Flow-based assembly of nucleic acid-loaded polymer nanoparticles
Zeyan Xu A , Joshua McCarrol B C D and Martina H. Stenzel A D *
A D *
A School of Chemistry, University of New South Wales, Sydney, NSW 2052, Australia.
B Children’s Cancer Institute, Lowy Cancer Research Centre, University of New South Wales, Sydney, NSW 2052, Australia.
C School of Women’s and Children’s Health, University of New South Wales, Sydney, NSW 2052, Australia.
D UNSW RNA Institute, University of New South Wales, Sydney, NSW 2052, Australia.
Abstract
Since the development of messenger RNA (mRNA)-based SARS-CoV-2 (COVID-19) vaccines, there is increased public awareness of the importance of nanoparticles, in this case lipid nanoparticles, to ensure safe delivery of an active compound. To ensure the formation of high-quality nanoparticles with reproducible results, these lipid nanoparticles are assembled with the nucleic acid drug using flow-based devices. Although flow assembly using lipid nanoparticles for nucleic acid delivery is well described in the literature, only a few examples use polymers. This is surprising because the field of polymers for nucleic acid delivery is substantial as hundreds of polymers for nucleic acid delivery have been reported in the literature. In this review, we discuss several aspects of flow-based assembly of nucleic acid-loaded polymer nanoparticles. Initially, we introduce the concept of chip-based or capillary-based systems that can be either used as single-phase or multiphase systems. Initially, researchers have to choose the type of mixing, which can be active or passive. The type of flow, laminar or turbulent, also significantly affects the quality of the nanoparticles. We then present the type of polymers that have so far been assembled with mRNA, small interfering RNA (siRNA) or plasmid DNA (pDNA) using flow devices. We discuss effects such as flow rate, concentration and polymer lengths on the outcome. To conclude, we highlight how flow assembly is an excellent way to generate well-defined nanoparticles including polyplexes in a reproducible manner.
Keywords: DNA, drug delivery, flow assembly, gene therapy, microfluidics, nanomedicine, polymers, RNA, self‐assembly.
Introduction
Nanoparticles are widely used for drug delivery.1 From initial concepts, this field has now matured and many drug-loaded nanoparticles have now entered clinical trials or are available on the market.2,3 The impact of nanoparticles in the medical field became evident during the SARS-CoV-2 (COVID-19) pandemic when lipid nanoparticles made it possible to deliver the mRNA (messenger ribonucleic acid) vaccines, which cannot be administered otherwise.4 Although nanoparticles can enhance the delivery of most drugs, no other class of drugs benefits from nanoparticles as much as nucleic acid-based therapeutics since nanoparticles can protect the drug while enhancing circulation time and cellular uptake.5,6 Lipid nanoparticles are a great success in vaccine delivery; however, other diseases require more tailor-made nanoparticle solutions and therefore this field is still evolving. When new nanoparticles are developed,7 researchers usually consider the interface between the nanoparticle and the biological environment as parameters such as surface chemistry, type of nanoparticles, size and shape and other physical properties influence the fate of the nanoparticle.8–10
Size is important in drug delivery.11 Smaller nanoparticles usually have longer circulation times in the blood stream as demonstrated with gold nanoparticles between 10 and 100 nm.12,13 Although reducing the size can be beneficial, nanoparticles below 10 nm are readily removed by renal filtration.14 Meanwhile, large particles above 100 nm can be quickly cleared by the mononuclear phagocytic system (MPS)15,16 or they accumulate in the liver and spleen.17 There are size sweet spots when it comes to tumour extravasation and penetration. Smaller nanoparticles, well below 50 nm, penetrate deeper into tumours,13,18 whereas extravasation, which relies on translocation through fenestration in the tumour vasculature, is often more efficient with nanoparticles above 50 nm in size. It is therefore evident that size plays a pivotal role in drug delivery, but there is also a paradox as each delivery step – circulation, extravasation, penetration and cell uptake – favours different nanoparticle sizes.19
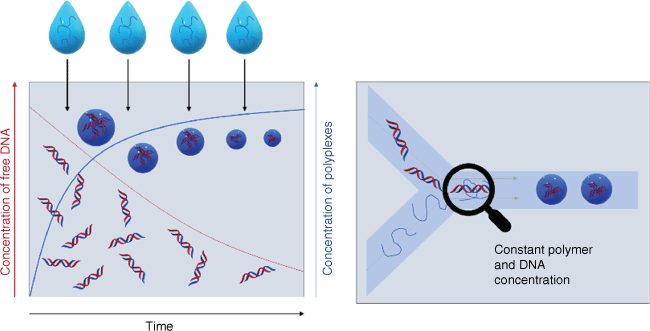
It is therefore essential not only to determine the average size of nanoparticles, but also evaluate the particle size distribution because a broad size distribution would result in unexpected biodistributions as the smaller fraction behaves differently to the larger fraction of the sample. There is often little attention to size distribution but reports on polydispersity indices (PDIs) measured using dynamic light scattering (DLS) of more than 0.2 are common. PDI is defined as the square of the standard deviation over the mean size, which means that a mean size of 50 nm and a PDI value of 0.2 has a standard deviation of 22 nm! Broad nanoparticle distributions are typically obtained when nanoparticles are prepared by hand in the laboratory, and, despite the use of syringe pumps and other control measures, broad distributions are ingrained in the preparation techniques. This is evident when mixing cationic polymers with negatively charged therapeutic nucleic acids. A plethora of different cationic polymers have been condensed with mRNA, plasmid DNA (pDNA), antisense oligonucleotides (ASOs) and similar therapeutics by electrostatic interaction.5 In a typical experiment, the polymer is added to the nucleic acid or vice versa, which means that during the addition process, the composition of the solution changes, which changes the nanoparticle properties in the next step (Fig. 1). These heterogeneities in this bulk mixing process can then result in a broad distribution of nanoparticle sizes and compositions. Electrostatic interactions are fairly strong and there is limited opportunity for rearrangement once the polyplex has been formed, in particular when the polymer or the therapeutic nucleic acids are large. PDI values are often not reported, but if they are, they are typically above 0.2.20–23 If we carefully scrutinise our own work, we find that PDIs are seldomly below 0.2 and, at times, extensive filtering is required to narrow the particle size distribution, which can have its own problems.22–25 Polyplex formation (often also called coacervate or polyion complex formation) is not only associated with a lack of uniformity, but also with challenges in reproducing nanoparticle formation. In the present review, we summarise the approaches to well-defined nucleic acid-loaded nanoparticles using flow-based devices that can serve as laboratory and industry-based assembly lines and provide nanoparticles of a certain size in a reliable manner. The focus here will be on polymers or dendrimers as the multitude of charges on the drug carrier make it particularly difficult to generate nanoparticles with small size distributions.5
Comparison of bulk mixing (left) and flow mixing (right) of nucleic acid and polymers. In bulk mixing, the polymer is continuously added to the solution and the amount of free nucleic acid therefore changes over time. The ratio of free DNA and polymer changes over time, and therefore the composition of the polyplexes. In flow mixing, the concentration of nucleic acid and polymer remains constant over the course of mixing.
Flow-based nanoparticle preparation devices
Low reproducibility of nanoparticle preparation, high polydispersity and a lack of avenues to scale up the process to the kilogram scale were identified as challenges that hamper nanomedicine development.26 The lack of reproducibility and the urgently needed high throughput were already a subject of discussion when the field of gene delivery was established. Early devices include a system that combined two syringes that were able to mix two solutions, one with DNA and the other with lipoplex, at a controlled rate. The PLEXER was able to produce particles of ~300 nm in a reproducible manner independently of the operator.27 The underpinning idea was refined over the next decade and the synthesis of lipid nanoparticles loaded with nucleic acid drugs can now be carried out at a large scale,28 exemplified by mRNA COVID-19 vaccines.29
Chip-based and capillary-based flow devices
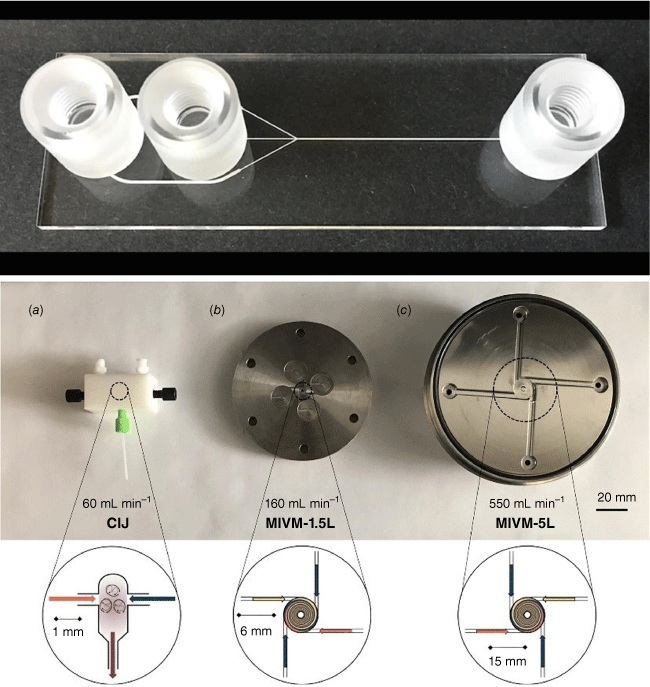
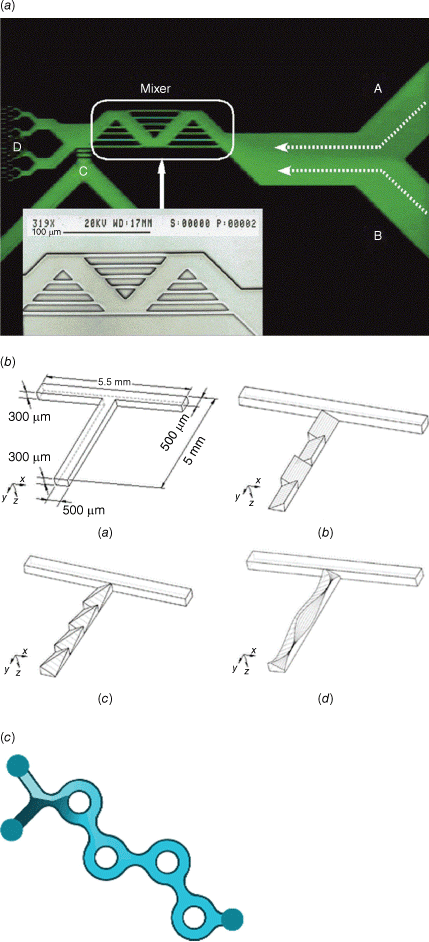
There are typically two types of flow devices, chip-based or capillary-based, with the commonality that two or more solutions are quickly combined.28 Chip-based systems are prepared from silicon, glass or occasionally polymer and they have all functions – solution combination, mixing and equilibrium line – integrated in one device.30 The channels are often square, owing to the preparation technique, with channel diameters of less than 1000 µm. Glass-based devices are the most robust as they can withstand organic solvents and high temperature, but they are usually more expensive and fragile. In the field of nucleic acid delivery where ambient temperatures and aqueous systems are used, cheaper polymer and silicone-derived microfluidic systems can be used. Capillary-based systems are commonly assembled from tubing made for example from Teflon and a mixer that can be made from a different material. The tubing diameter is often larger than that found in microfluidic devices although there are no limitations. The most famous example is the flash nanoprecipitation system by Johnson and Prud’homme who reported their system first in this journal in 2003.31 The central part is the design of the mixer that enables rapid combination of all solutions (Fig. 2).32 This system later became the foundation for the production of the COVID-19 vaccine.
Top: microfluidic devices as sold by Darwin Microfluidics, Herringbone Mixer – Glass Chip shown here; bottom: images of the three mixers: (a) confined impinging jet mixer (CIJ), (b) multi-inlet vortex mixer (MIVM)-1.5L, and (c) MIVM-5L, reproduced from Feng et al. (2019)32 with permission from Springer Nature under Creative Commons CC BY.
Single-phase flow system
Active and passive mixing
The central focus in flow-based devices is the development of suitable mixing approaches. Mixing of the solutions can either be achieved by active mixing or passive means (Fig. 3).33 Active mixing requires the integration of an externally controlled mixer such as acoustic, ultrasonic, thermal or dielectrophoretic force actuation, pressure perpetuation or electromagnetic micromixer.34 The mixer can also be integrated in the design, as demonstrated by a design that uses an in-line rotating impeller to mix solutions.35
Active and passive mixing with either laminar or chaotic mixing. Reproduced from Ober et al. (2015)35 with permission of PNAS. Pe, Péclet number; Q, net volumetric flow rate; , dimensionless rotation rate; l, d, , an are dimensions of the devices; , density; and , viscosity.
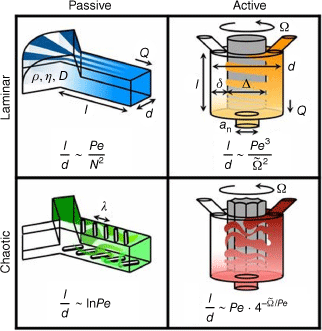
More commonly used in polymer laboratories, however, is passive mixing as many devices are commercially available, or they can be assembled from simple parts. The sole driving force is the pressure applied by the pump that determines the mixing rate. The solutions are mixed by applying a range of geometries, with Y- or T-shaped inlets the simplest designs (Fig. 4). The two solutions then mix at the interface. A way of increasing the size of this interface is hydrodynamic flow focusing (HFF) where the central jet has higher velocities than the sheathing fluid.36
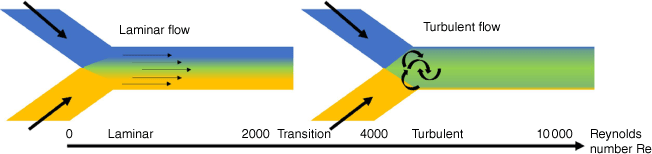
From laminar flow to turbulent flow
As the channels in which the two solutions are combined are narrow, laminar flow is usually observed. Laminar flow occurs when the two solutions flow in parallel without disruption, which contrasts with turbulent flow where random fluctuations are observed during mixing. Whether the flow is laminar or turbulent can be predicated by the Reynolds number (Re), which is given by:
where ρ is the density of the fluid (kg m–3), u is the flow speed (m s–1), L is the characteristic linear dimension (m) and μ is the dynamic viscosity of the fluid (kg m·s–1). Laminar flow transitions into turbulent flow at Reynolds numbers approximately above 1800, well above the values obtained for parallel flow in a microfluidic device (Fig. 5).37 Whether a device produces laminar or turbulent flow can be quickly observed by using two coloured solutions.38
The design of new microfluidic devices that enable faster mixing therefore became an integral part of this field. Passive micromixers that can create turbulent flows have built-in obstacles in the channels that create chaotic advection. This can be achieved by intersecting channels, zigzag channels, three-dimensional serpentine structures, channels with embedded barriers and slanted walls or twisted channels (Fig. 6).34 Among all these devices, the staggered herringbone design has made a significant impact in this field over the last 20 years since it was first reported in 2002.39 The V-shaped grooves, which are offset, cause transverse flow of the fluid, resulting in turbulent mixing (Fig. 7).40 Since then, a range of devices have been developed, but the bifurcating mixer is highlighted here as it part of the NanoAssemblr Platform that is widely used to assemble lipid nanoparticles.41 This commercial assembly platform is now widely used in many research laboratories as its ease of operation allows even the non-expert to generate drug-loaded nanoparticles in a reproducible manner. The mixing chamber is based on the patented Dean Vortex Bifurcating Mixers (Fig. 6), which uses a design that resembles the bifurcating pattern produced by liquid jets discharging into a quiescent fluid.42
(a) Photomicrograph and SEM of a microfluidic mixer with intersecting channels (A and B are entry channels, and C and D are outlet channels)43; (b) schematic diagrams (upside down) of (a) T-mixer, (b) inclined mixer, (c) oblique mixer, and (d) wavelike mixer.44 Reproduced from Jen et al. (2003)44 with permission of the Royal Society of Chemistry. (c) Dean Vortex bifurcating mixers as used in the NanoAssemblr Platform, reproduced with permission from Precision Nanosystems.
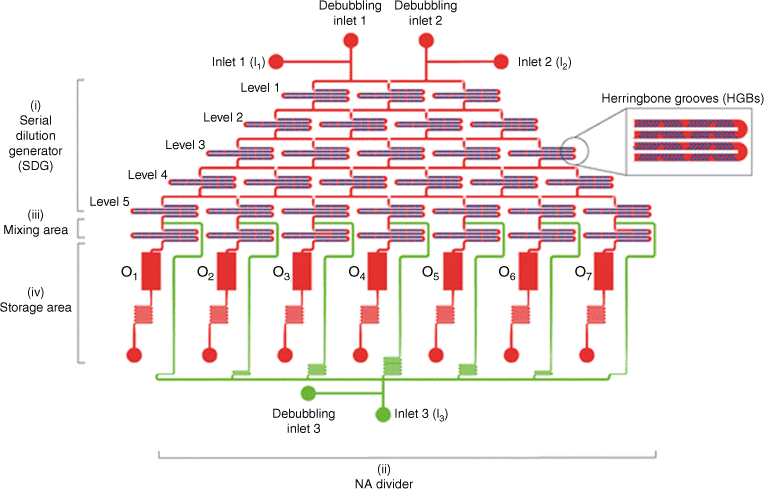
The microfluidic cartridge consists of (i) an upstream five-level serial dilution generator (SDG) integrated with herringbone grooves (HBGs), (ii) a nucleic acid (NA) divider, (iii) a mixing area, and (iv) a storage area. The polymer solution and the water-based medium are injected into the two main inlets (inlets 1 and 2, I1 and I2) through the SDG and moved to the downstream area. An additional inlet (I3) allows for the addition and distribution of the DNA solution to the seven mixing units, where the polymer and DNA are mixed to give rise to polyplexes. These are collected in the storage tanks. Every inlet has a debubbling port. The middle right panel is a magnified view of the staggered HBG units integrated into the cartridge channels. Reproduced from Protopapa et al. (2023)45 with permission of the Royal Society of Chemistry.
Single-flow devices for the preparation of drug-loaded nanoparticles
Both laminar and turbulent approaches are used to prepared drug-loaded nanoparticles such as polyplexes. What all these flow-based assembly approaches have in common is fast and continuous mixing on the nanoscale and the elimination of a concentration gradient as depicted in Fig. 1. Many devices have been developed to generate reliable nanoparticles for drug delivery and the choice of device depends on the specific drug loading process. Polyplexes are usually prepared by mixing an aqueous solution containing the cationic polymer with the aqueous nucleic acid solution. The ratio between both components such as the nitrogen-to-phosphorus (N/P) ratio, concentration, flow rate and many more parameters determine the outcome. The N/P ratio is especially important considering that nanoparticle formation is solely based on electrostatic interactions between both ingredients. Once formed, the polyplex size is locked in unless strong introduced competitive forces disrupt the interaction between the charges. Mixing using laminar flow relies on molecular diffusion, which can occasionally be insufficient when trying to obtain narrow size distributions. However, laminar flow is still sufficient to generate better-defined nanoparticles than bulk mixing alone.38,46,47 Sometimes, the nanoparticle size distribution is not improved compared with traditional preparation techniques to make polyplexes, but batch-to-batch variation is reduced, opening the door to large-scale production.48 By contrast, turbulent mixing was found to generate polyplexes with high control. Among these techniques, the staggered herringbone mixer is widely used to generate nanoparticles loaded with drugs such as doxorubicin49 or small interfering RNA (siRNA),50 generating nanoparticle sizes with high precision and PDIs as low as 0.02.50 The herringbone mixer enables the design of well-defined nanoparticles, but it does not shorten the optimisation workflow as each drug–drug carrier combination needs to be tested separately. To accelerate the process to identify suitable concentrations and, in the case of DNA delivery, suitable N/P ratios, a user-friendly microfluidic cartridge was prepared that can help screen seven N/P ratios in parallel (Fig. 7).45 Here, several herringbone mixers are assembled on one chip, enabling the production of seven solutions with different N/P ratios in one process.
Microfluidic-assisted nanoprecipitation (MF nanoprecipitation)
The formation of polyplexes based on water-soluble dendrimers and polymers is unique as, in contrast to other flow-based drug delivery approaches, no organic solvent is necessary. However, this only applies to soluble polymers and therefore excludes many degradable polymers such as polylactic acid (PLA). Nanoparticles based on hydrophobic polymers are in contrast prepared by mixing an organic solution containing the polymer and often the drug with an aqueous solution. Prerequisites are the miscibility of the organic solution and water and the immiscibility of the polymer in water. The relationship between these components is given by the ternary phase diagram of solute–solvent–non-solvent systems. When water is introduced into the system, the solubility of the polymer is reduced until at a specific water content, the binodal phase boundary is crossed, and thus nanoprecipitation occurs.51 This process is heavily used on the laboratory scale where the organic solution is slowly dropped into excess water, but assembling these nanoparticles using flow can improve size distribution and size control. Flow-based devices range from HFF devices (Fig. 4)52 and confined impinging jets mixers and to multi-inlet vortex mixers (Fig. 2).53,54 The assembly process is not only affected by the flow device and parameters such as flow rate but also by the complex phase diagram, which is determined by the nature of the solvent and the type of polymer. Therefore, mixing-induced nanoprecipitation using flow is complex.51 At the same time, the complexity opens the door to unusual nanostructured materials.51,55
Segmented flow–multiphase system
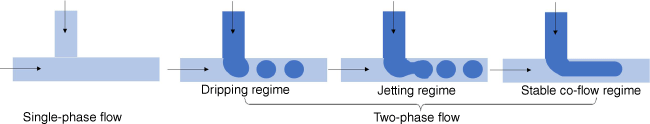
Segmented-flow microfluidic systems use two immiscible fluids or a gas and fluid that can aid the mixing process.56 With the addition of a new immiscible phase, droplets are formed that can assist the mixing process. Droplets are generated by an active method using valves or by a passive approach employing the inherent properties of the device and the solution. The droplet (dispersed) phase and the continuous phase can be combined from different direction such as by co-flowing, cross-flow (Fig. 8) or flow-focusing.57 The viscosity of each liquid, the interfacial tension between both, the flow rate and the geometry of the inlet determine the subsequent steps. Fairly high surface instabilities are required to form droplets, which are typically present when the interfacial tension γ is high, while viscosities µ and flow rate u are small in relation.58 This relationship is expressed by the dimensionless phase capillary number Ca = µ × u ÷ γ for the dispersed and the continuous phase, which can help to predict, among other influences, if droplets are formed or if the dispersed phase is not able to break up, resulting in the formation of jets (Fig. 8).58 These droplets can potentially coalesce and the introduction of stabilisers such as polymers or surfactants are required.59
Liquid–liquid systems, often named emulsion-based or droplet-based microfluidic system, are widely employed to generate nanoparticles for drug delivery.59–61 The size of the resulting nanoparticles is determined by the droplet size and can be predicted using the physical parameters of the system employed,58 but in general, the nanoparticles tend to be above 100 nm in size.62 The system is therefore useful to generate larger nanoparticles for nucleic acid delivery,63 although it is possible to obtain small nanoparticles.64 The clear advantage of this system is the ability to obtain non-spherical nanoparticles, but also the ability to create complex morphology when expanding the system from a two-phase to a multi-phase set-up.64 Moreover, droplet-based microfluidics offer the opportunity to entrap nucleic acids into neutral hydrogels.65 Whereas single-phase microfluidic systems rely on nanoprecipitation or electrostatic interaction paired with colloidal stabilisation to generate nanoparticles, the nature (size, shape) of the aqueous hydrogel droplets in droplet-based microfluidics is determined by the continuous organic phase. Droplet-based microfluidic systems, therefore, offer the opportunity to entrap these types of drugs in water-swollen polyethylene glycol (PEG) hydrogels.65
Comparison of techniques
It is difficult to single out a technique that is superior to the others as this depends on various parameters. The first consideration is the type of polymer and drug and their solubility. As many polymers and drugs are not water-soluble, researchers need to identify a set-up that allows nanoprecipitation such as MF or flash nanoprecipitation. Hydrogel nanoparticles are in contrast better produced in multiphase systems.
Another aspect that influences the choice is the amount of nanoparticles that need to be prepared. Whereas a coaxial turbulent jet mixer can produce 3.15 kg of nanoparticles per day, single-channel microfluidic devices can only generate a few milligrams in the same timeframe.66 However, this is often sufficient for further analysis in the development stage. The researcher might even prefer small-scale set-ups when trying to optimise the nanoparticle synthesis first.
Most important is, however, the knowledge and understanding required when choosing a system. Commercial assembly systems such as the NanoAssemblr Platform and other systems that are on the market are a great starting point for researchers who have no knowledge of fluid dynamics. The former can easily be operated with a push of a button. However, these systems come with a price tag that may not be affordable for a laboratory. In such cases, researchers can choose from a range of commercially available microfluidic devices that can be easily assembled with syringe pumps or, if the budget permits, high-accuracy microfluidic pumps. As polydimethylsiloxane (PDMS)-based or glass-based channels are transparent, it is also possible to observe the flow with the help of a microscope. This can help with troubleshooting as well as identifying areas with precipitated product. Researchers with knowledge in fluid dynamics will most likely enjoy the design of new mixing channels that can achieve better outcomes.
Preparation of nanoparticles loaded with nucleic acid using flow
Types of polymers employed to prepare DNA-based nanoparticles in flow
Flow-based assembly of nanoparticles has numerous benefits such as uniform mixing, ease of control and high efficiency,67 resulting in good control in terms of dispersity, average particle size distribution and morphology.68,69 Commonly, under bulk mixing conditions, polymers are dissolved in a solvent and then combined with another solution that contains the drug. Nanoparticle formation is then triggered by the formation of electrostatic interaction or by the addition of a non-solvent. The particle size, polydispersity and scaled-up production of nanoparticles may be affected by the mixing efficiency and time (Fig. 1). Although these issues apply to any nanoparticle synthesis, no other field is as much affected by heterogeneous mixing condition as the field of polymer nanoparticles. Polymers can easily be kinetically trapped, and strong forces between polymers and nucleic acid prevent the formation of nanoparticles that are in equilibrium with their environment. However, these characteristics can be controlled by microfluidics.70 Many mixing technologies exist for nanoparticle preparation by flow as outlined above. This review, however, focuses on the preparation of polymer nanoparticles for nucleic acid delivery.
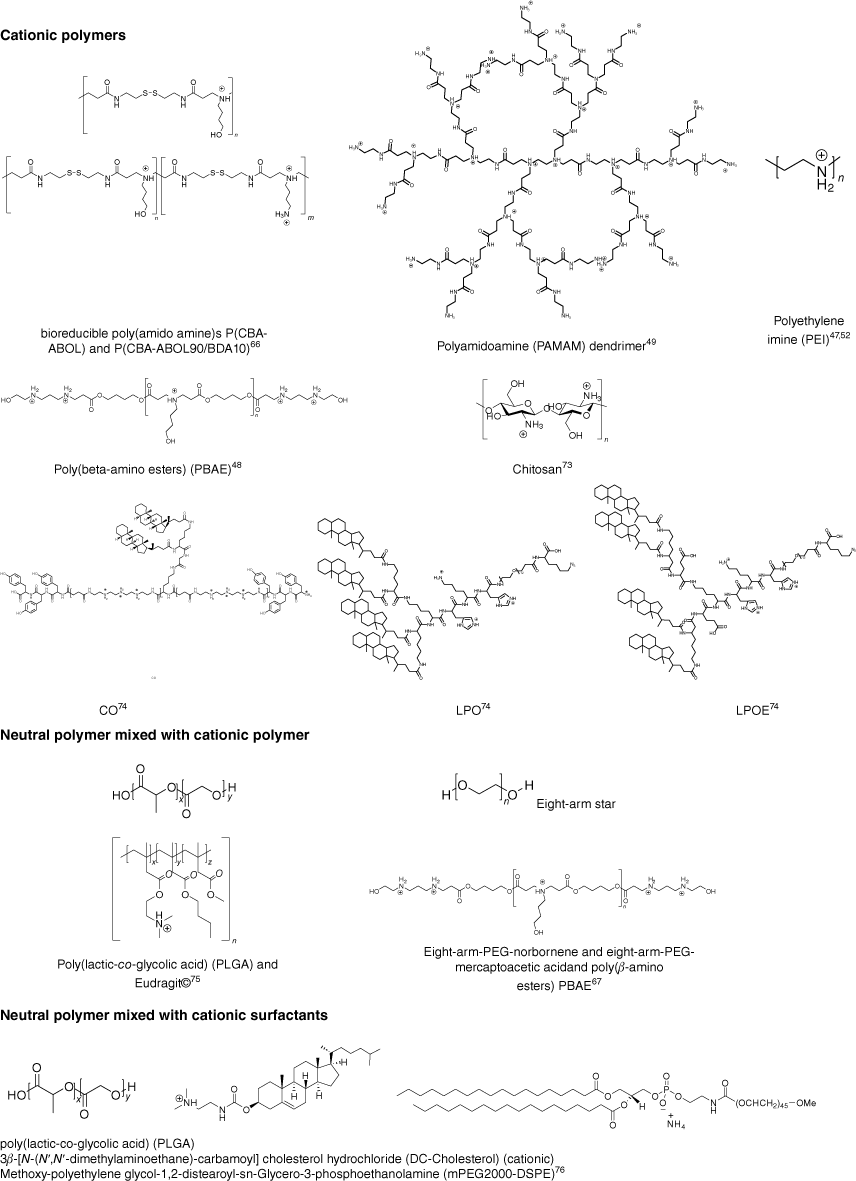
In general, there are two approaches to nucleic acid-loaded nanoparticles. Nanoparticles are either formed by compacting cationic polymers with nucleic acid to form polyplexes or by entrapping the negatively charged load into neutral, often hydrophobic polymers, usually with the help of a cationic polymers or cationic surfactant. Although many polymers have been explored for the delivery of nucleic acid,5 the number of reports on flow-based assembly is limited (Fig. 9). In general, three different approaches have been explored: mixing of two aqueous solution containing polymer and nucleic acid respectively, nanoprecipitation and droplet-based microfluidics (Table 1).
Polymers used for the preparation of nucleic acid-loaded nanoparticles using flow assembly; the cationic charges shown are dependent on the pH value (CO, LPO and LPOE are abbreviations given by Loy et al.71 as core oligomer, lipid anchored PEG12 oligomer, and lipid anchored PEG12 oligomer without glutamic acid respectively).
| Polymer | Drug | Technique | Flow mixing | Bulk mixing | Remarks | References | |||
|---|---|---|---|---|---|---|---|---|---|
| Size (nm) | PDI | Size (nm) | PDI | ||||||
| Cationic polymers | |||||||||
| PBAE | DNA | Mixing of aqueous solutions in flow focusing devices with pinch channel | No difference between MF and BM and no size data shown | Wilson et al. (2017) 47 | |||||
| P(CBA-ABOL) | pDNA | Water droplets in fluorocarbon oil emulsion-based microfluidic system | 100 | 0.15 | 166 | 0.34 | Grigsby et al. (2013) 63 | ||
| mRNA | 113.1 | 0.18 | 222 | 0.32 | |||||
| PEI | pDNA | Mixing of aqueous solutions in cartridge with seven chaotic serial dilution generator and dividers for variation of N/P ratios | 98 | 0.1 | 196 | >0.4 | N/P ratio 50 | Protopapa et al. (2023) 45 | |
| PEI | pDNA | Mixing of aqueous solutions in microfluidic hydrodynamic focusing device | 494 | 898 | N/P ratio 3.3, PDI not reported | Koh et al. (2009) 46 | |||
| PAMAM dendrimer | siRNA | Mixing of aqueous solution using funnel-shaped micromixer | 86 | 0.18 | 87 | >0.20 | N/P ratio 20 | Agnoletti et al. (2017) 48 | |
| CO + siRNA + LPO | siRNA | Mixing of aqueous solution in a single or a double meander channel microfluidic chip | 114.7 | 0.14 | 416.2 | 0.71 | Loy et al. (2021) 71 | ||
| CO + siRNA + LPOE | 141.9 | 0.23 | 128.2 | 0.47 | |||||
| Chitosan | pDNA | Mixing of aqueous solutions in in-line mixing system (AIMS) consisting of Y-mixer and pinch valves | 199 | 0.68 | 165 | 0.25 | Naeini et al. (2017) 72 | ||
| siRNA | 48 | 0.15 | 98 | 0.21 | |||||
| Cationic polymer and neutral polymers | |||||||||
| Eight-arm-PEG-norbornene and eight-arm-PEG-mercaptoacetic acid and poly(β-amino esters) PBAE | pDNA | Water in oil T-junction droplet break-up microfluidic device | 41 × 103–142 × 103 | Size of microspheres dependent on channel size | Deveza et al. (2015) 65 | ||||
| PLGA and Eudragit | pDNA | Microfluidic-assisted nanoprecipitation | 170 | 0.25 | 98 | 0.21 | Zoqlam et al. (2021) 73 | ||
| Cationic surfactant and neutral polymer | |||||||||
| PLGA DC-cholesterol (cationic) mPEG2000-DSPE (surfactant) | pDNA | Microfluidic-assisted nanoprecipitation in toroidal micromixer | 83 | 0.11 | 136 | 0.23 | Santhanes et al. (2022) 74 | ||
MF, mixing by microfluidics; BM, bulk mixing.
The simplest is the assembly of polyplexes, which are made using cationic polymers and nucleic acids. The nanoparticles are then formed as a result of electrostatic binding,75 which can be controlled by ionic strength and pH value, but also the nature of the cationic polymer.76,77 Typical cationic polymers such as PEI, PAMAM dendrimer and chitosan were used to prepare polyplexes by flow, but some more unusual structures were also explored, as summarised in Fig. 9. This is, of course, a far cry from the myriad of cationic polymer structures that have already been tested for nucleic acid delivery and highlights that the field of flow assembly is only emerging.5 The set-up appears fairly simple as two aqueous solutions are mixed either using turbulent flow such as in a microfluidic cartridge that contains herringbone grooves,45 or by laminar flow in a microfluidic hydrodynamic focusing device,47 which can have an advanced design and include single or double meandering channels.71 The latter publication also discusses in detail the intricacies of flow assembly such as the pump pressure profile and the design of a system that can be used for scaling up. Here, the size of the final nanoparticles is the result of the chosen microfluidic set-up, type of polymer and payload, concentrations and flow rate (Re number). PAMAM dendrimers were mixed with siRNA using a funnel-shaped micromixer with several inlets. COMSOL Multiphysics software simulation suggested a laminar flow whose pattern changed depending on the flow rate.48 (Fig. 9 and Table 1)
Switching from a single phase to an emulsion system allows the design of PEG-based hydrogel nanoparticles that can be filled with PBAE–pDNA (Fig. 9) complexes. The size of the hydrogel particles that were prepared using eight-arms stars, crosslinked with thiol-ene chemistry, was determined by the set-up of the T-junction droplet break-up microfluidic device.65 The resulting size of the aqueous PEG filled droplets was controlled by the channel dimensions and was adjusted between 41 and 142 µm. Significantly smaller particles in the nanometre range were obtained when bioreducible poly(amido amine)s P(CBA-ABOL) and P(CBA-ABOL90/BDA10)63 (Fig. 9 and Table 1) were loaded with mRNA and pDNA. The cationic polymer and the nucleic acid were directly mixed in the aqueous droplet.
Additional components like neutral hydrophobic polymers, which are added to the mixture that contains cationic polymers and DNA, can help control size and stability and therefore transfection efficiency. As soon as hydrophobic polymers are present, the use of organic solvents is essential. Nanoparticles are then formed when the organic solution is mixed with an aqueous solution; thus, microfluidic-assisted nanoprecipitation is employed. Although the use of organic solvent is usually undesirable, it is outweighed by the advantages hydrophobic and biodegradable polyesters such as poly(lactic-co-glycolic) acid (PLGA), polycaprolactone (PCL) and PLA provide.78 Microfluidic-assisted nanoprecipitation was used to form PLGA-Eudragit nanoparticles (Fig. 9 and Table 1). The PLGA-Eudragit polymers (hydrophobic PLGA polymer and cationic Eudragit polymer) were dissolved in acetone and mixed with water in the microfluid chip to form a core with positive charge that was able to entrap DNA by electrostatic interactions.73 As the resulting nanoparticles are hydrophobic and may have a tendency to aggregate, hydrophilic surfactants are added. This was achieved by introducing mPEG2000-DSPE into the aqueous phase (Fig. 9 and Table 1). In this study, PLGA was used as the hydrophobic polymer dissolved in the organic phase together with cationic DC-cholesterols. The two phases were mixed in a toroidal micromixer to form spherical DNA-loaded nanoparticles. The hydrophobic polymer and surfactant form the core while the amphiphilic mPEG2000-DSPE is inserted at the interface, with the PEG block providing steric stabilisation and water solubility. As the core is positively charged thanks to the presence of the cationic surfactant, DNA can be bound by electrostatic interaction. In the final nanoparticle, the PEG chains covered the DNA, which prevented particle aggregation.74
Comparison of bulk mixing with flow mixing
In the laboratory, nucleic acid-loaded nanoparticles are usually prepared by bulk mixing where the aqueous solution containing the cationic polymers is added to the DNA–siRNA solution or vice versa. As discussed earlier, local heterogeities often prevent the formation of well-defined nanoparticles with low dispersities while it is difficult to target a specific size (Fig. 1). Microfluidic techniques can improve the efficiency of mixing of these two solutions to produce DNA–siRNA-loaded nanoparticles of high quality compared with the bulk method (Table 1).79 For example, using a chaotic serial dilution generator (SDG), which is a stand-alone microfluidic cartridge that was designed to control the features of nanoparticles, smaller sized PEI-DNA polyplexes (145 nm) with lower dispersities (PDI < 0.2) were obtained compared with bulk mixing (176 nm, PDI > 0.5 at N/P ratio of 40).45 Using the same materials, Koh et al. employed a different microfluidic device, a stand-alone microfluidic cartridge (a chaotic SDG), to obtain similar results. The sizes of the polyplexes were bigger compared with those using the technique by Protopapa et al., but the flow assembly still led to smaller nanoparticles (494 nm) and lower PDIs than bulk mixing (898 nm).47 Although both research teams observed the same size and PDI reduction in flow mixing compared with bulk mixing, the differences in reported nanoparticle size can be assigned to the N/P ratio used. The formation of better nanoparticles was also reported in the study by Loy et al.71 who mixed the cationic oligomers CO, LPO and LPOE shown in Fig. 9 with siRNA (CO + siRNA + LPO and CO + siRNA + LPOE) using bulk mixing and two different microfluidic devices, a single meander channel microfluidic chip and a double meander channel microfluidic chip. The size and PDI of the two polyplexes assembled using flow (CO + siRNA + LPO 114.7 nm, PDI < 0.20 and CO + siRNA + LPOE 128.2 nm, PDI 0.468) are smaller than from bulk mixing (CO + siRNA + LPO 141.9 nm, PDI 0.23 and CO + siRNA + LPOE 416.2 nm, PDI > 0.2). This chosen microfluidic device was observed to be highly efficient to produce small sizes and uniform siRNA-loaded nanoparticles.71
However, improved outcome using flow assembly is not automatically guaranteed. Agnoletti et al. found that improvement compared with bulk mixing can be a matter of N/P ratio as well as flow ratios.48 Polyplexes obtained from PAMAM and siRNA only led to better outcomes compared with bulk mixing when the ratio between the flow rates of the two inlets was adjusted.48 Little size improvement was also reported using poly(beta-amino esters) (PBAE). Wilson et al. used a microfabricated PDMS–glass chip to generate nanoparticles, but they found little advancement in terms of size and PDI. However, the device allowed efficient scale up, good control over the assembly process and the ability to form nanoparticles that were stable under long-term storage.47
As mentioned in the previous section, nanoprecipitation is one of the most commonly used nanoparticle fabrication methods to reduce the size and PDI of the nanoparticles.80 Santhanes et al. used microfluidic nanoprecipitation and succeeded in generating smaller nanoparticles (83 nm) with lower dispersities (PDI < 0.2) than using bulk mixing nanoprecipitation.74 By contrast, Zoqlam et al. observed very different outcomes as the size and PDI of the nanoparticles generated using microfluidic nanoprecipitation were larger than with bulk mixing nanoprecipitation.73 It is, however, not possible to compare the quality of the devices as the differences may stem potentially from the polymers employed as Zoqlam et al. used PEG-based surfactants, which can prevent particle aggregation and aid in the mixing process.
Choice of the right microfluidic device is, however, essential to generate good nanoparticles, as shown by the study by Naeini et al.72 The authors developed an in-line mixing system (AIMS) that was improved over the course of the study. This system was used to compare chitosan–siRNA and chitosan–pDNA (Fig. 9) polyplex formation. Compared with bulk mixing, flow mixing at high Reynolds numbers was found to be a powerful tool to minimise the size and PDI of the resulting nanoparticles while being able to scale up production. With increasing siRNA concentration, the size of the flow-assembled chitosan–siRNA increased from 40 to 60 nm while the PDI of the polyplexes remained below 0.2. This contrasts with large-sized polyplexes (from 99 to 179 nm) with broad size distributions that were the result of bulk mixing. Different results were obtained when pDNA was used to prepare chitosan–pDNA polyplexes as the bulk mixing yielded better sizes than the nanoparticles prepared using microfluidic devices, which was attributed to the large size of pDNA.72
Effect of solution and flow changes
So far, it is apparent that smaller-sized polyplexes with smaller PDIs can be produced using microfluidic devices. However, there are additional elements that can affect the size and DPI of nanoparticles such as flow rate ratio, N/P ratio, the design of the microfluidic devices, the chemical structure of the polymer, the type of nucleic acid drug, concentration and so on. Thus, in this section, we discuss the influences on size and PDI of some of these factors.
First, microfluidic devices require different flow rates depending on the microfluidic mixer and the material and drug used. Another important parameter is the flow rate of each solution or the flow rate ratio, which is an expression of the relative rate of both flows.
Nucleic acid-containing polymer nanoparticles are often prepared using two aqueous solutions, one containing the drug, the other the water-soluble polymer (Table 1). However, occasionally hydrophobic polymers are involved and then the nanoparticles are often prepared by nanoprecipitation, which uses two different phases, aqueous and organic. The flow rate ratio is defined by the flow rate of the aqueous to the organic phase. For example, pDNA-loaded lipid–polymer nanoparticles were synthesised using flow rate ratios of 3:1 and 5:1 in a toroidal micromixer device. Smaller sizes, smaller PDIs and more stable nanoparticles were formed with lower flow rate ratios.74 However, in some studies, the opposite was observed and the size of nanoparticles decreased with increasing flow rate ratio as it changed from 1:1 to 5:1.81,82 This contradicting scenario may be caused by a combination of factors like differences in concentrations of the two different phases, which could affect the flow impedance as the viscosity is changed in the micromixer, but it should also be noted that in these studies, very different drug carriers and payloads were explored.83
The mixing process of two aqueous solutions is similarly affected by flow rate ratios. A specifically designed microfluidic device that can control the flow rate in the centre and outer inlet separately was used to assemble PAMAM dendrimer–siRNA complexes in (N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid, HEPES) buffer at flow rate ratios (centre inlet flow rate to outer flow rate) varying from 1:10 to 1:2.5.48 With increasing ratio, the size of the nanoparticles decreased, but the PDI increased, with the best nanoparticle being produced at a flow ratio of 1:5.
Other influential parameters are the channel design of the microfluidic device. For example, a T-junction droplet break up microfluidic device with input channel dimensions ranging from 50, 100, 150 to 200 µm, was used to demonstrate how the size of the nanoparticles is directly related to the dimensions of the input channel.65 Further, the Reynolds number Re is another powerful tool to control the flow regime in a microfluidic device.84 In the flow device used by Naeini et al., laminar flow was obtained when the Re number was below 2000, whereas a Re number above 4000 led to turbulent flow. A Re number between 2000 and 4000 represents a transitional flow regime between both laminar and turbulent flow (Fig. 8). With the help of an AIMS, the Re number could be dialled from 26 to 4000. With increasing Re number, the respective size and PDI of the size distribution of chitosan–siRNA and chitosan–pDNA polyplexes were observed to be reduced.72
The N/P ratio is the mole ratio of the amino groups (N in the cationic polymer) to the amount of phosphate groups (P) in the nucleic acid therapeutic.85 The N/P ratio is often discussed when preparing polyplexes using an aqueous solution of cationic polymer and an aqueous DNA solution as the N/P ratio is known to affect the size and PDI of the nanoparticles. For example, Koh et al. mixed PEI with pDNA and observed that a higher N/P ratio of 6.7 led to smaller and more uniform polyplexes compared with an N/P ratio of 3.3.46 Protopapa et al., who used the same polymer–drug combination, developed a stand-alone microfluidic cartridge that contained seven output ports to test seven different N/P ratios (N/P = 0, 10, 20, 30, 40, 50 and 60) at the same time. Although higher N/P ratios usually leads to smaller sizes and lower PDIs, the authors observed an optimum at a N/P ratio of 50.45
As with bulk mixing, the concentration of the solution plays an important role. In this case, it is important to distinguish between the different approaches. Mixing two aqueous solutions that contain polymer and drug respectively to generate polyplexes as shown in Fig. 1 is affected differently to microfluidic-assisted nanoprecipitation or droplet-assisted microfluidics.
An in-line mixing system (AIMS) of two aqueous solutions containing cationic oligomers CO and siRNA or alternatively pDNA showed that concentration may, however, not always influence the outcome. Whereas siRNA as payload led to size increases from 40 to 60 nm, when the siRNA concentration was increased from 0.1 to 0.4 mg mL–1, there was no equivalent effect when pDNA was loaded. Moreover, the size of the siRNA polyplexes across all concentrations was smaller than that for the pDNA polyplexes. As the larger pDNA diffuses more slowly than the small siRNA, polyplexes prepared using pDNA led to broader size distributions.42
When organic solvents and hydrophobic polymers are involved, as in the case of microfluidic-assisted nanoprecipitation, the concentrations of hydrophobic polymer influence the rate of precipitation. Furthermore, the ingredients added to the aqueous solution can be highly influential on the outcome. Different concentrations (0, 0.8, 3.4, 8.6, 17.2 µg mL–1) of pDNA dissolved in water together with the surfactant mPEG2000-DSPE were formulated with PLGA and cationic lipid DC-cholesterol that were both dissolved in organic solvent. With increasing concentration of pDNA, the size of the resulting nanoparticles decreased. The concentration of the surfactant has a similar effect as higher amounts lead to smaller sizes at higher flow rate ratios. However, at lower flow ratios, the system became unstable as evidenced by an increase in PDI.74
The nature of the polymer itself is a determining factor in the quality of the nanoparticles. Although there are plenty of studies on the relationship between polymer structure and nanoparticle properties available that use bulk mixing, there is limited information so far on flow-based mixing. Comparison of two cationic oligomers, LPO and LPOE, that differ in the length of the PEG chain (Fig. 9) and their complex with siRNA (CO + siRNA + LPO and CO + siRNA + LPOE) revealed that longer PEG ligands promote the formation of larger nanoparticles with larger PDIs.71 Also, the payload influences the outcome as shown when comparing chitosan complexes with siRNA or pDNA. Polyplexes based on siRNA had smaller sizes and smaller PDIs than pDNA at the same concentration and flow rate.72
Perspective and conclusion
Flow-based assembly of nanoparticles has become an established technique in laboratories and in industry. Researchers can choose from a wide range of set-ups, either chip-based or capillary-based. The mode of mixing is crucial as laminar flow will result in different outcomes to turbulent flow. Flow-based assembly of drug-loaded nanoparticles has clear advantages, but some challenges should also be mentioned.37 It is evident that the nanoparticle production is highly reproducible, with limited batch-to-batch variations. This allows efficient scaling up and the supply of high-quality nanoparticles for clinical applications. Flow production also leads to better-defined nanoparticles with narrower size distribution while the size can be tuned with the operating parameters. On the downside, it is not easy to switch all laboratory operations to flow without considering a range of challenges. For example, although polydimethylsiloxane devices can be quite cheaply produced, the material is sensitive to organic solvents. Glass, silicon, or polytetrafluoroethylene are in contrast more robust, but more costly. There are also some costs involved to set up a flow laboratory as pumps can be quite pricey whereas bulk mixing requires only a pipette and a beaker. There are also more demands on the researcher as flow assembly requires more specialised skills and ideally an understanding of fluid flow. Having established one system may not be sufficient to meet the demands of the laboratory as each nanoparticle may require adjustment. One aspect that may also deter researchers from using flow is the minimum sample size, which could be an important aspect when trying to assemble costly nucleic acids in flow. Although researchers can assemble small volumes down to only a few microlitres in bulk mixing, even the smallest microfluidic devices require larger sample sizes, notwithstanding that with cheaper pumps that have a pressure ramp, there is always a fraction that needs to be discarded. It also needs to be considered that flow design is not timesaving per se and complex channel design is required to parallelise the process to allow testing of different parameters in one system. In general, chip design is still the biggest drawback as current chips have only a single purpose and they cannot be adjusted to suit other purposes.86 In this case, modular systems that can be changed and adjusted offer more flexibility. However, the advantages of flow assembly certainly outweigh potential obstacles and flow assembly is therefore an essential tool that will help accelerate nanomedicine research.
Acknowledgements
The authors thank the Australian Research Council and the UNSW RNA Institute for funding.
References
1 Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov 2021; 20: 101-124.
| Crossref | Google Scholar |
2 Halwani AA. Development of pharmaceutical nanomedicines: from the bench to the market. Pharmaceutics 2022; 14: 106.
| Crossref | Google Scholar |
3 Shan X, Gong X, Li J, Wen J, Li Y, Zhang Z. Current approaches of nanomedicines in the market and various stage of clinical translation. Acta Pharm Sin B 2022; 12: 3028-3048.
| Crossref | Google Scholar |
4 Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater 2021; 6: 1078-1094.
| Crossref | Google Scholar |
5 Kumar R, Santa Chalarca CF, Bockman MR, Bruggen CV, Grimme CJ, Dalal RJ, et al. Polymeric delivery of therapeutic nucleic acids. Chem Rev 2021; 121: 11527-11652.
| Crossref | Google Scholar |
6 Paunovska K, Loughrey D, Dahlman JE. Drug delivery systems for RNA therapeutics. Nat Rev Genet 2022; 23: 265-280.
| Crossref | Google Scholar |
7 Zhang L, Nguyen TLU, Bernard J, Davis TP, Barner-Kowollik C, Stenzel MH. Shell-cross-linked micelles containing cationic polymers synthesized via the RAFT process: toward a more biocompatible gene delivery system. Biomacromolecules 2007; 8: 2890-2901.
| Crossref | Google Scholar |
8 Hui Y, Yi X, Hou F, Wibowo D, Zhang F, Zhao D, et al. Role of nanoparticle mechanical properties in cancer drug delivery. ACS Nano 2019; 13: 7410-7424.
| Crossref | Google Scholar |
9 Donahue ND, Acar H, Wilhelm S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv Drug Delivery Rev 2019; 143: 68-96.
| Crossref | Google Scholar |
10 Stenzel MH. The trojan horse goes wild: the effect of drug loading on the behavior of nanoparticles. Angew Chem Int Ed Engl 2021; 60: 2202-2206.
| Crossref | Google Scholar |
11 Hoshyar N, Gray S, Han H, Bao G. The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomed 2016; 11: 673-692.
| Crossref | Google Scholar |
12 Choi CHJ, Zuckerman JE, Webster P, Davis ME. Targeting kidney mesangium by nanoparticles of defined size. Proc Natl Acad Sci USA 2011; 108: 6656-6661.
| Crossref | Google Scholar |
13 Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WCW. Mediating tumor targeting efficiency of nanoparticles through design. Nano Lett 2009; 9: 1909-1915.
| Crossref | Google Scholar |
14 Soo Choi H, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, et al. Renal clearance of quantum dots. Nat Biotechnol 2007; 25: 1165-1170.
| Crossref | Google Scholar |
15 Gustafson HH, Holt-Casper D, Grainger DW, Ghandehari H. Nanoparticle uptake: the phagocyte problem. Nano Today 2015; 10: 487-510.
| Crossref | Google Scholar |
16 Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharmaceutics 2008; 5: 505-515.
| Crossref | Google Scholar |
17 Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol 2015; 33: 941-951.
| Crossref | Google Scholar |
18 Cabral H, Matsumoto Y, Mizuno K, Chen Q, Murakami M, Kimura M, et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat Nanotechnol 2011; 6: 815-823.
| Crossref | Google Scholar |
19 Yu W, Liu R, Zhou Y, Gao H. Size-tunable strategies for a tumor targeted drug delivery system. ACS Cent Sci 2020; 6: 100-116.
| Crossref | Google Scholar |
20 Ulkoski D, Scholz C. Impact of cationic charge density and pegylated poly(amino acid) tercopolymer architecture on their use as gene delivery vehicles. Part 2: DNA protection, stability, cytotoxicity, and transfection efficiency. Macromol Biosci 2018; 18: 1800109.
| Crossref | Google Scholar |
21 Bauer M, Tauhardt L, Lambermont-Thijs HML, Kempe K, Hoogenboom R, Schubert US, et al. Rethinking the impact of the protonable amine density on cationic polymers for gene delivery: a comparative study of partially hydrolyzed poly(2‐ethyl‐2‐oxazoline)s and linear poly(ethylene imine)s. Eur J Pharm Biopharm 2018; 133: 112-121.
| Crossref | Google Scholar |
22 Jiang Y, Stenzel M. Drug delivery vehicles based on albumin-polymer conjugates. Macromol Biosci 2016; 16: 791-802.
| Crossref | Google Scholar |
23 Joshi N, Liu DL, Dickson KA, Marsh DJ, Ford CE, Stenzel MH. An organotypic model of high-grade serous ovarian cancer to test the anti-metastatic potential of ROR2 targeted polyion complex nanoparticles. J Mater Chem B 2021; 9: 9123-9135.
| Crossref | Google Scholar |
24 Jiang Y, Wong CK, Stenzel MH. An oligonucleotide transfection vector based on hsa and pdmaema conjugation: effect of polymer molecular weight on cell proliferation and on multicellular tumor spheroids. Macromol Biosci 2015; 15: 965-978.
| Crossref | Google Scholar |
25 Jiang Y, Lu H, Khine YY, Dag A, Stenzel MH. Polyion complex micelle based on albumin-polymer conjugates: multifunctional oligonucleotide transfection vectors for anticancer chemotherapeutics. Biomacromolecules 2014; 15: 4195-4205.
| Crossref | Google Scholar |
26 Đorđević S, Gonzalez MM, Conejos-Sánchez I, Carreira B, Pozzi S, Acúrcio RC, et al. Current hurdles to the translation of nanomedicines from bench to the clinic. Deliv and Transl Res 2022; 12: 500-525.
| Crossref | Google Scholar |
27 Hirota S, de Ilarduya CT, Barron LG, Szoka Jr FC. Simple mixing device to reproducibly prepare cationic lipid–DNA complexes (lipoplexes). Biotechniques 1999; 27: 286-290.
| Google Scholar |
28 Maeki M, Kimura N, Sato Y, Harashima H, Tokeshi M. Advances in microfluidics for lipid nanoparticles and extracellular vesicles and applications in drug delivery systems. Adv Drug Deliv Rev 2018; 128: 84-100.
| Crossref | Google Scholar |
29 Jung HN, Lee SY, Lee S, Youn H, Im HJ. Lipid nanoparticles for delivery of rna therapeutics: current status and the role of in vivo imaging. Theranostics 2022; 12: 7509-7531.
| Crossref | Google Scholar |
31 Johnson BK, Prud’homme RK. Flash nanoprecipitation of organic actives and block copolymers using a confined impinging jets mixer. Aust J Chem 2003; 56: 1021-1024.
| Crossref | Google Scholar |
32 Feng J, Markwalter CE, Tian C, Armstrong M, Prud’homme RK. Translational formulation of nanoparticle therapeutics from laboratory discovery to clinical scale. J Transl Med 2019; 17: 200.
| Crossref | Google Scholar |
33 Zhang H, Zhu Y, Shen Y. Microfluidics for cancer nanomedicine: from fabrication to evaluation. Small 2018; 14: 1800360.
| Crossref | Google Scholar |
34 Lee CY, Chang CL, Wang YN, Fu LM. Microfluidic mixing: a review. Int J Mol Sci 2011; 12: 3263-3287.
| Crossref | Google Scholar |
35 Ober TJ, Foresti D, Lewis JA. Active mixing of complex fluids at the microscale. Proc Natl Acad Sci USA 2015; 112: 12293-12298.
| Crossref | Google Scholar |
36 Lu M, Ozcelik A, Grigsby CL, Zhao Y, Guo F, Leong KW, et al. Microfluidic hydrodynamic focusing for synthesis of nanomaterials. Nano Today 2016; 11: 778-792.
| Crossref | Google Scholar |
37 Liu D, Zhang H, Fontana F, Hirvonen JT, Santos HA. Current developments and applications of microfluidic technology toward clinical translation of nanomedicines. Adv Drug Deliv Rev 2018; 128: 54-83.
| Crossref | Google Scholar |
38 Debus H, Beck-Broichsitter M, Kissel T. Optimized preparation of PDNA/poly(ethylene imine) polyplexes using a microfluidic system. Lab Chip 2012; 12: 2498-2506.
| Crossref | Google Scholar |
39 Stroock AD, Dertinger SKW, Ajdari A, Mezic’ I, Stone HA, Whitesides GM. Chaotic mixer for microchannels. Science 2002; 295: 647-651.
| Crossref | Google Scholar |
40 Williams MS, Longmuir KJ, Yager P. A practical guide to the staggered herringbone mixer. Lab Chip 2008; 8: 1121-1129.
| Crossref | Google Scholar |
41 Ali MS, Hooshmand N, El-Sayed M, Labouta HI. Microfluidics for development of lipid nanoparticles: paving the way for nucleic acids to the clinic. ACS Appl Bio Mater 2021;
| Crossref | Google Scholar |
42 Reynolds WC, Parekh DE, Juvet PJD, Lee MJD. Bifurcating and blooming jets. Ann Rev Fluid Mechanics 2003; 35: 295-315.
| Crossref | Google Scholar |
43 He B, Burke BJ, Zhang X, Zhang R, Regnier FE. A picoliter-volume mixer for microfluidic analytical systems. Anal Chem 2001; 73: 1942-1947.
| Crossref | Google Scholar |
44 Jen C-P, Wu C-Y, Lin Y-C, Wu C-Y. Design and simulation of the micromixer with chaotic advection in twisted microchannels. Lab Chip 2003; 3: 77-81.
| Crossref | Google Scholar |
45 Protopapa G, Bono N, Visone R, D’Alessandro F, Rasponi M, Candiani G. A new microfluidic platform for the highly reproducible preparation of non-viral gene delivery complexes. Lab Chip 2023; 23: 136-145.
| Crossref | Google Scholar |
46 Koh CG, Kang X, Xie Y, Fei Z, Guan J, Yu B, et al. Delivery of polyethylenimine/DNA complexes assembled in a microfluidics device. Mol Pharmaceutics 2009; 6: 1333-1342.
| Crossref | Google Scholar |
47 Wilson DR, Mosenia A, Suprenant MP, Upadhya R, Routkevitch D, Meyer RA, et al. Continuous microfluidic assembly of biodegradable poly(beta-amino ester)/DNA nanoparticles for enhanced gene delivery. J Biomed Mater Res Part A 2017; 105: 1813-1825.
| Crossref | Google Scholar |
48 Agnoletti M, Bohr A, Thanki K, Wan F, Zeng X, Boetker JP, et al. Inhalable siRNA-loaded nano-embedded microparticles engineered using microfluidics and spray drying. Europ J Pharm Biopharm 2017; 120: 9-21.
| Crossref | Google Scholar |
49 Zhigaltsev IV, Belliveau N, Hafez I, Leung AKK, Huft J, Hansen C, et al. Bottom–up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing. Langmuir 2012; 28: 3633-3640.
| Crossref | Google Scholar |
50 Belliveau NM, Huft J, Lin PJC, Chen S, Leung AKK, Leaver TJ, et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol Ther Nucleic Acids 2012; 1: e37.
| Crossref | Google Scholar |
51 Chen T, Peng Y, Qiu M, Yi C, Xu Z. Recent advances in mixing-induced nanoprecipitation: from creating complex nanostructures to emerging applications beyond biomedicine. Nanoscale 2023; 15: 3594-3609.
| Crossref | Google Scholar |
52 Karnik R, Gu F, Basto P, Cannizzaro C, Dean L, Kyei-Manu W, et al. Microfluidic platform for controlled synthesis of polymeric nanoparticles. Nano Lett 2008; 8: 2906-2912.
| Crossref | Google Scholar |
53 Liu Y, Yang G, Zou D, Hui Y, Nigam K, Middelberg APJ, et al. Formulation of nanoparticles using mixing-induced nanoprecipitation for drug delivery. Ind Eng Chem Res 2020; 59: 4134-4149.
| Crossref | Google Scholar |
54 Saad WS, Prud’homme RK. Principles of nanoparticle formation by flash nanoprecipitation. Nano Today 2016; 11: 212-227.
| Crossref | Google Scholar |
55 Yan X, Bernard J, Ganachaud F. Nanoprecipitation as a simple and straightforward process to create complex polymeric colloidal morphologies. Adv Colloid Interface Sci 2021; 294: 102474.
| Crossref | Google Scholar |
56 Ma J, Lee SM-Y, Yi C, Li C-W. Controllable synthesis of functional nanoparticles by microfluidic platforms for biomedical applications – a review. Lab Chip 2017; 17: 209-226.
| Crossref | Google Scholar |
58 Nunes JK, Tsai SSH, Wan J, Stone HA. Dripping and jetting in microfluidic multiphase flows applied to particle and fibre synthesis. J Phys D: Appl Phys 2013; 46: 114002.
| Crossref | Google Scholar |
59 Moragues T, Arguijo D, Beneyton T, Modavi C, Simutis K, Abate AR, et al. Droplet-based microfluidics. Nat Rev Methods Primers 2023; 3: 32.
| Crossref | Google Scholar |
60 Amirifar L, Besanjideh M, Nasiri R, Shamloo A, Nasrollahi F, de Barros NR, et al. Droplet-based microfluidics in biomedical applications. Biofabrication 2022; 14: 022001.
| Crossref | Google Scholar |
61 Tian F, Cai L, Liu C, Sun J. Microfluidic technologies for nanoparticle formation. Lab Chip 2022; 22: 512-529.
| Crossref | Google Scholar |
62 He F, Zhang M-J, Wang W, Cai Q-W, Su Y-Y, Liu Z, et al. Designable polymeric microparticles from droplet microfluidics for controlled drug release. Adv Mater Technol 2019; 4: 1800687.
| Crossref | Google Scholar |
63 Grigsby CL, Ho Y-P, Lin C, Engbersen JFJ, Leong KW. Microfluidic preparation of polymer-nucleic acid nanocomplexes improves nonviral gene transfer. Sci Rep 2013; 3: 3155.
| Crossref | Google Scholar |
64 Riahi R, Tamayol A, Shaegh SAM, Ghaemmaghami AM, Dokmeci MR, Khademhosseini A. Microfluidics for advanced drug delivery systems. Curr Opin Chem Eng 2015; 7: 101-112.
| Crossref | Google Scholar |
65 Deveza L, Ashoken J, Castaneda G, Tong X, Keeney M, Han L-H, et al. Microfluidic synthesis of biodegradable polyethylene-glycol microspheres for controlled delivery of proteins and DNA nanoparticles. ACS Biomater Sci Eng 2015; 1: 157-165.
| Crossref | Google Scholar |
66 Zhang L, Chen Q, Ma Y, Sun J. Microfluidic methods for fabrication and engineering of nanoparticle drug delivery systems. ACS Appl Bio Mater 2020; 3: 107-120.
| Crossref | Google Scholar |
67 Luo G, Du L, Wang Y, Lu Y, Xu J. Controllable preparation of particles with microfluidics. Particuology 2011; 9: 545-558.
| Crossref | Google Scholar |
68 Zhao C-X, He L, Qiao SZ, Middelberg APJ. Nanoparticle synthesis in microreactors. Chem Eng Sci 2011; 66: 1463-1479.
| Crossref | Google Scholar |
69 Soni G, Yadav KS. Applications of nanoparticles in treatment and diagnosis of leukemia. Mater Sci Eng C 2015; 47: 156-164.
| Crossref | Google Scholar |
70 Khan IU, Serra CA, Anton N, Vandamme TF. Production of nanoparticle drug delivery systems with microfluidics tools. Expert Opin Drug Deliv 2015; 12: 547-562.
| Crossref | Google Scholar |
71 Loy DM, Krzysztoń R, Lächelt U, Rädler JO, Wagner E. Controlling nanoparticle formulation: a low-budget prototype for the automation of a microfluidic platform. Processes 2021; 9: 129.
| Crossref | Google Scholar |
72 Tavakoli Naeini A, Soliman OY, Alameh MG, Lavertu M, Buschmann MD. Automated in-line mixing system for large scale production of chitosan-based polyplexes. J Colloid Interface Sci 2017; 500: 253-263.
| Crossref | Google Scholar |
73 Zoqlam R, Morris CJ, Akbar M, Alkilany AM, Hamdallah SI, Belton P, et al. Evaluation of the benefits of microfluidic-assisted preparation of polymeric nanoparticles for DNA delivery. Mater Sci Eng C 2021; 127: 112243.
| Crossref | Google Scholar |
74 Santhanes D, Wilkins A, Zhang H, John Aitken R, Liang M. Microfluidic formulation of lipid/polymer hybrid nanoparticles for plasmid DNA (pdna) delivery. International J Pharm 2022; 627: 122223.
| Crossref | Google Scholar |
75 Ita K. Polyplexes for gene and nucleic acid delivery: progress and bottlenecks. Eur J Pharm Sci 2020; 150: 105358.
| Crossref | Google Scholar |
76 Tros de Ilarduya C, Sun Y, Düzgüneş N. Gene delivery by lipoplexes and polyplexes. Eur J Pharm Sci 2010; 40: 159-170.
| Crossref | Google Scholar |
77 Elouahabi A, Ruysschaert J-M. Formation and intracellular trafficking of lipoplexes and polyplexes. Mol Ther 2005; 11: 336-347.
| Crossref | Google Scholar |
78 Miladi K, Sfar S, Fessi H, Elaissari A. Nanoprecipitation process: from particle preparation to in vivo applications. In: Vauthier C, Ponchel G, editors. Polymer nanoparticles for nanomedicines: a guide for their design, preparation and development. Cham, Switzerland: Springer International Publishing; 2016. pp. 17–53.
79 Zhang L, Chen Q, Ma Y, Sun J. Microfluidic methods for fabrication and engineering of nanoparticle drug delivery systems. ACS Appl Bio Mater 2019; 3: 107-120.
| Crossref | Google Scholar |
80 Brocchini S, James K, Tangpasuthadol V, Kohn J. A combinatorial approach for polymer design. J Am Chem Soc 1997; 119: 4553-4554.
| Crossref | Google Scholar |
81 Roces CB, Christensen D, Perrie Y. Translating the fabrication of protein-loaded poly (lactic-co-glycolic acid) nanoparticles from bench to scale-independent production using microfluidics. Drug Deliv Transl Res 2020; 10: 582-593.
| Crossref | Google Scholar |
82 Kastner E, Kaur R, Lowry D, Moghaddam B, Wilkinson A, Perrie Y. High-throughput manufacturing of size-tuned liposomes by a new microfluidics method using enhanced statistical tools for characterization. Int J Pharm 2014; 477: 361-368.
| Crossref | Google Scholar |
83 Wild A, Leaver T, Taylor RJ. Bifurcating mixers and methods of their use and manufacture. Google Patents; 2018. US10076730B2. Available at https://patents.google.com/patent/US10076730B2/en
85 Bono N, Ponti F, Mantovani D, Candiani G. Non-viral in vitro gene delivery: it is now time to set the bar! Pharmaceutics 2020; 12: 183.
| Crossref | Google Scholar |
86 Wu J, Fang H, Zhang J, Yan S. Modular microfluidics for life sciences. J Nanobiotechnology 2023; 21: 85.
| Crossref | Google Scholar |