Experiments with two varieties.Crossref | GoogleScholarGoogle Scholar |
Thormann I, Parra-Quijano M, Rubio Teso ML, Endresen DTF, Dias S, Iriondo JM, Maxted N (2016) Predictive characterization methods for accessing and using CWR diversity. In ‘Enhancing crop genepool use. Capturing wild relative and landrace diversity for crop improvement’. (Eds N Maxted, ME Dulloo, BV Ford-Lloyd) pp. 64–77. (CABI International: Wallingford, UK)
Thuiller W, Georges D, Engler R (2014) Biomod2: Ensemble platform for species distribution modelling. The R Foundation, Vienna. Available at: http://cran.r-project.org/web/packages/biomod2/index.html (accessed 16 March 2015)
Till I (1987) Variability in expression of cyanogenesis in white clover (Trifolium repens L.). Heredity 59, 265–271.
| Variability in expression of cyanogenesis in white clover (Trifolium repens L.).Crossref | GoogleScholarGoogle Scholar |
Till-Bottraud I, Kakes P, Dommee B (1988) Variable phenotypes and stable distribution of the cyanotypes of Trifolium repens L. in southern France. Acta Oecologica 9, 393–404.
Ulyatt MJ (1981) The feeding value of herbage: can it be improved? New Zealand Journal of Agricultural Science 15, 200–205.
Viette M, Tettamanti C, Saucy F (2000) Preference for acyanogenic white clover (Trifolium repens) in the vole Arvicola terrestris. II. Generalization and further investigations. Journal of Chemical Ecology 26, 101–122.
| Preference for acyanogenic white clover (Trifolium repens) in the vole Arvicola terrestris. II. Generalization and further investigations.Crossref | GoogleScholarGoogle Scholar |
Visscher PK, Seeley TD (1982) Foraging strategy of honey bee colonies in a temperate deciduous forest. Ecology 63, 1790–1801.
| Foraging strategy of honey bee colonies in a temperate deciduous forest.Crossref | GoogleScholarGoogle Scholar |
Wiersema JH, Remsen D (2018) GRIN Taxonomy. Checklist dataset. US National Plant Germplasm System. National Genetic Resources Program Germplasm Resources Information Network (GRIN). National Germplasm Resources Laboratory, US Department of Agriculture Agricultural Research Service, Beltsville, MD, USA. https://doi.org/10.15468/ao14pp (accessed via GBIF.org 19 May 2019)



