Molecular characterisation of PAL gene family reveals their role in abiotic stress response in lucerne (Medicago sativa)
Yating Feng A B # , Qiaoli Huang B # , Rui Zhang A B , Junyi Li A B , Kai Luo A B * and Yinhua Chen A C *
A B # , Qiaoli Huang B # , Rui Zhang A B , Junyi Li A B , Kai Luo A B * and Yinhua Chen A C *
A Key Laboratory of Sustainable Utilization of Tropical Biological Resources of Hainan Province, Haikou 570228, P.R. China.
B School of Tropical Crops, Hainan University, Haikou 570228, P.R. China.
C School of Life and Pharmaceutical Sciences, Hainan University, Haikou 570228, P.R. China.
Handling Editor: Marta Santalla
Crop & Pasture Science 73(3) 300-311 https://doi.org/10.1071/CP21558
Submitted: 1 July 2021 Accepted: 8 October 2021 Published: 9 February 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Phenylalanine ammonia lyase (PAL) is the first enzyme in the phenylpropanoid pathway and plays a critical role in plant growth, development and stress defence. However, there have been few reports of the PAL gene family in lucerne (also known as alfalfa, Medicago sativa L.), one of the most important forage legume species worldwide. In this study, we report that PAL in lucerne is encoded by a family of seven genes: MsPAL1–MsPAL7. Furthermore, a comprehensive genome-wide bioinformatics analysis of the MsPAL gene family is presented, including chromosomal locations, phylogenetic relationships, gene structures and conserved motifs. The cis-elements and potential biological functions of these genes were investigated, revealing the potential roles of MsPAL members in response to various stresses. RT-qPCR results showed that the expression of MsPAL6 was significantly upregulated under both salinity- and waterlogging-stress conditions. Other MsPAL members such as MsPAL1 and MsPAL2 were downregulated under saline conditions and upregulated significantly after waterlogging stress. Our findings provide useful information for further practical analyses and for the genetic improvement of abiotic stress tolerance of lucerne.
Keywords: abiotic stress, lucerne (Medicago sativa), bioinformatics analysis, expression profiling, functional verification, gene family, phenylalanine ammonia-lyase (PAL), phylogenetic.
Introduction
Phenylalanine ammonia lyase (PAL; EC 4.3.1.5) is the first and main regulatory enzyme in the phenylpropanoid pathway and is conserved in virtually all eukaryotes. PAL was first isolated from Hordeum vulgare (Koukol and Conn 1961), and has since been found across various organisms including plants, liverworts, viruses, algae, yeasts and fungi (MacDonald and D’Cunha 2007). PAL catalyses the conversion of phenylalanine to cinnamic acid, and the synthesis of secondary metabolites depends on PAL activity, because it connects primary metabolism to secondary metabolism (Dong et al. 2016). Therefore, PAL is involved in the biosynthesis of a wide range of secondary metabolites including lignin, flavonoids, tannins, and many other less-studied benzene compounds and phenolic glycosides that play many essential physiological roles in plant growth, development and adaptations (MacDonald and D’Cunha 2007; Vogt 2010). Numerous studies have suggested the essential role of PAL in protecting plants from environmental stressors such as drought (Guo and Wang 2009; Khakdan et al. 2018), salinity (Gao et al. 2008), extreme temperatures (Olsen et al. 2008), nutrient deficiency (Olsen et al. 2008; Gho et al. 2020) and waterlogging (Nguyen et al. 2016). This makes it an excellent inducer that triggers reactions aimed at increasing plant resistance to multiple stresses.
The indispensable role of PAL in phenylpropanoid biosynthesis means that PAL genes have been extensively studied. PAL exists as different isoforms, apparently encoded by a multigene; moreover, the number of members differs among species. For instance, the number of PAL gene copies is four in tobacco (Nicotiana tabacum) (Reichert et al. 2009) and Arabidopsis (Raes et al. 2003), five in Populus trichocarpa (Hamberger et al. 2007), six in Medicago truncatula (Ren et al. 2019), and nine in rice (Oryza sativa) (Hamberger et al. 2007). PAL genes in several plant species have also been functionally characterised. For example, in Arabidopsis, expression studies of the four AtPAL genes have shown that only AtPAL1 and AtPAL2 are induced in response to decreased nitrogen and low temperatures (Olsen et al. 2008). Six PAL genes were recently identified, characterised and expressed in M. truncatula; these genes were also differentially expressed in different tissues and in response to different abiotic stresses (Ren et al. 2019). Similarly, three Pyrus bretschneideri PAL genes were induced in response to abiotic stress to varying degrees (Mamat et al. 2019). Together, these studies indicate that the general function of PAL genes is clear; however, the functional differences of individual PAL genes are not well understood in many species.
Lucerne (also known as alfalfa, Medicago sativa L.) is one of the most important perennial leguminous herbaceous species, and it is planted on >40 Mha worldwide. This species is referred to as the ‘king of herbs’ because of its high yield, nitrogen fixation ability and nutrient profiles (Mouttet et al. 2014). In China, lucerne cultivation areas are distributed mainly in the northern and north-eastern regions, where salinisation is dramatically increasing (Ashrafi et al. 2014). Moreover, the problem of poor drainage in those areas can lead to both waterlogging and increased salinity in the root-zone. Among legumes, lucerne is moderately saline tolerant; however, its production decreases at salinities >2 dS m−1 (Maas and Hoffman 1977). In addition, this species is susceptible to waterlogging stress, and even very short periods of waterlogging can lead to severe damage (Evans 1998; Humphries and Auricht 2001). The PAL gene family plays an important role in plant growth; therefore, its evolutionary mechanism and expression patterns should be explored, laying a foundation for further research on the functions of genes involved in tolerance to multiple stresses such as salinity and waterlogging. Therefore, we analysed the PAL gene family by searching the whole genome of lucerne to provide initial insight into the functions of PAL genes in lucerne under salt and waterlogging stress.
Materials and methods
Database mining and identification of the lucerne PAL genes
Lucerne genome sequences were downloaded from the Figshare website (Chen et al. 2020). Searching for PAL genes in the lucerne genome, 27 protein sequence encoding PAL genes from Arabidopsis, rice, M. truncatula and soybean (Glycine max) were retrieved from Phytozome v12 (Goodstein et al. 2012). These sequences were used to identify homologous peptides from lucerne by means of Basic Local Alignment Search Tool algorithms (BLASTP) at the CADL Genome Blast Server (https://www.alfalfatoolbox.org/). With the help of the Pfam database (El-Gebali et al. 2019), all of the putative MsPAL proteins identified from the hidden Markov Model (HMM) profile were searched for confirmation that they contained the Lyase_aromatic domains (PF00221). Then, the redundant sequences were removed by using the decrease redundancy tool (web.expasy.org/decrease_redundancy). Multiple sequence alignment was performed by using the DNAMAN program (Lynnon Biosoft, Quebec, Canada) with default parameters.
Phylogenetic relationship, conserved motifs and gene structure analysis
In order to investigate the evolutionary relationships among lucerne, Arabidopsis, rice, M. truncatula and soybean, multiple sequence alignment of PAL proteins was performed with ClustalW (Yuan et al. 1999), and a phylogenetic tree was constructed by using the maximum likelihood phylogenetic method in MEGA5.0 with 1000 bootstrap replicates. The conserved motifs in full-length PAL proteins were identified using Multiple Expectation Maximisation for Motif Elicitation (MEME) (http://meme-suite.org/), with the maximum number of motifs as 20 (Timothy et al. 2009). By comparing DNA sequences with their corresponding coding sequences, the gene structures of MsPAL were drawn using the Gene Structure Display Server (GSDS 2.0) (http://gsds.gao-lab.org/) (Hu et al. 2015).
In silico sequence analysis and gene expression patterns
The amino acid sequences of putative lucerne PAL proteins were analysed to calculate the molecular weight (MW), grand average of hydropathicity (GRAVY) and theoretical isoelectric point (pI) using the ProtParam tool (http://web.expasy.org/compute_pi/), and to predict subcellular localisation using WoLF PSORT (Horton et al. 2007).
Chromosomal locations, tandem duplication and synteny analysis
Chromosomal location information was obtained from the gff file of the lucerne genome database (https://figshare.com/). The chromosomal localisation was displayed by using TBtools software (Chen et al. 2020). Collinear blocks were evaluated by TBTools (Chen et al. 2020), and the alignments with E-value ≤1E−10 were considered as significant matches (Tang et al. 2008). Tandem duplicated genes were defined as adjacent homologous MsPAL genes on a single chromosome, with no more than one intervening gene (Min et al. 2019). For synteny analysis, the synteny block of PAL between lucerne and other three species, Arabidopsis, soybean and M. truncatula, was obtained from Plant Genome Duplication database (http://chibba.pgml.uga.edu/duplication/) (Lee et al. 2013). Then, the genes of the synteny blocks were identified and analysed, and connected by solid lines. Additionally, nonsynonymous (Ka) and synonymous (Ks) substitution rates were calculated to explore the mechanism of gene divergence after duplication; Ka and Ks were computed by using TBtools software (Chen et al. 2020).
Promoter cis-element and tissue-specific expression analysis
The 2.0 kb sequence upstream from the translation start site of the seven MsPAL genes was obtained from the CADL Genome Blast Sever. The PlantCARE online database was used to analyse the putative hormone- or stress- responsive cis-acting regulatory elements (Lescot et al. 2002). The transcriptome data of two alfalfa subspecies was provided by the Noble Research Institute, which were previously generated and analyzed by O’Rourke et al.(2015). The transcriptome data included six tissues: leaf, flower, root, nodule, pre-elongated stem and elongated stem. All of the expression data were then gene-wise normalised using the MeV v4.9 software (http://www.mybiosoftware.com/).
Plant materials and stress treatments
Lucerne variety Gannong No. 9 was used in this study. It has been widely cultivated in the northern and north-eastern regions of China, and was provided by the Official Herbage and Turfgrass Seed Testing Centre, Ministry of Agriculture and Rural Affairs (Lanzhou, China). Seeds were scarified, sterilised and germinated at 20°C in the dark. After 3 days, uniform seedlings were grown in an artificial climate chamber under controlled light-cycle conditions of 16 h daylight and 8 h night at 25°C. Two different plant cultivation methods were used. For salinity treatment, seedlings were grown in 36-well plates supported by a plastic container and then hydroponically grown in half-strength Murashige and Skoog nutrient solution at pH 5.8. For waterlogging treatment, seedlings were grown in plastic pots containing a 2:1 mixture (v/v) of potting soil and vermiculite.
After cultivating for 14 days, salinity and waterlogging stress treatments were performed on the seedlings parallel to an untreated control. The salinity treatment involved adding 250 mM NaCl for 12 and 24 h. The waterlogging treatment was imposed by filling the pots with water up to 2–3 cm above the soil surface for 3, 12 and 24 h. For each of our three biological replicates, roots were combined from 10 seedlings, then rapidly frozen in liquid nitrogen and stored at −80°C.
RNA isolation and RT-qPCR analysis
Total RNA extraction was performed according to the manufacturer’s instructions (Sangon Biotech, Shanghai, China). Isolated total RNA (∼1 μg) was used to generate cDNA, using a reverse transcriptase kit (catalogue no. M1631; Thermo Fisher, MA, USA). The primer pairs were examined by melting curve, agarose gel electrophoresis and sequencing PCR products (Supplementary Material Table S1). RT-qPCR was performed on a 7500 Real Time PCR System (Thermo Fisher) with a total reaction volume of 20 mL, containing 2 mL cDNA templates, 0.4 mL each of 10 mM forward and reverse primers, 10 mL qPCR Master Mix, 0.4 mL ROX and 6.8 mL sterilised ddH2O. The PCR conditions were set as follows: 95°C for 30 s; 40 cycles of 95°C for 5 s, 60°C for 34 s, 95°C for 15 s and 60°C for 1 min. Relative fold expression changes of the seven MsPAL genes were normalised to Actin1 genes and calculated using the 2−ΔΔCt method described by Schmittgen and Livak (2008): (ΔΔCt = (Cttarget gene − CtActin1) under stress condition − (Cttarget gene − CtActin1) under control condition). The average fold change was plotted with the standard error. Three biological replicates for each group were run, and each reaction was performed with two technical replicates. Data were analysed by analysis of variance followed by Duncan’s new multiple range test, using SPSS Statistics ver. 20.0 (IBM, Armonk, NY, USA). The significance level was set at P = 0.05.
Results
Identification and phylogenetic relationship analysis of PAL family members in lucerne
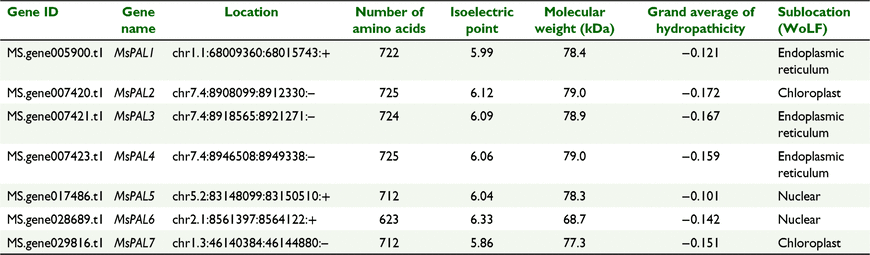
In total, 102 putative PAL protein sequences were identified by querying the lucerne genome database via HMM profile and BLAST searches, with Arabidopsis, rice, M. truncatula and soybean sequences used as queries. All of them were confirmed to contain Lyase_aromatic domains (PF00221) according to searches of the Pfam database. After removing the pseudogenes, genes for seven unique PAL proteins were retained for further phylogenetic and functional analysis, and named MsPAL1– MsPAL7. We conducted an additional in silico analysis to identify and systematise the MsPAL genes, including their chromosome locations, length of coding DNA sequence (CDS), number of amino acids, pI, MW, GRAVY and sublocation (Table 1). Furthermore, a detailed sequence alignment of the MsPAL proteins is shown in Fig. 1. The conserved enzymatic active site Ala-Ser-Gly was found in all of the MsPAL proteins.
|
The classification of, and relationships between, MsPAL proteins and PAL proteins from Arabidopsis, rice, M. truncatula and soybean was studied by constructing an unrooted neighbour-joining phylogenetic tree (Fig. 2a), which showed division into two distinct groups, dicots and monocots, suggesting that the PAL gene family may have existed before the dicot and monocot divergence. As expected, the PAL proteins of legume families fell into the dicot class, and MsPAL proteins clustered more closely to the PAL proteins from M. truncatula.
Conserved motif and gene structure of MsPAL members
The conserved motifs of the MsPAL proteins were predicted by using the MEME web server to determine the protein structure (Fig. 2b). In total, 20 predicted motifs of 34 PAL genes were identified, ranging from eight to 50 amino acids in length (Fig. S1). In general, PAL proteins with similar motif compositions tended to cluster together, indicating potential functional similarities among the PAL proteins. Many motifs, except motifs 9, 12, 15 and 16, were found in every PAL protein. Among the PAL proteins in lucerne, MsPAL6 had the fewest motifs, indicating that the PAL domain may be incomplete for this protein.
The exon–intron structures of the PAL genes in lucerne, Arabidopsis, rice, M. truncatula and soybean were determined by matching cDNA sequences to genomic sequences (Fig. 2c). The number, length and location of exons and introns differed across different PAL genes. The intron positions of orthologous PAL genes in lucerne, M. truncatula and soybean and their insertions with symmetric exons were well conserved, and all of them had one intron in the middle. Overall, the whole genes were longer in the legume species than in rice (Fig. 2c).
Chromosomal distribution and synteny analysis of MsPAL members
The seven MsPAL genes were assigned to five chromosomes of the lucerne genome: chromosomes 1.1, 1.3, 2.1, 5.2 and 7.4. Chromosome 7.4 contained three genes, and the other chromosomes each contained one gene (Table 1, Fig. S2). The evolutionary process of PAL genes was further explored by constructing comparative syntenic maps between lucerne and the other three species Arabidopsis, M. truncatula and soybean (Fig. 3; Table S2). Among them, one pair (MsPAL7/AtPAL1) existed in both the lucerne and Arabidopsis genomes, 10 pairs (MsPAL2/Glyma.03G181600, MsPAL2/Glyma.10G058200, MsPAL2/Glyma.13G145000, MsPAL2/Glyma.19G182300, MsPAL5/Glyma.02G309300, MsPAL7/Glyma.03G181600, MsPAL7/Glyma.10G058200, MsPAL7/Glyma.13G145000 and MsPAL7/Glyma.19G182300, MsPAL1/Glyma.20G180800) in both the lucerne and soybean genomes, and 10 pairs (MsPAL2/MtPAL1, MsPAL2/MtPAL3, MsPAL2/MtPAL5, MsPAL5/MtPAL4, MsPAL6/MtPAL1, MsPAL6/MtPAL3, MsPAL6/MtPAL5, MsPAL7/MtPAL1, MsPAL7/MtPAL3 and MsPAL7/MsPAL5) in both the lucerne and M. truncatula genomes. No gene pair existed in both the lucerne and rice genomes. Our results suggest that these genes may have been involved in the evolution of the PAL family. In addition, MsPAL3 and MsPAL4 did not map to any of the syntenic blocks with other plant PAL genes, indicating that their chromosomes have undergone extensive rearrangements and fusions that possibly led to selective gene loss. To understand the evolutionary constraints acting on the PAL gene family, the Ka/Ks ratios of the PAL gene pairs were calculated (Table S2), and all orthologous PAL gene pairs had Ka/Ks <1, suggesting that the lucerne MsPAL gene family might have experienced strong purifying selection during evolution.
Expression profiles of MsPAL genes in different tissues
The expression profiles of all seven MsPAL genes in six tissues were analysed by using AGED (O’Rourke et al. 2015) (Fig. 4). The heatmap showed that the expression patterns of the two lucerne subspecies were highly similar, indicating that these varieties may present the same transcript abundance. Among the seven MsPAL genes, three (MsPAL1, MsPAL6 and MsPAL7) were highly expressed (value >4) in the flowers and post-elongation stem internodes, and five (MsPAL3, MsPAL4, MsPAL6 and MsPAL7) were expressed at relatively low levels in roots. For expression in leaf, MsPAL6 was relatively high in the two subspecies, but MsPAL2–4 had high expression levels in M. sativa subsp. Falcata. Only MsPAL7 showed high expression levels (value >4) in the elongating stem internodes. By contrast, expression of all of the MsPAL genes was relatively low in the nodules.
Characterisation of putative cis-regulatory elements in the MsPAL genes
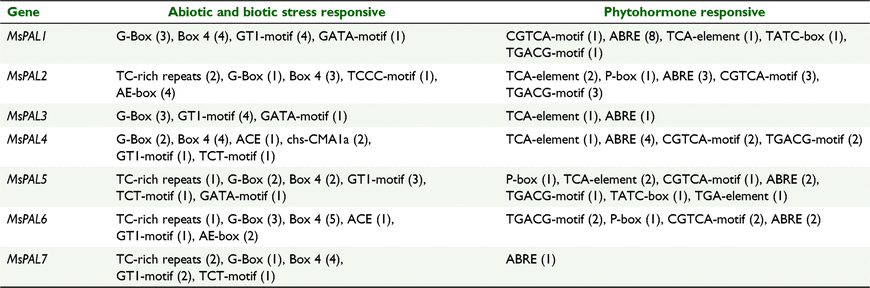
Using the PlantCARE database, we predicted possible cis-acting elements of seven MsPAL genes, and 17 kinds of cis-acting elements were used in this study, including 10 that were stress-responsive (G-Box, Box 4, GT1-motif, GATA-motif, TC-rich repeats, TCCC-motif, AE-box, ACE, chs-CMA1a and TCT-motif) and seven that were hormone-responsive (CGTCA-motif, ABRE, TCA-element, TATC-box, TGACG-motif, P-box and TGA-element) (Table 2). The ABRE, related to the abscisic acid response, was the most common motif in MsPAL gene promoters. These results suggest that MsPAL genes may play an important role in the response to various abiotic and biotic stresses. The number of cis-elements in the promoter regions of the MsPAL genes was variable. For example, the MsPAL5 promoter has 13 cis-elements, whereas MsPAL3 contains only five cis-elements (Table 2).
|
Expression levels of MsPAL genes under salinity and waterlogging stress
We used RT-qPCR to analyse the expression profiles of PAL family genes in lucerne under salinity and waterlogging treatments. Expression of MsPAL3, MsPAL4 and MsPAL6 was significantly upregulated after salinity treatment. Among these three upregulated homologous genes, MsPAL6 had the highest expression, increasing by nearly 71-fold at 24 h compared with the control. By contrast, expression of MsPAL1, MsPAL2, MsPAL5 and MsPAL7 exhibited the opposite trend after salinity treatment (Fig. 5a). As shown in Fig. 5b, expression of MsPAL1, MsPAL2, MsPAL4, MsPAL5 and MsPAL6 increased significantly under waterlogging stress. Moreover, expression of MsPAL3 was strongly upregulated at 3 h but then decreased, whereas MsPAL7 was first downregulated at 3 h and 12 h but then peaked at 24 h.
Discussion
Phenylalanine ammonia lyase is the first enzyme in the phenylpropanoid pathway and catalyses the deamination of L-phenylalanine into trans-cinnamic acid; as such, PAL is involved in the first step in which carbon flux is committed from primary metabolism into phenylpropanoid metabolism (Vogt 2010). PAL is also regulated developmentally and environmentally by transcriptional regulation (Zhao and Dixon 2011). A better understanding of the PAL gene family at the molecular level could be useful for elucidating mechanisms underlying stress resistance and for developing new plant varieties with increased resistance. To this end, we report here the identification and initial characterisation of lucerne PAL genes.
With the help of high-throughput sequencing technology, PAL genes, including those of both monocots and dicots, have now been functionally (Shang et al. 2012) characterised (Shang et al. 2012; Hou et al. 2013; Ren et al. 2019; Yan et al. 2019). The copy number of PAL genes varies widely across plant species but mostly ranges from three to nine. Some plant species such as tomato (Solanum lycopersicum) have a large PAL family, with several dozen members, among which many are inactive (Chang et al. 2008). Southern blot analysis of lucerne indicated a small PAL multigene family, which may be slightly more complex than that in bean (Phaseolus vulgaris) (Gowri et al. 1991). The recent availability of the M. sativa genome has provided opportunity to identify PAL genes in this species (Chen et al. 2020; Shen et al. 2020). Seven PAL members were identified, which is consistent with results obtained for other plant species such as cucumber (Cucumis sativus, seven PAL members; Shang et al. 2012). Researchers have found that the size of the lucerne genome (∼1200 Mb) is approximately three times that of the cucumber genome (367 Mb; Huang et al. 2009). Similarly, the genome size of tomato (4434 Mb) is >35 times that of Arabidopsis (125 Mb), despite the numbers of PAL genes being the same (Fukasawa-Akada et al. 1996; Raes et al. 2003). These results suggest that genome size may not be positively correlated with number of members of the PAL gene family. In previous studies, members of the PAL gene family were found to be widely dispersed across the genome. For instance, AtPAL1 and AtPAL3 are located on chromosomes 2 and 5, respectively, and AtPAL2 and AtPAL4 are located on the short arm and long arm of chromosome 3, respectively (Huang et al. 2009). In M. truncatula, the six PAL genes are located on four chromosomes (Ren et al. 2019). In the present study, chromosome distribution analysis demonstrated that the seven MsPAL family members are unevenly distributed across five chromosomes (Table 1; Fig. S2), and all of them share a high degree of sequence similarity (Fig. 1). This suggests that one or more tandem duplication events of PAL genes occurred during the evolution of lucerne.
The molecular evolution of the PAL gene family has been analysed in several plant species including cucurbit (Dong et al. 2016), M. truncatula (Ren et al. 2019) and Rosaceae spp. (Li et al. 2020). Those studies showed that PAL members clustered into at least two subfamilies according to phylogenetic analysis. As the phylogenetic tree of the plants in the present study indicated, the PAL genes from monocots and dicots could be divided into two distinct clades (Fig. 2a), suggesting that the PAL genes may have diverged or undergone functional specialisation before monocots and dicots split (165 million years ago (Mya)), which is consistent with findings of previous studies (Dong and Shang 2013; Ren et al. 2019). In the dicot subgroup, the PAL genes from lucerne were most closely related to those of M. truncatula, indicating that the expansion of the MsPAL gene family might have occurred before the speciation of lucerne and M. truncatula. A recent genomic analysis of cultivated lucerne revealed that the genome was estimated to have diverged from M. truncatula ∼8 Mya (Shen et al. 2020). Chen et al. (2020) reported that cultivated lucerne and M. truncatula might have diverged ∼5.3 Mya. Furthermore, the core domain of MsPAL genes was found to be highly conserved with that of MtPAL genes through conservative motif analysis and multisequence alignment (Figs 1 and 2b), indicating high conservation of gene structure during evolution. Moreover, synteny analysis showed at least 10 putative MsPAL orthologous genes in the examined leguminous plant species, and only one MsPAL orthologous gene in Arabidopsis (Fig. 3; Table S2). Considering that gene orthologs often have similar functions (Altenhoff et al. 2009), we infer that the functions of MsPAL genes are more similar to those of the PAL genes of soybean and M. truncatula.
Gene structure and conserved sequence construction may be intimately related to the diversity of gene functions (Cao et al. 2018). In the present study, there were two exons within most of the 34 PAL members (88.2% of cases), including all the PAL members in lucerne, soybean and M. truncatula. According to a structure analysis of Bambusa oldhamii, Juglans regia and Rosaceae spp., the number of exons per gene differed (Hsieh et al. 2010; Li et al. 2019; Yan et al. 2019). We speculate that the members of the PAL gene family in the legume plant family might have been more conserved during evolution. Moreover, our results reveal that most PAL proteins with similar exon/intron structures and motif compositions cluster together, which is consistent with reported results of a whole-genome analysis of other plant PAL families (Chen et al. 2016; Li et al. 2019; Yan et al. 2019). For example, MsPAL2–4 and MtPAL5 and MtPAL6 clustered together, and had the same number of exons and similar motif structures. Four PAL genes (MsPAL1, MtPAL2, Glyma.10G209800.1 and Glyma.20G180800.1) with relatively long exon–intron structures were also grouped within the same terminal branches. This also indicated potential functional similarities among the PAL proteins.
Increasing lines of evidence suggest that overexpression of tissue-specific genes can promote tissue remodelling and functional improvement. For example, AtPAL1 and AtPAL2 are highly expressed in Arabidopsis roots (Wanner et al. 1995) and are involved in the biosynthesis of stress-induced flavonoids (Olsen et al. 2008). In pear (Pyrus sp.), PbPAL1 and PbPAL2 are expressed mostly in the stems and roots, and potentially function in processes related to lignin synthesis and stone cell development (Li et al. 2019). Although all of the MsPAL genes were expressed in all of the tissues analysed, different expression patterns were observed (Fig. 4). The findings that the MsPAL genes identified in this study might exert diverse functions during various physiological and cellular processes could provide guidance toward the functional identification of candidate genes for specific traits to target for genetic engineering in lucerne. The varied expression of MsPAL genes in the six plant tissues may be due to their diverse promoter regions. Identical promoter regions with conserved cis-regulatory elements could be the result of segmental duplication. In the present study, we discovered many cis-acting components related to hormone and stress responses, such as abscisic acid treatment-responsive ABREs (Hobo et al. 2010), TC-rich repeat elements, and anaerobic-responsive AREs (Geffers et al. 2001) in the 2.0 kb upstream regulatory sequences of the MsPAL family members (Table 2). These results suggest that most MsPAL genes could be induced by hormones and might contribute to defence against various abiotic and biotic stresses in lucerne. Furthermore, there were different combinations of cis-elements in each MsPAL gene, indicating that they might be associated with different environmental stimuli.
PAL represents a potential node for the regulation of the whole phenylpropanoid pathway and is also a gateway between primary metabolism and natural product biosynthesis. Therefore, PAL genes rapidly response during the early stages of biotic stress conditions. For example, salinity stress was shown to induce the expression of key genes encoding PAL in two Salvia species and consequently increased PAL activity and total phenolic accumulation (Valifard et al. 2015). Increased PAL activity of Jatropha curcas could be a response to the cellular damage induced by higher salt concentrations (Gao et al. 2008). We found that the expression levels of three MsPAL genes were significantly upregulated in response to 250 mM NaCl treatment (Fig. 5a), suggesting that the induced PAL genes could be related to the implication of increased PAL activity in the lucerne response to salinity stress. Our results are also consistent with the findings of a previous study showing that salt stress enhanced the expression of the SlPAL5 gene in tomato (Guo and Wang 2009). Phenolic components are known to function as very efficient regulators of reactive oxygen species (ROS) scavenging, thus alleviating membrane lipid peroxidation caused by stress (Du et al. 2010). Increased ROS generation under waterlogging conditions can trigger an increase in PAL activity and consequently catalyse the formation of secondary metabolites such as tannins and anthocyanins (Smith et al. 2003; Wettberg et al. 2010). Our results prove that MsPAL genes in lucerne are responsive to waterlogging stress and that a majority of MsPAL genes are upregulated under waterlogging conditions, which is in agreement with the results of the cis-element analysis (Fig. 5b, Table 2). In wheat (Triticum aestivum), waterlogging was shown to increase expression of TaPAL1–3 and TaPAL5 in the internodes and TaPAL6 in the leaves (Nguyen et al. 2016). Our results showed that the expression of many MsPAL members increased under waterlogging conditions, which also confirmed the results of the cis-element analysis (Fig. 5b, Table 2). Moreover, all MsPAL genes were induced or inhibited to varying degrees under salinity and waterlogging treatments. For instance, MsPAL1 and MsPAL2 were downregulated under saline conditions, whereas these genes were upregulated significantly after waterlogging stress. The contrasting responses of some MsPAL genes to salinity and waterlogging suggested that they may have different response mechanisms to stress. Notably, both MsPAL4 and MsPAL6 displayed increased expression under both salinity and waterlogging conditions and could be excellent candidates for further functional characterisation and application in legume breeding programs.
Conclusions
Overall, our results showed the existence of at least seven PAL family genes in lucerne, an important legume forage species worldwide. The phylogenetic relationships, conserved motifs, gene structure, tissue expression and cis-elements of MsPAL genes were evaluated, and the potential biological functions of these genes were elucidated. RT-qPCR showed that the expression of MsPAL4 and MsPAL6 significantly increased after both salt and waterlogging stress, which indicated that these PAL family members may be the genes coding the main operative proteins under general conditions and play vital roles in the response to abiotic stress. Our observations provide the first insight into the function of PAL family genes when lucerne is subjected to salinity and waterlogging stress. Considering our results with those of previous reports, we stress the crucial role of PAL genes in lucerne. Our results provide new information about molecular traits to improve understanding of plant defence mechanisms.
Supplementary material
Supplementary material is available online.
Data availability
The data that support this study are available in the article and accompanying online supplementary material.
Conflicts of interest
No competing financial interests exist.
Declaration of funding
This work was supported by the Natural Science Foundation of Hainan Province (Grant No. 2019RC052); the Scientific Research Funds for Hainan University [Grant No. KYQD(ZR)1965, KYQD(ZR)1966].
Author contributions
Study concept and design: LK and CY Performance of the experiments: FY, HQ, ZR and LJ. Data analysis: FY and H.Q. Article writing: FY. All authors reviewed and approved the article before its submission.
References
Altenhoff AM, Dessimoz C, Eisen J (2009) Phylogenetic and functional assessment of orthologs inference projects and methods. PLOS Computational Biology 5, e1000262| Phylogenetic and functional assessment of orthologs inference projects and methods.Crossref | GoogleScholarGoogle Scholar | 19148271PubMed |
Ashrafi E, Razmjoo J, Zahedi M, Pessarakli M (2014) Selecting alfalfa cultivars for salt tolerance based on some physiochemical traits. Agronomy Journal 106, 1758–1764.
| Selecting alfalfa cultivars for salt tolerance based on some physiochemical traits.Crossref | GoogleScholarGoogle Scholar |
Branca A, Paape TD, Zhou P, Briskine R, Farmer AD, Mudge J, Bharti A, Woodward JE, May GD, Gentzbittel L, Ben C, Denny R, Sadowsky MJ, Ronfort J, Bataillon T, Young ND, Tiffin P (2011) Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proceedings of the National Academy of Sciences of the United States of America 108, E864–E870.
| Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula.Crossref | GoogleScholarGoogle Scholar | 21949378PubMed |
Cao YP, Li SM, Han YH, Meng DD, Jiao CY, Abdullah M, Li DH, Jin Q, Lin Y, Cai YP (2018) A new insight into the evolution and functional divergence of FRK genes in Pyrus bretschneideri. Royal Society Open Science 5, 1–19.
Chang A, Lim M-H, Lee S-W, Robb EJ, Nazar RN (2008) Tomato phenylalanine ammonia-lyase gene family, highly redundant but strongly underutilized. Journal of Biological Chemistry 283, 33591–33601.
| Tomato phenylalanine ammonia-lyase gene family, highly redundant but strongly underutilized.Crossref | GoogleScholarGoogle Scholar |
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant 13, 1194–1202.
| TBtools: an integrative toolkit developed for interactive analyses of big biological data.Crossref | GoogleScholarGoogle Scholar | 32585190PubMed |
Chen H, Zeng Y, Yang Y, Huang L, Tang B, Zhang H, Hao F, Liu W, Li Y, Liu Y, Zhang X, Zhang R, Zhang Y, Li Y, Wang K, He H, Wang Z, Fan G, Yang H, Bao A, Shang Z, Chen J, Wang W (2020) Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nature Communications 11, 2494
| Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa.Crossref | GoogleScholarGoogle Scholar | 32427850PubMed |
Chen YL, Dai W, Sun BM, Zhao Y, Ma Q (2016) Genome-wide identification and comparative analysis of the TUBBY-like protein gene family in maize. Genes & Genomics 38, 25–36.
| Genome-wide identification and comparative analysis of the TUBBY-like protein gene family in maize.Crossref | GoogleScholarGoogle Scholar |
Dong CJ, Shang QM (2013) Genome-Wide characterization of phenylalanine ammonia-lyase gene family in watermelon (Citrullus lanatus). Planta 238, 35–49.
| Genome-Wide characterization of phenylalanine ammonia-lyase gene family in watermelon (Citrullus lanatus).Crossref | GoogleScholarGoogle Scholar | 23546528PubMed |
Dong C-J, Cao N, Zhang Z-G, Shang Q-M (2016) Phenylalanine ammonia-lyase gene families in cucurbit species: structure, evolution, and expression. Journal of Integrative Agriculture 15, 1239–1255.
| Phenylalanine ammonia-lyase gene families in cucurbit species: structure, evolution, and expression.Crossref | GoogleScholarGoogle Scholar |
Du C-X, Fan H-F, Guo S-R, Tezuka T, Li J (2010) Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 71, 1450–1459.
| Proteomic analysis of cucumber seedling roots subjected to salt stress.Crossref | GoogleScholarGoogle Scholar | 20580043PubMed |
El-Gebali S, Mistry J, Bateman A, Bateman A, Eddy S, Luciani A, Potter S, Qureshi M, Richardson L, Salazar G, Smart A, Sonnhammer E, Hirsh L, Paladin L, Piovesan D, Tosatto S, Finn R (2019) The Pfam protein families database in 2019. Nucleic Acids Research 47, D427–D432.
| The Pfam protein families database in 2019.Crossref | GoogleScholarGoogle Scholar | 30357350PubMed |
Evans P (1998) Lucerne. In ‘Soilguide: a handbook for understanding and managing agricultural soils’. (Ed. G Moore) Bulletin 4343. (Agriculture Western Australia: Perth, WA, Australia)
Fukasawa-Akada T, Kung SD, Watson JC (1996) Phenylalanine ammonia-lyase gene structure, expression, and evolution in Nicotiana. Plant Molecular Biology 30, 711–722.
| Phenylalanine ammonia-lyase gene structure, expression, and evolution in Nicotiana.Crossref | GoogleScholarGoogle Scholar | 8624404PubMed |
Gao S, Ouyang C, Wang S, Xu Y, Tang L, Chen F (2008) Effects of salt stress on growth, antioxidant enzyme and phenylalanine ammonia-lyase activities in Jatropha curcas L. seedlings. Plant Soil and Environment 54, 374–381.
| Effects of salt stress on growth, antioxidant enzyme and phenylalanine ammonia-lyase activities in Jatropha curcas L. seedlings.Crossref | GoogleScholarGoogle Scholar |
Geffers R, Sell S, Cerff R, Hehl R (2001) The TATA box and a Myb binding site are essential for anaerobic expression of a maize GapC4 minimal promoter in tobacco. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1521, 120–125.
| The TATA box and a Myb binding site are essential for anaerobic expression of a maize GapC4 minimal promoter in tobacco.Crossref | GoogleScholarGoogle Scholar |
Gho Y-S, Kim S-J, Jung K-H (2020) Phenylalanine ammonia-lyase family is closely associated with response to phosphate deficiency in rice. Genes & Genomics 42, 67–76.
| Phenylalanine ammonia-lyase family is closely associated with response to phosphate deficiency in rice.Crossref | GoogleScholarGoogle Scholar |
Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar Daniel S (2012) Phytozome: a comparative platform for green plant genomics. Nucleic acids research 40, D1178–86.
| Phytozome: a comparative platform for green plant genomics.Crossref | GoogleScholarGoogle Scholar | 22110026PubMed |
Gowri G, Paiva NL, Dixon RA (1991) Stress responses in alfalfa (Medicago sativa L.) 12. Sequence analysis of phenylalanine ammonia-lyase (PAL) cDNA clones and appearance of PAL transcripts in elicitor-treated cell cultures and developing plants. Plant Molecular Biology 17, 415–429.
| Stress responses in alfalfa (Medicago sativa L.) 12. Sequence analysis of phenylalanine ammonia-lyase (PAL) cDNA clones and appearance of PAL transcripts in elicitor-treated cell cultures and developing plants.Crossref | GoogleScholarGoogle Scholar | 1715786PubMed |
Guo J, Wang M-H (2009) Characterization of the phenylalanine ammonia-lyase gene (SlPAL5) from tomato. Molecular Biology Reports 36, 1579–1585.
| Characterization of the phenylalanine ammonia-lyase gene (SlPAL5) from tomato.Crossref | GoogleScholarGoogle Scholar | 18791854PubMed |
Hamberger B, Ellis M, Friedmann M, de Azevedo Sousa C, Barbazuk B, Douglas C (2007) Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene families. Botany 2007, 1182–1201.
| Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene families.Crossref | GoogleScholarGoogle Scholar |
Hobo T, Asada M, Kowyama Y, Hattori T (1999) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. The Plant Journal 19, 679–689.
| ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent.Crossref | GoogleScholarGoogle Scholar | 10571853PubMed |
Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Research 35, W585–W587.
| WoLF PSORT: protein localization predictor.Crossref | GoogleScholarGoogle Scholar | 17517783PubMed |
Hou XM, Shao FJ, Ma YM, Lu SF (2013) The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: genome-wide characterization, molecular cloning and expression analysis. Molecular Biology Reports 40, 4301–4310.
| The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: genome-wide characterization, molecular cloning and expression analysis.Crossref | GoogleScholarGoogle Scholar |
Hsieh L-S, Ma G-J, Yang C-C, Du L-P (2010) Cloning, expression, site-directed mutagenesis and immunolocalization of phenylalanine ammonia-lyase in Bambusa oldhamii. Phytochemistry 71, 1999–2009.
| Cloning, expression, site-directed mutagenesis and immunolocalization of phenylalanine ammonia-lyase in Bambusa oldhamii.Crossref | GoogleScholarGoogle Scholar | 21035152PubMed |
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297.
| GSDS 2.0: an upgraded gene feature visualization server.Crossref | GoogleScholarGoogle Scholar | 25504850PubMed |
Huang SW, Li RQ, Zhang ZH, Li L, Gu XF, Fan W, Lucas WJ, Wang XW, Xie BY, Ni PX, Ren YY, Zhu H, Li J, Lin K, Jin WW, Fei ZJ, Li G, Staub J, Kilian A, Vossen E, Wu Y, Guo J, He J, Jia ZQ, Ren Y, Tian G, Lu Y, Ruan J, Qian W, Wang M, Huang Q, Li B, Xuan Z, Cao J Huang SW, Li RQ, Zhang ZH, Li L, Gu XF, Fan W, Lucas WJ, Wang XW, Xie BY, Ni PX, Ren YY, Zhu H, Li J, Lin K, Jin WW, Fei ZJ, Li G, Staub J, Kilian A, Vossen E, Wu Y, Guo J, He J, Jia ZQ, Ren Y, Tian G, Lu Y, Ruan J, Qian W, Wang M, Huang Q, Li B, Xuan Z, Cao J Huang SW, Li RQ, Zhang ZH, Li L, Gu XF, Fan W, Lucas WJ, Wang XW, Xie BY, Ni PX, Ren YY, Zhu H, Li J, Lin K, Jin WW, Fei ZJ, Li G, Staub J, Kilian A, Vossen E, Wu Y, Guo J, He J, Jia ZQ, Ren Y, Tian G, Lu Y, Ruan J, Qian W, Wang M, Huang Q, Li B, Xuan Z, Cao J (2009) The genome of the cucumber, Cucumis sativus L. Nature Genetics 41, 1275–1281.
| The genome of the cucumber, Cucumis sativus L.Crossref | GoogleScholarGoogle Scholar |
Humphries AW, Auricht GC (2001) Breeding lucerne for Australia’s southern dryland cropping environments. Australian Journal of Agricultural Research 52, 153–169.
| Breeding lucerne for Australia’s southern dryland cropping environments.Crossref | GoogleScholarGoogle Scholar |
Khakdan F, Alizadeh H, Ranjbar M (2018) Molecular cloning, functional characterization and expression of a drought inducible phenylalanine ammonia-lyase gene (ObPAL) from Ocimum basilicum L. Plant Physiology and Biochemistry 130, 464–472.
| Molecular cloning, functional characterization and expression of a drought inducible phenylalanine ammonia-lyase gene (ObPAL) from Ocimum basilicum L.Crossref | GoogleScholarGoogle Scholar | 30077922PubMed |
Koukol J, Conn EE (1961) The metabolism of aromatic compounds in higher plants. IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare. Journal of Biological Chemistry 236, 2692–2698.
| The metabolism of aromatic compounds in higher plants. IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare.Crossref | GoogleScholarGoogle Scholar |
Lee T-H, Tang H, Wang X, Paterson AH (2013) PGDD: a database of gene and genome duplication in plants. Nucleic Acids Research 41, D1152–D1158.
| PGDD: a database of gene and genome duplication in plants.Crossref | GoogleScholarGoogle Scholar | 23180799PubMed |
Lescot M, Patrice D, Gert T, Kathleen M, Yves M, Yves V, Pierre R, Stephane R (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327.
| PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences.Crossref | GoogleScholarGoogle Scholar | 11752327PubMed |
Li G, Wang H, Cheng X, Su X, Zhao Y, Jiang T, Jin Q, Lin Y, Cai YP (2019) Comparative genomic analysis of the PAL genes in five Rosaceae species and functional identification of Chinese white pear. PeerJ 7, e8064
| Comparative genomic analysis of the PAL genes in five Rosaceae species and functional identification of Chinese white pear.Crossref | GoogleScholarGoogle Scholar | 31824757PubMed |
Li J, An YY, Wang LJ (2020) Transcriptomic analysis of Ficus carica peels with a focus on the key genes for anthocyanin biosynthesis. International Journal of Molecular Sciences 21, 1–19.
| Transcriptomic analysis of Ficus carica peels with a focus on the key genes for anthocyanin biosynthesis.Crossref | GoogleScholarGoogle Scholar |
Maas EV, Hoffman GJ (1977) Crop salt tolerance-current assessment. Journal of the Irrigation and Drainage Division 103, 115–134.
| Crop salt tolerance-current assessment.Crossref | GoogleScholarGoogle Scholar |
MacDonald MJ, D’Cunha GB (2007) A modern view of phenylalanine ammonia lyase. Biochemistry and Cell Biology 85, 273
| A modern view of phenylalanine ammonia lyase.Crossref | GoogleScholarGoogle Scholar | 17612622PubMed |
Mamat A, Ayup M, Zhang XL, Ma K, Mei C, Yan P, Liqun Han LQ, Wang JX (2019) Pulp lignification in Korla fragrant pear. European Journal of Horticultural Science 84, 263–273.
| Pulp lignification in Korla fragrant pear.Crossref | GoogleScholarGoogle Scholar |
Min XY, Wu HL, Zhang Z, Wei XY, Jin XY, Ndayambaza B, Wang YR, Liu WX (2019) Genome-wide identification and characterization of the aquaporin gene family in Medicago truncatula. Journal of Plant Biochemistry and Biotechnology 28, 320–335.
| Genome-wide identification and characterization of the aquaporin gene family in Medicago truncatula.Crossref | GoogleScholarGoogle Scholar |
Mouttet R, Escobar-Gutiérrez A, Esquibet M, Gentzbittel L, Mugniéry D, Reignault P, Sarniguet C, Castagnone-Sereno P (2014) Banning of methyl bromide for seed treatment: could Ditylenchus dipsaci again become a major threat to alfalfa production in Europe? Pest Management Science 70, 1017–1022.
| Banning of methyl bromide for seed treatment: could Ditylenchus dipsaci again become a major threat to alfalfa production in Europe?Crossref | GoogleScholarGoogle Scholar | 24482310PubMed |
Nguyen T-N, Son S, Jordan MC, David B, Belay T (2016) Lignin biosynthesis in wheat (Triticum aestivum L.): its response to waterlogging and association with hormonal levels. BMC Plant Biology 16, 28
| Lignin biosynthesis in wheat (Triticum aestivum L.): its response to waterlogging and association with hormonal levels.Crossref | GoogleScholarGoogle Scholar | 26811086PubMed |
O’Rourke JA, Fu FL, Bucciarelli B, Yang SS, Samac DA, Lamb JFS, Monteros MJ, Graham MA, Gronwald JW, Krom N, Li J, Dai X, Zhao PX, Vance CP (2015) The Medicago sativa gene index 1.2: a web-accessible gene expression atlas for investigating expression differences between Medicago sativa subspecies. BMC Genomics 16, 502
| The Medicago sativa gene index 1.2: a web-accessible gene expression atlas for investigating expression differences between Medicago sativa subspecies.Crossref | GoogleScholarGoogle Scholar | 26149169PubMed |
Olsen KM, Lea US, Slimestad R, Verheul M, Lillo C (2008) Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental triggered flavonoid synthesis. Journal of Plant Physiology 165, 1491–1499.
| Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental triggered flavonoid synthesis.Crossref | GoogleScholarGoogle Scholar | 18242769PubMed |
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiology 133, 1051–1071.
| Genome-wide characterization of the lignification toolbox in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 14612585PubMed |
Reichert AI, He X-Z, Dixon RA (2009) Phenylalanine ammonia-lyase (PAL) from tobacco (Nicotiana tabacum): characterization of the four tobacco PAL genes and active heterotetrameric enzymes. The Biochemical Journal 424, 233–242.
| Phenylalanine ammonia-lyase (PAL) from tobacco (Nicotiana tabacum): characterization of the four tobacco PAL genes and active heterotetrameric enzymes.Crossref | GoogleScholarGoogle Scholar | 19725811PubMed |
Ren W, Wang Y, Xu A, Zhao YG (2019) Genome-wide identification and characterization of the phenylalanine ammonia-lyase (PAL) gene family in Medicago truncatula. Legume Rese Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. Legume Research 42, 461–466.
| Genome-wide identification and characterization of the phenylalanine ammonia-lyase (PAL) gene family in Medicago truncatula. Legume Rese Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase.Crossref | GoogleScholarGoogle Scholar |
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nature Protocols 3, 1101–1108.
| Analyzing real-time PCR data by the comparative CT method.Crossref | GoogleScholarGoogle Scholar | 18546601PubMed |
Shang QM, Liang L, Dong CJ (2012) Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L. Planta 236, 1093–1105.
| Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L.Crossref | GoogleScholarGoogle Scholar | 22572777PubMed |
Shen C, Du H, Chen Z, Lu H, Zhu F, Chen H, Meng X, Liu Q, Liu P, Zheng L, Li X, Dong J, Liang C, Wang T (2020) The chromosome-level genome sequence of the autotetraploid alfalfa and resequencing of core germplasms provide genomic resources for alfalfa research. Molecular Plant 13, 1250–1261.
| The chromosome-level genome sequence of the autotetraploid alfalfa and resequencing of core germplasms provide genomic resources for alfalfa research.Crossref | GoogleScholarGoogle Scholar | 32673760PubMed |
Smith A H, Imlay JA, Mackie RI (2003) Increasing the oxidative stress response allows Escherichia coli to overcome inhibitory effects of condensed tannins. Applied and Environmental Microbiology 69, 3406–3411.
| Increasing the oxidative stress response allows Escherichia coli to overcome inhibitory effects of condensed tannins.Crossref | GoogleScholarGoogle Scholar | 12788743PubMed |
Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (2008) Synteny and collinearity in plant genomes. Science 320, 486–488.
| Synteny and collinearity in plant genomes.Crossref | GoogleScholarGoogle Scholar | 18436778PubMed |
Timothy B, Mikael B, Buske FA, Martin F, Charles EG, Luca C, Ren JY, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208.
| MEME SUITE: tools for motif discovery and searching.Crossref | GoogleScholarGoogle Scholar |
Valifard M, Mohsenzadeh S, Niazi A, Moghadam A (2015) Phenylalanine ammonia lyase isolation and functional analysis of phenylpropanoid pathway under salinity stress in Salvia species. Australian Journal of Crop Science 9, 656–665.
Vogt T (2010) Phenylpropanoid biosynthesis. Molecular Plant 3, 2–20.
| Phenylpropanoid biosynthesis.Crossref | GoogleScholarGoogle Scholar | 20035037PubMed |
Wanner LA, Li GQ, Ware D, Somssich IE, Davis KR (1995) The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Molecular Biology 27, 327–338.
| The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 7888622PubMed |
Wettberg EJV, Stanton ML, Whittall JB (2010) How anthocyanin mutants respond to stress: the need to distinguish between stress tolerance and maximal vigour. Evolutionary ecology research 12, 457–476.
| How anthocyanin mutants respond to stress: the need to distinguish between stress tolerance and maximal vigour.Crossref | GoogleScholarGoogle Scholar |
Yan F, Li HZ, Zhao P (2019) Genome-Wide Identification and Transcriptional Expression of the PAL Gene Family in Common Walnut (Juglans Regia L.). Genes 10, 1–16.
| Genome-Wide Identification and Transcriptional Expression of the PAL Gene Family in Common Walnut (Juglans Regia L.).Crossref | GoogleScholarGoogle Scholar |
Yuan J, Amend A, Borkowski J, Demarco R, Bailey W, Liu Y, Xie G, Blevins R (1999) MULTICLUSTAL: a systematic method for surveying Clustal W alignment parameters. Bioinformatics 15, 862–863.
| MULTICLUSTAL: a systematic method for surveying Clustal W alignment parameters.Crossref | GoogleScholarGoogle Scholar | 10705440PubMed |
Zhao Q, Dixon RA (2011) Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends in Plant Science 16, 227–233.
| Transcriptional networks for lignin biosynthesis: more complex than we thought?Crossref | GoogleScholarGoogle Scholar | 21227733PubMed |



