APCalign: an R package workflow and app for aligning and updating flora names to the Australian Plant Census
Elizabeth H. Wenk A * , William K. Cornwell
A * , William K. Cornwell  A , Anne Fuchs
A , Anne Fuchs  B , Fonti Kar
B , Fonti Kar  A , Anna M. Monro
A , Anna M. Monro  B , Hervé Sauquet
B , Hervé Sauquet  A C , Ruby E. Stephens
A C , Ruby E. Stephens  A D and Daniel S. Falster
A D and Daniel S. Falster  A
A
A
B
C
D
Abstract
Here we present ‘APCalign’, an R package and accompanying browser-sourced application to align and update scientific names for Australian vascular plants to the most likely currently accepted name in the Australian Plant Census (APC) or a name in the Australian Plant Names Index (APNI). Scientific names are the label assigned to unique taxon concepts by the scientific community, but this common terminology is most useful if a taxon concept is consistently referred to by the same name. These links can be broken because of either spelling mistakes or taxonomic changes. Automated tools are required to resolve taxon lists, aligning and updating long lists of possibly erroneous scientific names to the most likely currently accepted names. It is essential that tools specific to the APC/APNI be developed, because these lists specify an endorsed national-level nomenclature used in government legislation and include the uniquely Australian concept of phrase names, absent in global taxonomic datasets. To align input names to names within the APC or APNI, ‘APCalign’ works progressively through a sequence of checks that combine different permutations of the input name, exact versus fuzzy matches, matches that consider the entire name input versus a subset of words, and character strings that indicate a name can be resolved only to a genus or family. The aligned names are then, when possible, updated to a currently accepted taxon concept within the APC. This package should facilitate all research outputs that require diverse scientific name lists to be merged or outdated lists to be updated.
Keywords: Australian Plant Census, biodiversity informatics, bioinformatics, conservation biology, plant taxonomy, R-package, taxon concept, vascular plants.
Introduction
Taxonomic names and lists are essential in modern e-research infrastructure, enabling us to link concepts about what taxa exist to information about where they occur, how they are related to one another, and what traits they have. However, these linkages are easily broken or missed, owing to inconsistencies in the delineation and naming of taxon concepts among different datasets (Franz and Peet 2009; Sandall et al. 2023), where a taxon concept can be described as ‘a circumscribed set of organisms’ (https://github.com/tdwg/tnc/issues/1) (Whitbread 2018). The scientific name applied at a given point in time indicates a snapshot of our understanding of evolutionary relationships within a taxonomic group. However, names change, as understanding of what morphological or genetic features circumscribe a specific taxon concept is refined. Scientific names are also easily misspelt or mistyped. When dealing with large numbers of taxa, such errors are hard to spot. To maintain the unique link among a scientific name, the associated taxon concept, and associated data, diverse users must be able to automatically align and update a list of possibly inaccurate or outdated scientific names to the currently accepted scientific name.
A range of software tools have been developed to resolve taxonomic mismatches (to the extent possible), each referencing one or more taxonomic datasets (Grenié et al. 2023; Schellenberger Costa et al. 2023); however, as yet, none are specific to the current Australian National Species Lists (auNSL) for vascular plants (https://biodiversity.org.au/nsl/). Typically, these tools input a list of species names, returning a list of said names matched to the most likely currently accepted name. Working out the best or most likely match for a species name can be a computationally intensive task; still, modern tools have found several approaches to do this with reasonable speed and measurable accuracy. Different tools also have different features (Table 1) and are presented in a range of formats, from more accessible browser-based tools, such as the Taxonomic Name Resolution Service (TNRS; Boyle et al. 2013) to more specialised R packages, such as the ‘taxize’ R-package (Chamberlain and Szöcs 2013; Chamberlain et al. 2022). Whereas all existing global tools do handle names of Australian taxa, the taxon lists and taxonomy in these global resources can differ from our National Species Lists, because they are managed by different authorities, each with their own missions and guidance councils (Garnett et al. 2020; Schellenberger Costa et al. 2023).
| Item | APCalign | taxize | TNRS | kewr | |
|---|---|---|---|---|---|
| Batch processing | X | X | X | X | |
| Fuzzy matching | X | X | X | ||
| Adjustable fuzzy matching | X | ||||
| Aligns names | X | X | X | X | |
| Updates names | X | X | X | ||
| Updates synonyms | X | X | X | X | |
| Handles phrase names | X | ||||
| Identifies taxon rank of name | X | X | X | ||
| Handles infraspecific taxa | X | X | X | X | |
| Handles higher taxonomy | X | X | X | X | |
| Handles taxonomic splits | X | ||||
| Retains field note tags | X | ||||
| Available via R package | X | X | X | X | |
| Available via browser | X | X | X | ||
| Can handle global datasets | X | X | X |
In Australia, the national taxonomic standard for vascular plants is the Australian Plant Census (APC), supported by a comprehensive list of plant names documented in references, the Australian Plant Names Index (APNI). The APC is endorsed by the Council of Heads of Australasian Herbaria (CHAH), with taxonomy being agreed on by consensus. The APC is updated on a regular basis, following a review of both newly described plant taxa and taxonomic revisions, first by a working group comprising representatives from all major Australasian herbaria, followed by ratification by CHAH. The working group reviews taxon concepts extracted from recent taxonomic publications, ensuring Australia’s botanical community considers newly published names and changed taxon circumscriptions robust. The names documented in APNI and accepted by APC may be out-of-step with those used in international checklists, because each checklist may deem different scientific names or taxonomies as ‘current’. Having a national checklist is also essential for Australia because of the inclusion of large numbers of phrase names within the APC/APNI that are excluded from international checklists. Phrase names are assigned to unique taxa that are yet to be formally described, a nomenclatural process that is particular to Australian plant taxonomy (Barker 2005). These names follow a specific convention, namely, the generic name, a rank indicator, a geographic or morphological descriptor, the collector’s name and a number representing an herbarium voucher for the new taxon concept. Moreover, as our official national list, the APC is the natural (sometimes mandated) focus for documenting and linking data for Australian vascular plants.
Here, we provide a new software tool, ‘APCalign’, that can quickly match plant names to accepted taxon concepts in the APC and to names in APNI. ‘APCalign’ can function both as an R package for scientific and technical users, and in a browser interface for lay users. ‘APCalign’ offers a two-step process for creating a list of names matched to the APC/APNI. In the first step, algorithms align each name input to the best match of any name within the APC/APNI, allowing matches to the infraspecies, species, genus, or family level, as is appropriate. In the second step, aligned names that have been matched to a scientific name within the APC are updated to a currently accepted taxon concept within the APC. Our implementation builds on best practices for global taxonomic resources, as well as additional enhancements (Table 1). It includes a well-considered and well-tested sequence of algorithm matching, including both complex string matches and fuzzy-matching, to maximise meaningful links. The taxonomic updating functions handle taxonomic uncertainties. For scientific names linked to taxon concepts accepted by APC, the final table adds distribution and native status. In total, 11 functions are exported by ‘APCalign’ (Table 2), including functions to download taxonomic resources, functions to clean, align and update names to a name recorded in the APC/APNI, and functions to tabulate information about taxon distribution, diversity, and native status. By working directly with Australia’s National Species Lists for vascular plants, ‘APCalign’ fills an important gap in our biodiversity e-resources.
| Function | Description | Arguments | Output | |
|---|---|---|---|---|
| Data access | ||||
| load_taxonomic_resources | Loads two taxonomic datasets for Australia’s vascular plants, the APC and APNI, into the global environment. It accesses taxonomic data from a dataset using the provided version number or the default version. The function creates several data frames by filtering and selecting data from the loaded lists. | stable_or_current_data = ‘stable’ | A list of dataframes that include the full APC and APNI, and subsets based on taxon rank and taxonomic status. | |
| version = default_version() | ||||
| reload = FALSE | ||||
| standardise_taxon_rank | Takes a character vector of Latin taxon ranks as input and returns a character vector of taxon ranks using standardised English terms. | taxon_rank | A character vector of English taxon ranks. | |
| Standardising name syntax | ||||
| standardise_names | Standardises taxon names by performing a series of text substitutions to remove common inconsistencies in taxonomic nomenclature. The function takes a character vector of taxon names as input and returns a character vector of taxon names using standardised taxonomic syntax as output. In particular it standardises taxon rank abbreviations and qualifiers (subsp., var., f.), as people use many variants of these terms. It also standardises or removes a few additional filler words used within taxon names (affinis becomes aff.; s.l. and s.s. are removed). | taxon_names | A character vector of standardised taxon names. | |
| strip_names | Removes taxon rank abbreviations and qualifiers (‘subsp.’, ‘var.’, ‘f.’, and ‘ser’), special characters (e.g. ‘-’, ‘.’, ‘(‘, ’)’, ‘?’), and extra whitespace. The resulting vector of names is also converted to lowercase. | taxon_names | A character vector of stripped taxonomic names, with taxon rank abbreviations and qualifiers, extra whitespace, and special characters removed, and all letters converted to lowercase. | |
| strip_names_extra | Strip taxonomic names of ‘sp.’ and hybrid symbols. This function assumes that a character function has already been run through the function ‘strip_names’. | taxon_names | A character vector of stripped taxonomic names, with taxon rank abbreviations and qualifiers, additional filler words, special characters and extra whitespace removed, and all letters converted to lowercase. | |
| Taxonomic alignment and updating | ||||
| align_taxa | Finds taxonomic alignments in the APC or APNI. It uses the internal function ‘match_taxa()’ to attempt to match input strings to taxon names in the APC/APNI. It sequentially searches for matches against more than 20 different string patterns, prioritising exact matches (to accepted names as well as synonyms, orthographic variants) over fuzzy matches. It prioritises matches to taxa in the APC over names in the APNI. It identifies string patterns in input names that suggest a name can only be aligned to a genus (hybrids that are not in the APC/APNI; graded species; taxa not identified to species), and indicates these names only have a genus-rank match. | original_name, | A data frame with rows representing each taxon and with columns documenting the alignment made, the reason for the alignment, and a selection of taxon name mutations to which the original name was compared. | |
| output = NULL, | ||||
| quiet = FALSE, | ||||
| full = FALSE, | ||||
| resources = load_taxonomic_resources(), | ||||
| fuzzy_abs_dist = 3, | ||||
| fuzzy_rel_dist = 0.2, | ||||
| fuzzy_matches = TRUE, | ||||
| imprecise_fuzzy_matches = FALSE, | ||||
| APNI_matches = TRUE, | ||||
| identifier = NA_character_ | ||||
| update_taxonomy | Uses the APC to update the taxonomy of names aligned to a taxon concept listed in the APC to the currently accepted name for the taxon concept. The aligned_data data frame that is input must contain five columns, ‘original_name’, ‘aligned_name’, ‘taxon_rank’, ‘taxonomic_dataset’, and ‘aligned_reason’. The aligned name is a plant name that has been aligned to a taxon name in the APC or APNI by the ‘align_taxa()’ function. | aligned_data, | A data frame with rows representing each taxon and columns documenting taxon metadata | |
| taxonomic_splits = ‘most_likely_species’, | ||||
| output = NULL, | ||||
| quiet = FALSE, | ||||
| resources = load_taxonomic_resources() | ||||
| create_taxonomic_update_lookup | Takes a list of Australian plant names that need to be reconciled with current taxonomy and generates a lookup table of the best-possible scientific name match for each input name. It uses first the function ‘align_taxa()’, then the function ‘update_taxonomy()’ to achieve the output. | taxa, | A data frame with rows representing each taxon and columns documenting taxon metadata. It includes the original species names, the aligned species names, and additional taxonomic information such as taxon IDs and genera. | |
| stable_or_current_data = ‘stable’, | ||||
| version = default_version(), | ||||
| taxonomic_splits = ‘most_likely_species’, | ||||
| full = FALSE, | ||||
| fuzzy_abs_dist = 3, | ||||
| fuzzy_rel_dist = 0.2, | ||||
| fuzzy_matches = TRUE, | ||||
| APNI_matches = TRUE, | ||||
| imprecise_fuzzy_matches = FALSE, | ||||
| identifier = NA_character_, | ||||
| resources = load_taxonomic_resources(), | ||||
| output = NULL, | ||||
| quiet = FALSE | ||||
| Distribution and establishment means | ||||
| create_species_state_origin_matrix | Processes the geographic data available in the APC and returns state level native, introduced and more complicated origins status for all taxa. | resources = load_taxonomic_resources() | A data frame with rows representing each species and columns for taxon name and each state. The values in each cell represent the origin of the species in that state. | |
| native_anywhere_in_australia | Checks if the given species is native anywhere in Australia according to the APC. Note that this will not detect within-Australia introductions, e.g. whether a species is from Western Australia and is invasive on the eastern coast. | species, resources = load_taxonomic_resources() | A data frame with rows representing each taxon and two columns: ‘species’, which is the same as the unique values of the input ‘species’, and ‘native_anywhere_in_aus’, a vector indicating whether each species is native anywhere in Australia, introduced by humans from elsewhere, or unknown with respect to the APC resource. | |
| state_diversity_counts | Calculates state-level diversity for native, introduced, and more complicated species origins based on the geographic data available in the APC. | state, | A data frame with three columns: ‘origin’ indicating the origin of the species, ‘state’ indicating the Australian state or territory, and ‘num_species’ indicating the number of species for that origin and state. | |
| resources = load_taxonomic_resources() | ||||
For additional information, see https://traitecoevo.github.io/APCalign/articles/function_notes.html.
The package is coded in R (ver. 4.3.1, https://www.R-project.org/), an open-source language widely used in the ecological sciences (R Core Team 2024). Within R, we leveraged open source packages, including diverse packages from tidyverse (dplyr, purr, readr, rlang, stringr) (Wickham et al. 2019), arrow (Richardson et al. 2024), curl (Ooms et al. 2024), httr (Wickham 2023), jsonlite (Ooms 2014), stringdist (van der Loo 2014), stringi (Gagolewski 2022), and testthat (Wickham 2011). All analyses within the paper use APCalign v1.0.0, with APC/APNI data downloaded 29 April 2024 (ver. 0.0.5.9000).
Loading taxonomic resources
The taxonomic resources underpinning ‘APCalign’ are the datasets within Australia’s National Species List (auNSL) pertaining to terrestrial vascular plants, the Australian Plant Census (APC) and the Australian Plant Names Index (APNI). The APC is a compilation of endorsed taxon concepts, including both currently accepted taxon concepts and taxon concepts with an alternative taxonomic status, such as synonyms or orthographic variants. Nearly all taxon concepts with a taxonomic status other than ‘accepted’ can be matched to an accepted taxon concept through an identifier (‘acceptedNameUsageID’). For ‘APCalign’, taxon concepts (and their associated scientific names) within the APC are divided into accepted taxon concepts and taxon concepts with an alternative taxonomic status, such as taxonomic synonym, basionym, orthographic variant, and misapplied statuses. Taxon concepts within the APC that have a taxonomic status other than ‘accepted’ are collectively referred to hereafter as ‘APC-synonym’. The APNI is a dataset of all scientific names documented as having been applied to a native or naturalised plant growing in Australia, including both the ~110,000 names included in the APC and ~24,500 names not linked to an APC taxon concept. For ‘APCalign’, only the subset of APNI names not also in APC is considered.
The function ‘load_taxonomic_resources()’ loads copies of the APC and APNI, either a default version (the latest archived version), the current data downloaded directly from the APC, or a specified version number. On loading, the function creates several data frames by filtering and selecting data from the loaded tables, and dividing the resources on the basis of the taxon concepts’ taxonomic status (for APC) and the taxon rank of the taxon concepts (for APC) or scientific names (for APNI). A separate function, (‘standardise_taxon_rank()’, then standardises taxon ranks, replacing Latin rank with their English equivalents.
Taxonomic alignment and updating
Overview
Aligning the original scientific names to the best match within the APC/APNI requires multiple considerations, including (1) specific syntax patterns that identify the rank to which the name can most appropriately be aligned, (2) whether matches are best made to the entire string or a subset of words, (3) when matches should be made to names from the APC versus APNI, and (4) whether fuzzy matches are appropriate. Users can select whether (1) matches are sought in both the APC and APNI (default) or just the APC, (2) whether fuzzy matching should be enabled (default) or disabled, (3) the initial fuzzy-matching threshold (how fuzzy of a match is permitted, with a default threshold of no more than three individual and 20% changed characters), and (4) whether a second round of imprecise fuzzy matching should be enabled or disabled (default).
Scientific names matched to a taxon concept within the APC are then updated to a currently accepted taxon concept within the APC and additional metadata columns propagated depending on the taxon rank of the taxon concept. For instance, a user input of ‘Dryandra arborea’ would be updated to ‘Banksia arborea’, and the APC-identifiers for both the taxon concept and the genus are then added. Users can select how to update names for taxon concepts where taxonomic splits introduce ambiguity as to which currently accepted taxon concept is correct.
‘APCalign’ includes one function to standardise taxon names (‘standardise_names()’, two functions to simplify names (‘strip_names()’ and ‘strip_names_extra()’), one function to align input names with names in the APC/APNI (‘align_names()’), and one function to update names to those for currently accepted taxon concepts (‘update_taxonomy()’) (Table 2). However, most users will instead use the function ‘create_taxonomic_update_lookup()’ which uses these individual functions to standardise, align, and update the original names in a single step (Tables 2, 3). Example inputs, outputs and code are available at the package website at https://traitecoevo.github.io/APCalign/.
| Original name variant | Match type | Taxonomic datasets | Taxon rank of aligned name | |
|---|---|---|---|---|
| 1. Detect perfect matches. | ||||
| ‘scientific_name’ | Exact | APC-accepted | Species/infraspecific | |
| ‘scientific_name’ | Exact | APC-synonyms | Species/infraspecific | |
| ‘cleaned_name’ | Exact | APC-accepted | Species/infraspecific | |
| ‘cleaned_name’ | Exact | APC-synonyms | Species/infraspecific | |
| 2. Align 2-word strings that indicate an unknown species within a genus (or family). | ||||
| ‘cleaned_name’ | Exact | APC-accepted, APC-synonyms, APNI | Genus | |
| ‘cleaned_name’ | Fuzzy | APC-accepted | Genus | |
| ‘cleaned_name’ | Fuzzy | APC-synonyms | Genus | |
| ‘cleaned_name’ | Exact | APC-accepted | Family | |
| 3. Find strings that indicate a name is an intergrade between two taxa. These names can only be aligned to a genus. | ||||
| ‘cleaned_name’ | Exact | APC-accepted, APC-synonyms, APNI | Genus | |
| ‘cleaned_name’ | Fuzzy | APC-accepted | Genus | |
| ‘cleaned_name’ | Fuzzy | APC-synonyms | Genus | |
| ‘cleaned_name’ | Fuzzy | APNI | Genus | |
| ‘cleaned_name’ | No match | NA | NA | |
| 4. Find strings that indicate a name reflects a data collector’s indecision about which of two (or more) taxa is the appropriate taxon. These names can only be aligned to a genus. | ||||
| ‘cleaned_name’ | Exact | APC-accepted, APC-synonyms, APNI | Genus | |
| ‘cleaned_name’ | Fuzzy | APC-accepted | Genus | |
| ‘cleaned_name’ | Fuzzy | APC-synonyms | Genus | |
| ‘cleaned_name’ | Fuzzy | APNI | Genus | |
| ‘cleaned_name’ | No match | NA | NA | |
| 5. Check if strings are taxon names, lacking authorship. | ||||
| ‘stripped_name’ | Fuzzy | APC-accepted | Species/infraspecific | |
| ‘stripped_name’ | Fuzzy | APC-synonyms | Species/infraspecific | |
| ‘cleaned_name’ | Exact | APNI | Species/infraspecific | |
| 6. Find strings that indicate a name that indicates an affinity to a specific taxon or suggestion to compare it to a specific taxon, but the name itself is not that taxon. Such names, unless documented in APC (i.e. matches 1 or 5 above) can only be aligned to genus. | ||||
| ‘cleaned_name’ | Exact | APC-accepted, APC-synonyms, APNI | Genus | |
| ‘cleaned_name’ | Fuzzy | APC-accepted | Genus | |
| ‘cleaned_name’ | Fuzzy | APC-synonyms | Genus | |
| ‘cleaned_name’ | Fuzzy | APNI | Genus | |
| ‘cleaned_name’ | No match | NA | NA | |
| 7. Further checks if strings are taxon names, lacking authorship, now with imprecise fuzzy matching. | ||||
| ‘stripped_name’ | Imprecise fuzzy | APC-accepted | Species/infraspecific | |
| ‘stripped_name’ | Imprecise fuzzy | APC-synonyms | Species/infraspecific | |
| 8. Find strings that indicate a name that is a hybrid between two taxa. Such names, unless documented in APC (i.e. matches 1 or 5 above) can only be aligned to genus. | ||||
| ‘cleaned_name’ | Exact | APC-accepted, APC-synonyms, APNI | Genus | |
| ‘cleaned_name’ | Fuzzy | APC-accepted | Genus | |
| ‘cleaned_name’ | Fuzzy | APC-synonyms | Genus | |
| ‘cleaned_name’ | Fuzzy | APNI | Genus | |
| ‘cleaned_name’ | No match | NA | NA | |
| 9. Check if the first three words in the name string match with a taxon name, allowing notes to be discarded. Also useful for aligning phrase names. | ||||
| words 1–3 of ‘stripped_name_2’ | Exact | APC-accepted | Species/infraspecific | |
| words 1–3 of ‘stripped_name_2’ | Exact | APC-synonyms | Species/infraspecific | |
| words 1–3 of ‘stripped_name_2’ | Fuzzy | APC-accepted | Species/infraspecific | |
| words 1–3 of ‘stripped_name_2’ | Fuzzy | APC-synonyms | Species/infraspecific | |
| 10. Check if the first two words in the name string match with a taxon name, allowing notes and invalid infraspecific names to be discarded. Also useful for aligning phrase names. | ||||
| words 1–2 of ‘stripped_name_2’ | Exact | APC-accepted | Species | |
| words 1–2 of ‘stripped_name_2’ | Exact | APC-synonyms | Species | |
| words 1–2 of ‘stripped_name_2’ | Fuzzy | APC-accepted | Species | |
| words 1–2 of ‘stripped_name_2’ | Fuzzy | APC-synonyms | Species | |
| 11. Further checks if name strings are APNI taxon names, lacking authorship, now with fuzzy matching or considering just the first three or two words in the string. | ||||
| ‘stripped_name’ | Fuzzy | APNI | Species/infraspecific | |
| ‘stripped_name’ | Imprecise fuzzy | APNI | Species/infraspecific | |
| words 1–3 of ‘stripped_name_2’ | Exact | APNI | Species/infraspecific | |
| words 1–2 of ‘stripped_name_2’ | Exact | APNI | Species | |
| 12. Check if the first word in the name string matches with a taxon name, allowing an alignment to the genus level or family level. | ||||
| first word of ‘original_name’ | Exact | APC-accepted | Genus | |
| first word of ‘original_name’ | Exact | APC-synonyms | Genus | |
| first word of ‘original_name’ | Exact | APNI | Genus | |
| first word of ‘original_name’ | Exact | APC-accepted | Family | |
| first word of ‘original_name’ | Exact | APC-synonyms | Family | |
| first word of ‘original_name’ | Fuzzy | APC-accepted | Genus | |
| first word of ‘original_name’ | Fuzzy | APC-synonyms | Genus | |
| first word of ‘original_name’ | Fuzzy | APC-accepted | Family | |
| first word of ‘original_name’ | Fuzzy | APC-synonyms | Family | |
Names are sequentially tested until successful alignment is achieved. The APC is divided into two subsets, one including all accepted taxon concepts (‘APC-accepted’) and a second with taxon concepts with a taxonomic status other than ‘accepted’ (‘APC-synonyms’). For additional information, see https://traitecoevo.github.io/APCalign/articles/updating-taxon-names.html.
NA, not applicable.
Standardising names
A preprocessing step before aligning input names to names in the APC/APNI is to create six derivations of the original name (for the list of submitted names) or the canonical name (from the taxonomic resources), because different algorithms seek matches against different derivations. For instance, for the APC, matches are sought to scientific names with authorship, scientific names without authorship (termed canonical names), and subsets of words from the canonical name. To aid with name alignments, three standardisation and simplification functions are applied both to the taxonomic resources and the submitted original names. First, for the original names, taxon rank abbreviations and qualifiers are standardised (to: sp., var., subsp., f., aff., ser.), and variants on sensu lato, sensu stricto, and excess white space are removed; this is performed by the function ‘standardise_names()’ (Table 2). The output, termed the ‘cleaned_name’, is then passed through one of two additional functions: ‘strip_names()’, which removes punctuation, var., subsp., f., and ser., or both ‘strip_names’ and ‘strip_names_extra()’, the latter which removes the filler words cf., sp., and x) (Table 2). From the output of ‘strip_names_ extra’, one can then extract the first one, two or three words, and be confident that they are informative terms pertaining to the taxon name. All canonical names in the APC and APNI are also passed through ‘strip_names()’ and ‘strip_names_ extra()’ to create derivations of the complete scientific names to permit comparisons between the input names and the names in the taxonomic resources.
Aligning with APC/APNI names
The function ‘align_taxa()’ then uses the six name derivations to indicate the following: (1) the lowest taxon rank to which the original name could be aligned (family, genus, species, or infraspecies); (2) the taxonomic dataset to which the original name could be aligned (APC or APNI); (3) for names aligned to the APC, whether the taxonomic status of the aligned name is accepted or a ‘synonym’ (broadly defined as a true synonym or a basionym, orthographic variant, etc.); and (4) the best possible match to a taxon concept (in APC) or other scientific name (in APNI) (Table 3).
Perfect matches to scientific names in the APC (with or without authorship) are sought first. If a perfect match is not detected, each original name, or derivation thereof, is automatically, sequentially passed through as many of the additional 50 algorithms as is required until an alignment can be made (Tables 3, 4). Each of these algorithms uses a unique combination of (1) names from the APC/APNI, (2) entire versus partial strings (1, 2 or 3 words), (3) perfect versus fuzzy matches and (4) specific fuzzy-match settings. In addition, specific characters are identified that reliably indicate when an original name can never be resolved more specifically than a genus-level (or even family-level) name. The sequence of these match-steps was calibrated to ensure that perfect matches take precedence over fuzzy matches, APC matches over APNI matches, and that taxon names that cannot be aligned to a species are matched only against a list of genus and family names. The sequence of algorithms was carefully curated to maximise the number of accurate alignments by repeatedly testing the more than 47,000 unique original names from the AusTraits plant-trait database (Falster et al. 2021). Particular considerations incorporated into the algorithms are listed below.
| original_name | aligned_name | accepted_name | suggested_name | genus | taxon_rank | taxonomic_ dataset | taxonomic_ status | scientific_name | aligned_reason | update_reason | number_of_ collapsed_ taxa | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acacia aneura | Acacia aneura | Acacia aneura | Acacia aneura [alternative possible names: Acacia minyura (pro parte misapplied) | Acacia paraneura (pro parte misapplied) | Acacia quadrimarginea (misapplied)] | Acacia | Species | APC | Accepted | Acacia aneura F.Muell. ex Benth. | Exact match of taxon name to an APC-accepted canonical name once punctuation and filler words are removed (29 January 2024) | Aligned name accepted by APC | 1 | |
| Argyrodendron Whyanbeel | Argyrodendron sp. Whyanbeel (B.P.Hyland RFK1106) | Argyrodendron sp. Whyanbeel (B.P.Hyland RFK1106) | Argyrodendron sp. Whyanbeel (B.P.Hyland RFK1106) | Argyrodendron | Species | APC | Accepted | Argyrodendron sp. Whyanbeel (B.P.Hyland RFK1106) Qld Herbarium | Exact match of the first two words of the taxon name to an APC-accepted canonical name (29 January 2024) | Aligned name accepted by APC | NA | |
| Atherosperma moschatum integrifolium | Atherosperma moschatum subsp. integrifolium | Atherosperma moschatum subsp. integrifolium | Atherosperma moschatum subsp. integrifolium | Atherosperma | Subspecies | APC | Accepted | Atherosperma moschatum subsp. integrifolium (A.Cunn. ex Tul.) Schodde | Fuzzy match of taxon name to an APC-accepted canonical name once punctuation and filler words are removed (29 January 2024) | Aligned name accepted by APC | NA | |
| Athrotaxis laxiflolia | Arthrotaxis laxifolia | Athrotaxis × laxifolia | Athrotaxis × laxifolia | Athrotaxis | Species | APC | Accepted | Athrotaxis × laxifolia Hook. | Fuzzy match of taxon name to an APC-synonymous canonical name once punctuation and filler words are removed (29 January 2024) | Orthographic variant | NA | |
| Banksia integrifolia | Banksia integrifolia | Banksia integrifolia | Banksia integrifolia | Banksia | Species | APC | Accepted | Banksia integrifolia L.f. | Exact match of taxon name to an APC-accepted canonical name once punctuation and filler words are removed (29 January 2024) | Aligned name accepted by APC | NA | |
| Commersonia rosea | Commersonia rosea | Androcalva rosea | Androcalva rosea | Androcalva | Species | APC | Accepted | Androcalva rosea (S.A.J.Bell & L.M.Copel.) C.F.Wilkins & Whitlock | Exact match of taxon name to an APC-synonymous canonical name once punctuation and filler words are removed (29 January 2024) | Basionym | NA | |
| Dryandra sp. | Dryandra sp. | NA | Banksia sp. | Banksia | Genus | APC | Genus accepted | NA | Exact match of taxon name ending with ‘sp.‘ to an APC genus (29 January 2024) | NA | NA | |
| Fabaceae sp. | Fabaceae sp. | NA | Fabaceae sp. | NA | Family | APC | Family accepted | NA | Exact match of taxon name ending with ‘sp.‘ to an APC-accepted family (29 January 2024) | NA | NA | |
| Galactia striata | Galactia sp. [Galactia striata] | NA | Galactia sp. [Galactia striata] | Galactia | Genus | APC | Genus accepted | NA | Exact match of the first word of the taxon name to an APC-accepted genus (29 January 2024) | NA | NA | |
| Grevillea umbellulata subsp. acerosa | Grevillea umbellulata subsp. acerosa | Grevillea umbellulata | Grevillea umbellulata | Grevillea | Subspecies | APC | Accepted | Grevillea umbellulata Meisn. | Exact match of taxon name to an APC-synonymous canonical name once punctuation and filler words are removed (29 January 2024) | Taxonomic synonym | NA | |
| Hibbertia stricta | Hibbertia stricta | Hibbertia stricta | Hibbertia stricta [alternative possible names: Hibbertia australis (pro parte misapplied) | Hibbertia devitata (pro parte misapplied) | Hibbertia glebosa (pro parte misapplied) | Hibbertia glebosa subsp. glebosa (pro parte misapplied) | Hibbertia glebosa subsp. oblonga (pro parte misapplied) | Hibbertia platyphylla (pro parte misapplied) | Hibbertia platyphylla subsp. platyphylla (pro parte misapplied) | Hibbertia riparia (pro parte misapplied) | Hibbertia australis (misapplied) | Hibbertia glebosa (misapplied) | Hibbertia glebosa subsp. glebosa (misapplied) | Hibbertia gracilipes (misapplied) | Hibbertia platyphylla subsp. platyphylla (misapplied) | Hibbertia riparia (misapplied)] | Hibbertia | Species | APC | Accepted | Hibbertia stricta (DC.) R.Br. ex F.Muell. | Exact match of taxon name to an APC-accepted canonical name once punctuation and filler words are removed (29 January 2024) | Aligned name accepted by APC | 1 |
For details on Hibbertia stricta, see Toelken and Miller (2012).
NA, not applicable.
The first four algorithms detect perfect matches to scientific names in the APC (with or without authorship) (Table 3). Exact matches to APNI names are performed only much later in the series of algorithm steps, as APNI includes many scientific names that are slight spelling variants of the names of taxon concepts documented by the APC, but which have not yet been matched to an APC taxon concept. An explicit decision was made to match these names to a similar name in the APC instead of an exact match in APNI, as otherwise many apparent typographic errors were aligned with a name only present in APNI, rather than an accepted taxon concept in the APC that differed by a single character.
The following three string patterns reliably indicate that a name can be aligned only to a genus: (1) ‘genus sp.’, indicating a plant has been identified only to genus level; (2) ‘–’, indicating an intergrade, and (3) ‘/’, indicating indecision between two names (Table 3). If these string patterns are detected, the name is reformatted as ‘genus sp. [originally submitted name]’. The function ‘align_taxa()’ includes an argument ‘identifier’ that allows either a fixed string or a vector of strings (or a column) to be added to each reformatted name: ‘genus sp. [originally submitted name; identifier]’. This ensures that, for instance, multiple records of ‘Eucalyptus sp.’ that are documented at different locations or by different people have an identifier linked that clarifies that they are not necessarily identical taxon concepts, but rather members of the genus Eucalyptus that the dataset collector could not identify to species level.
There are additional string patterns that can occur either as part of a name documented by the APC/APNI or which indicate a name that can be resolved only to genus level. These include hybrids (detected by the presence of ‘x’), species affinis (detected by variants of ‘aff’), conferret (meaning ‘compare to’, detected by ‘cf’). If an input name includes these strings, a match is first sought against the APC/APNI; if that fails, genus-level matches are sought. Overall, for all names for which a genus-resolution match is appropriate, the first word of the name is matched sequentially against APC-accepted genera, then ‘APC-synonymous’ genera and APNI genera, followed by fuzzy matches to genera from each of these lists, and, finally, to APC-accepted family names.
Minor typographic errors are commonplace in scientific names in ecological datasets, most often owing to incorrect vowel combinations or omitting/incorrectly including double-consonants. The fuzzy-matching algorithm uses the ‘stringdist’ function within the eponymous R-package, setting ‘method = ‘dl’’ (van der Loo 2014). The ‘dl’ method uses the Damerau–Levenshtein distance, which calculates the number of insertion, deletion, substitution, and transposition operations required to transform one string into the other, offering a ‘distance’ measure that closely reflects spelling errors humans make. Fuzzy matching allows such errors to be corrected, but fuzzy-match algorithms must be carefully calibrated to correct mistakes without making erroneous matches. Therefore, two levels of fuzzy matches were performed by the algorithms. For the first, the default is set to a conservative maximum change threshold of three characters and 20% of characters, which is sufficient to catch most minor typographic errors. The first letter of each word in the genus, species epithet and, when relevant, infraspecific epithet, remain fixed, because erroneous changes to the first letter of the species epithet are uncommon in datasets, yet allowing the first letter to be changed repeatedly introduced erroneous matches in test datasets. Arguments in the function ‘align_taxa()’ allow the degree of fuzziness to be changed or to disable all fuzzy matches. Later in the algorithm sequence, alignments are sought through less precise fuzzy matches, which allow changes of up to five characters or 25% of the characters. This occasionally corrects legitimate typographic errors, but more frequently makes erroneous alignments and is therefore turned off as a default. Each of these function arguments can be adjusted by users, or, if omitted, the defaults are applied.
Additional matches (exact or fuzzy) can be accomplished by extracting just particular words from the original name string. Such matches are made on name strings that have been processed by the functions ‘strip_names’ followed by ‘strip_names_extra()’. The following three categories of names are readily aligned when just two or three words in the stripped name are considered: phrase names, accepted species names with infraspecific epithets that do not exist in the APC/APNI, and names where the ‘name’ includes both a valid scientific name and field notes. Phrase names are accurately aligned through these algorithms, because the first word (usually the genus) and the second and third words (usually a place name or descriptive term; or a place name or descriptive term + voucher number) are generally accurately recorded in the original name, but the exact syntax and the inclusion/omission of the collector’s name and initials rarely follows the exact APC convention (Barker 2005). For instance, these matches successfully align ‘Argyrodendron Whyanbeel’ to ‘Argyrodendron sp. Whyanbeel (B.P.Hyland RFK1106)’, the latter being an APC-accepted phrase name (Table 4).
Finally, at the end of the match algorithm sequence, any taxon names that have not been aligned are assumed to be able to be aligned only to genus or family level. For these, the first word in the original name is matched to lists of genus-level APC/APNI names and family-level APC names.
The output from the ‘align_taxa()’ function is a table that includes an aligned name, the taxonomic dataset for the aligned name, and the taxon rank of the aligned name.
Updating taxonomic names
The function ‘update_taxonomy()’ performs a series of manipulations, including (1) updating names that are ‘APC synonyms’ to their currently accepted taxon concept (‘accepted_name’), (2) offering a ‘suggested_name’ for each ‘aligned_name’ (especially important for ‘aligned_names’ for which there is no ‘accepted name’, as occurs if the ‘aligned_name’ is genus rank or aligned to an APNI name; the ‘suggested_name’ is identical to the ‘accepted_name’ whenever an ‘accepted_name’ exists), (3) adding taxon name identifiers to each row, and (4) offering suggested alternative names for taxon names that are ambiguous owing to taxon concept splits. The function ‘update_taxonomy()’ first splits the table of aligned names into subtables on the basis of the taxonomic dataset and taxon rank specified. This permits different subfunctions to be used to update the taxon names in each subtable, because different rules for updating names and adding taxon concept/scientific name metadata are applied to each group (Table 5).
| Function name | Category of aligned name processed | Update to ‘aligned_name’ | Format of ‘suggested_name’ | Columns filled in | ||||
|---|---|---|---|---|---|---|---|---|
| Taxonomic dataset | Taxon rank | Accepted name A | Scientific name A | Genus A | ||||
| update_taxonomy_APC_genus | APC | Genus | To APC-accepted genus | genus sp. [notes] | No | No | Yes | |
| update_taxonomy_APNI_genus | APNI | Genus | None | genus sp. [notes] | No | No | No | |
| update_taxonomy_APC_family | APC | Family | None | family sp. [notes] | No | No | No | |
| update_taxonomy_APC_species_and_infraspecific_taxa | APC | Species B | APC-accepted species name | Yes | Yes | Yes | ||
| taxonomic_splits = ‘most_likely_species’ | To APC-accepted taxon concept | Most likely APC-accepted species name [alternative possible names] | Yes | Yes | Yes | |||
| taxonomic_splits = ‘return_all’ | To APC-accepted taxon concept | All possible APC-accepted species name (extra rows added) | Yes | Yes | Yes | |||
| taxonomic_splits = ‘collapse_to_higher_taxon’ | Collapsed to APC-accepted genus | genus sp. [collapsed names] | No | No | Yes | |||
| update_taxonomy_APNI_species_and_infraspecific_taxa | APNI | Species B | None to ‘aligned_name’; genus to APC-accepted genus if possible | APNI listed species name | No | Yes | Sometimes | |
| (Not aligned) | (Not aligned) | (Not aligned) | None | None | No | No | No | |
These functions are called within the function ‘update_taxonomy’ and are not exported. For additional information, see https://traitecoevo.github.io/APCalign/articles/updating-taxon-names.html.
The accepted name column is filled in only for species (and infraspecific taxon concepts) that occur in the APC. For taxon concepts whose taxonomic status is ‘accepted’, their ‘accepted_name’ is identical to their ‘aligned_name’. Names that have been aligned to the list of ‘APC synonyms’ are linked to taxon concepts with a taxonomic status other than ‘accepted’. The APC includes a column ‘accepted_name_usage_ID’ that allows ‘APC-synonym’ names to be matched to the currently accepted taxon concept; this column makes it possible to update ‘APC synonym’-aligned names to the appropriate APC-accepted name (and corresponding taxon concept).
The ‘suggested_name’ column offers a best-practice name for all input names that can be aligned at least at the family level. In order of preference, the suggested name is an APC-accepted taxon concept, a taxon concept from the ‘APC-synonym’ list that has not been connected to an accepted taxon concept in the APC (a rare situation), a documented APNI canonical name (the aligned name), or a genus resolution-aligned or family resolution-aligned name. For genus-resolution names, where the genus is a ‘synonym’ (broadly defined) of an ‘APC-accepted’ genus, the genus is updated to the currently accepted genus name.
When possible, unique identifiers provided as part of the auNSL resources are attached to each taxon concept (for species and infraspecific taxon concepts aligned to the APC), scientific names (for all species and infraspecific names aligned to either the APC or APNI), and the genus taxon concept (where the aligned genus is accepted by the APC).
For APC-accepted taxon concepts, taxonomic splits offer an additional complication. A taxonomic split occurs when a previously unified taxon concept has been divided into multiple unique taxon concepts. When the aligned name is the name of the older taxon concept, it is ambiguous whether this name refers to the older broader taxon concept or the current, more narrowly circumscribed taxon concept. The ‘update_taxonomy()’ function includes an argument ‘taxonomic_splits’ that offers three different outputs for these scenarios. (1) The default is to return the name of the ‘most_likely’ taxon concept, the taxon concept that existed in the past as a broad taxon concept and continues to be used today as a narrower taxon concept. The list of alternative taxon-concept names is included in square brackets following the most likely name (Table 4). (2) ‘Collapse_to_higher_taxon’ aligns such names to the genus, implying that is the best resolution that can be achieved for these ambiguous taxon concepts. The list of currently accepted taxon-concept names is included in square brackets following the genus. (3) ‘Return_all’ leads to a longer output table, with all currently accepted taxon concepts returned as separate rows.
Distribution and establishment means
The APC also specifies state-level distribution for the accepted taxa, including information about their native or introduced status; this piece of data is termed ‘establishment means’ in DarwinCore (Wieczorek et al. 2012). Information about state-level establishment means is a result of a complex consultation process involving the state herbaria. A static version of this is available (Martín-Forés et al. 2023), along with R scripts. A very similar product is available here, but in a more user-friendly R package with automatic file handling that also dynamically updates as APC moves forward.
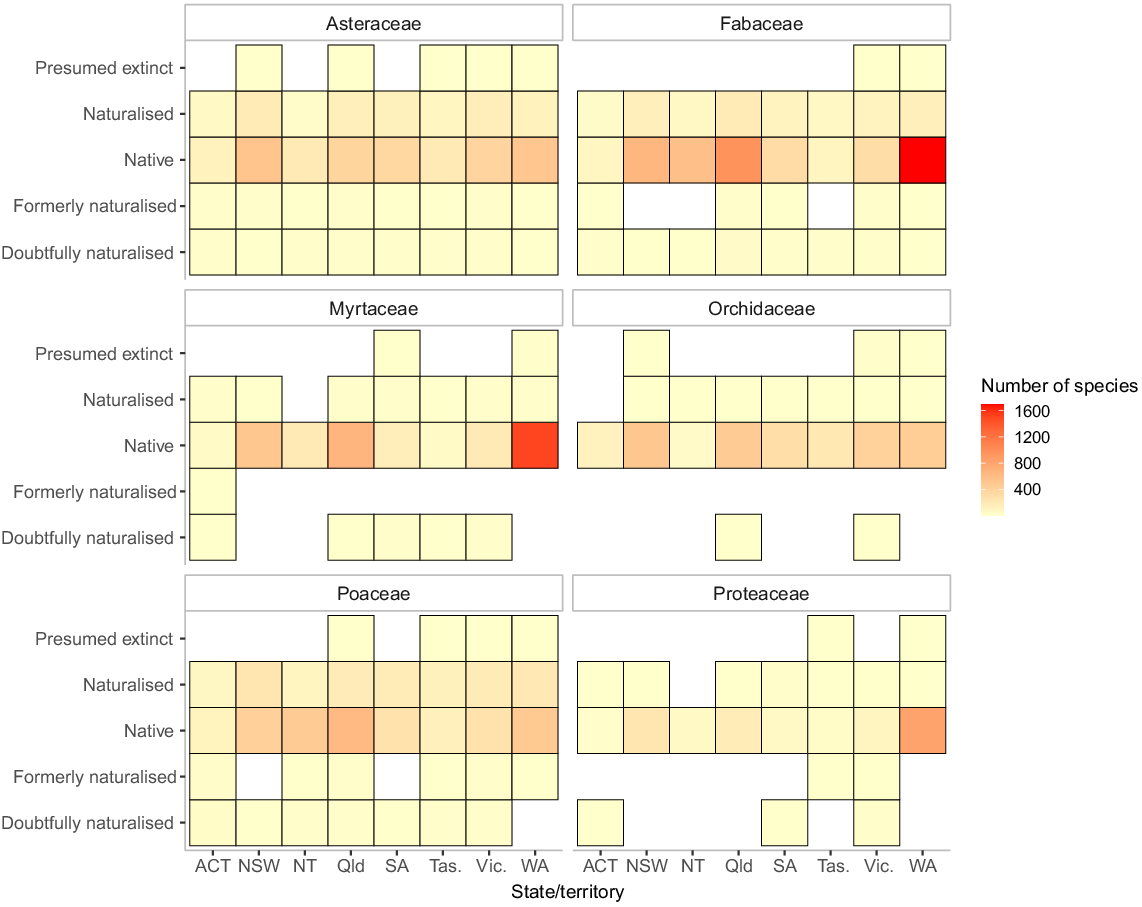
One complex feature of the Australian flora is internal out-of-range introductions, with taxa from one part of the continent becoming naturalised in another (Fig. 1). Some of these taxa become very successful outside of their ‘native’ range. A specific type of introduction that may become more important under future climate scenarios is where taxa from mainland Australia have been very recently introduced to Tasmania and other islands. The full complexity of taxon distribution and native status documented in the APC data is available via two functions, namely ‘create_species_state_origin_matrix()’ and ‘native_anywhere_in_australia()’. The function ‘create_species_state_origin_matrix()’ can be especially informative for specific taxa (Fig. 1). For the reduced species lookup table, treating all of Australia as an entity, there is a function ‘native_anywhere_in_australia()’. A related function, ‘state_diversity_counts()’ calculates state-level diversity for native taxa, introduced taxa, and taxa with more complicated origins (Table 2).
Distribution of the number of vascular plant species in several major Australian families, split by family, state/territory, and establishment means. Note that individual species may appear in different states with different statuses, and the striking variation in geographic diversity patterns across families.
Performance and comparison with existing tools
The ‘APCalign’ algorithms were explicitly developed to standardise and align diverse lists of taxonomic names submitted to the AusTraits plant-trait database (Falster et al. 2021). These taxon lists were derived from ecological field studies, taxonomic revisions, online floras, experimental datasets, and government biodiversity reports. Each category of dataset offered unique challenges that were considered in algorithm development. In particular, aligning the many phrase names in common use in Australian taxonomic literature required distinct tools not yet available in any other existing taxonomic resolution tools worldwide. Although the APC prescribes an exact syntax for phrase names, such as the absence of spaces between the collector’s initials (Barker 2005), phrase names in different resources rarely align perfectly with the APC and fuzzy matches cannot reliably match names with missing collectors or voucher numbers. The solution within ‘APCalign’ was, late in the algorithm sequence, to perform matches on just the first two or three ‘real’ words.
To test the performance of ‘APCalign’ across diverse datasets, 47,698 unique taxon names contained within the 362 datasets in AusTraits v4.2.0 (Falster et al. 2021) were submitted to ‘APCalign’. The distinct names in the AusTraits dataset include minor syntax variants within the dataset (e.g. ‘subsp.’ versus ‘ssp.’), more significant syntax variants (absence of taxon designations such as ‘subsp.’ and ‘var.’; incomplete phrase names), spelling mistakes (in the original datasets submitted to AusTraits), taxonomic synonyms, and incomplete names (e.g. Acacia sp. [long leaves]). In total, 95.7% of the AusTraits names had perfect matches to names in the APC (whether accepted or synonyms, with or without authorship) (Fig. 2); the effort to create alignment algorithms focused on the final 4.3% of names. Most of the additional matches were names where a conservative fuzzy match (changes of fewer than three characters and 20% of characters) aligned the name to the APC (1.8%) or names that were documented in the APNI (0.4%). However, the algorithms for other alignments captured important classes of name. In particular, many ecological datasets include names that indicate a taxon that the observer could not resolve to species, with names that follow some variant of ‘genus sp.’ or ‘genus species A/species B’ or ‘genus sp. (notes)’.
Frequency of different name-handling classes found during alignment of 47,698 names from 362 raw inputs to the AusTraits compilation (Falster et al. 2021). Each cell is sized to represent the proportion of taxon names within AusTraits that were matched by a specific algorithm, with the broad algorithm categories being indicated in the legend and all algorithms included in Table 3. Although the vast majority of names were ‘perfect’ matches, almost all APCalign algorithms were triggered by some of the names submitted.
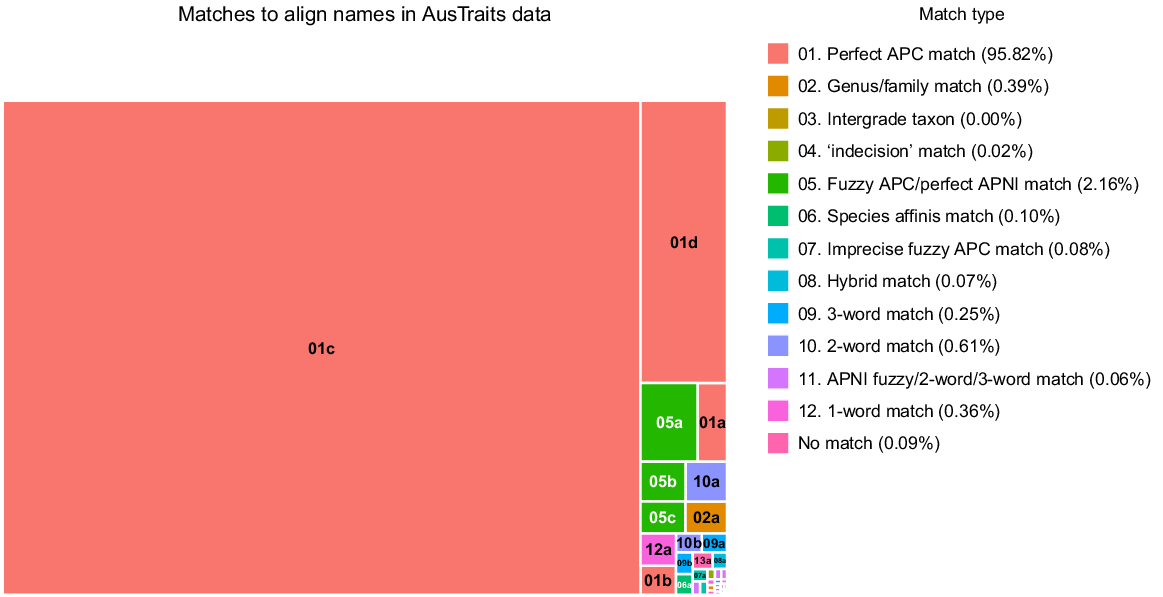
A sequence of different algorithms was used to identify all genus- and family-level names and confirm that the name was listed within the APC or APNI. In all, 0.7% of all taxon names could be resolved only to genus- or family-level names. The matches that looked at only the first two or three words, ignoring all infraspecific taxon designations and punctuation, also were shown to be essential to accurately align many taxon names documented in AusTraits datasets. These matches aligned an additional 0.8% of taxa to an accepted name, mostly aligning (1) two or three words from the submitted name string where the name string included notes and (2) phrase names. Just 240 names (0.1%) could not be aligned by any of the matching algorithms; manual inspection of these names indicated that alignments to an APC/APNI name were possible for just 32 of these, mostly to the family level. Importantly, of the 99.9% of names that were aligned, there were <0.1% that the AusTraits team deemed to be incorrect alignments during comprehensive manual checks of names matched by anything other than the exact matches to species names and conservative fuzzy-matching algorithm, and spot checks (~25%) of names matched by the conservative fuzzy-matching algorithm; these were almost entirely ‘imprecise fuzzy matches’, which are known to often introduce errors and are, by default, turned off within the ‘APCalign’ functions. When turned on, 0.1% of taxon names were aligned by the imprecise fuzzy matches, but about half of these were subsequently reverted (manually), indicating that they are a useful tool for exploring possible name alignments, but not for mass processing when each aligned name will not be reviewed. When names of non-native, non-naturalised taxa are submitted to APCalign, if the genus is not present in the APC/APNI the name is usually unmatched (54 of the unmatched taxa from AusTraits). If the genus, but not species, is in APC/APNI, the name will be aligned only to the genus level.
We also compared the alignment algorithms of ‘APCalign’ with those of three existing global tools, namely, (taxonomic name resolution service (‘TNRS’); Boyle et al. 2013), ‘taxize’ (Chamberlain and Szöcs 2013; Chamberlain et al. 2022), and ‘kewr’ (Walker 2021). ‘taxize’ describes itself as a taxonomic toolbelt, simultaneously searching 20 separate global taxonomic resources to find matches, whereas ‘kewr’ matches names to six different taxonomic resources. There is a significant overlap between these resources. ‘TNRS’ uses the World Flora Online (Borsch et al. 2020) and the World Checklist of Vascular Plants (Govaerts et al. 2021). These R-based tools all allow lists of names to be read in, offering the rapid alignment of entire datasets. Across the Tree of Life, many such packages exist, referencing either global or regional taxonomic datasets (Boyle et al. 2013; Grenié et al. 2023).
Of the tools compared, ‘APCalign’, ‘TNRS’, and ‘taxize’ include fuzzy-matching algorithms, required to correct the minor typographic errors that proliferate in scientific names (Table 1). Meanwhile, ‘kewr’ does not perform syntax standardisation or fuzzy matches and, therefore, returns ‘NA’ when an exact match is not detected. Another feature of ‘APCalign’ that is shared by ‘TNRS’ and ‘taxize’ is an output column that identifies the resolution of the output name, indicating, if, for instance, a name can be matched to a genus or family, but not a species (Table 1). The outputs from each tool vary according to the encoded matching algorithms. Although ‘TNRS’ and ‘taxize’ also incorporated fuzzy matching, a distinctive feature of the ‘APCalign’ fuzzy-matching function is that the first letter of each word cannot change. This occasionally misses a valid alignment, but without this feature, there are many additional false alignments made. ‘APCalign’ also allows the fuzzy-matching level to be adjusted, should a user wish to institute more or less conservative fuzzy matching.
Another feature of ‘APCalign’, not shared by the other tools, was detecting a series of symbols/punctuation that indicated that the name could be aligned only to a genus, not a species. These include ‘x’ (a hybrid), ‘aff.’ (given taxon shows an affinity to the named taxon) ‘cf.’ (given taxon should be compared to the named taxon), ‘--’ (taxon is an intergrade between two taxa), and ‘/’ (two taxon names submitted, because the author is unsure which is correct). The comparison tools all prioritise looking at just the first words in the string, which leads to the submitted name being aligned to the first of the names listed, whereas ‘APCalign’ automatically flags these names as being appropriate to align only to a genus. The initially submitted string is then inserted in square brackets following the genus, so the information is not lost (e.g. ‘genus sp. [genus species × genus species]’). In APCalign, the two-word and three-word matches are made only after names with such syntax patterns are flagged as being suitable to align only to genus resolution, and hence are not passed through these matching algorithms.
One ‘APCalign’ feature that is not available in any of the other outputs is the addition of a custom identifier for names resolved to the genus or family level. This does not improve alignment or increase our taxonomic interpretation of a specific name, but does offer a means to indicate that a taxon name that is resolvable only to a particular genus or family represents an unidentified species within a given dataset or location. For AusTraits, this is essential, because many datasets have submitted the name ‘Acacia sp.’ and a mechanism is required to indicate that these do not all refer to the same taxon concept, but instead to a multitude of different Acacia species that the dataset contributors could not identify to species.
An important feature of ‘APCalign’ shared by ‘TNRS’ and ‘taxize’ is the ability to update outdated taxon concepts to the names associated with their currently accepted taxon concept. Because lists of taxonomic names quickly become outdated, it is time-consuming to manually determine which names must be updated, then change them one by one. Because taxonomic concepts are a moving target, different taxon lists will consider different names to be ‘current’. As such, although ‘TNRS’ and ‘APCalign’ agree on the currently correct name for the majority of taxa, there are instances where ‘TNRS’ returned a recently updated name, whereas ‘APCalign’ reported an older name that is still considered current by the APC. This asynchrony will always exist across taxonomic lists, because each is updated by a different panel who, for some taxa, will make divergent judgements on the application of each name and review name changes at different times. All detected names that were updated by ‘TNRS’, but not ‘APCalign’, were manually checked and all were names that had been changed in the past 5 years. However, APCalign is the only resource that offers suggestions of alternative matches for scientific names that are ambiguous as a result of a previous taxonomic split, because the APC is one of the few taxonomic lists (regional or global in scope) that documents splits.
The goal of ‘APCalign’ is to align submitted names to the currently accepted name within Australia’s two vascular plant taxonomic resources, APC and APNI. ‘TNRS’ offers a similar aim, but at a global scale, and is therefore aligned to a global taxonomic resource. In contrast, ‘taxize’ and ‘kewr’, although seeking to standardise names, have the objective of matching an input taxon name to associated taxon names across many resources. For instance, whereas the ‘APCalign’ and ‘TNRS’ workflows aim to replace all outdated taxonomy with a currently accepted name, the outputs of ‘taxize’ and ‘kewr’ are developed to display ‘here are all the names, in all these resources, that are linked to the string that was input’, both leaving more name-selection decisions up to the user and providing a snapshot of the complexity of selecting a ‘best’ taxonomic name across all global resources. APCalign will perform poorly when presented with scientific names of species occurring only outside Australia or horticultural species, because these are not included in APCalign’s taxonomic reference lists, the APC and APNI.
Options for different users
Although ‘APCalign’ was created with researchers who are well-versed in R programming in mind, we also wanted to cater to non-R users. For non-R users who want to align and update plant taxonomic names, we recommend using the ‘APCalign-app’ web interface (https://unsw.shinyapps.io/APCalign-app/). ‘APCalign-app’ provides an easy entry point to the core functionality of the R package by replacing the multi-step, code workflow with a simple interface. The interface is preset with intuitive default settings that can be bypassed. This allows users to feasibly supply their taxonomic names, either by typing into an input box or uploading a .csv file and obtain aligned and updated names that can be downloaded. We hope that this mobile-enabled web interface can improve the consistency of plant taxonomic names for all users in the botanical community.
Conclusions
Australian researchers are fortunate to have the resources provided as part of the Australian National Species List for vascular plants, regularly updated to incorporate taxonomic revisions and the names of newly described taxa. This paper presents a new interface to the dynamic APC, which is the consensus taxonomy for native and naturalised Australian plants. It is essential that research projects, biodiversity assessments, and, indeed, nurseries in Australia, use the scientific names for vascular plants accepted by the APC, to facilitate communication at the national level.
Country or continental efforts will always be complementary to global efforts. Although there exist global taxon-matching tools with similar functionalities, none references the APC. Our comparison with the global tool ‘TNRS’ highlighted the importance of a local tool, as only the APC includes phrase names and aligns names to those specified within the Australian National Species Lists for vascular plants. Unlike global taxonomic alignment tools, ‘APCalign’ also considers taxonomic splits, allowing the user to decide what scientific name they consider appropriate for taxon concepts that have been subdivided during taxonomic revisions. The Brazil Flora 2020 effort has a similar country-specific tool with a shiny interface (http://www.plantminer.com/), specific to the Brazilian flora. An added benefit of working with a continent- or country-specific list is that fuzzy matching and synonym correction are less prone to false positive errors with smaller lists than with longer lists.
Although ‘APCalign’ was released as a complete R package only in late 2023, the underlying code has been tested on multiple datasets during its development, including the 47,000+ taxon names submitted to AusTraits, 12,600+ taxon names submitted to the AIslands project on the floras of Australia’s islands (J. Schrader, pers. comm.), and 15,700+ taxon names within the New South Wales BioNet Atlas (Department of Planning and Environment; https://www.environment.nsw.gov.au/topics/animals-and-plants/biodiversity/nsw-bionet). These projects and others have each merged disparate datasets collected across more than a century, successfully outputting harmonised compilations of updated taxon names. Taxonomy will always be a moving target, but we have built the ‘APCalign’ package to be stable as the APC continues to evolve and improve. We hope that others can use the tool to synchronise their research with the APC, leveraging the actively developed, high-quality taxonomic resource that the APC provides.
Data availability
The code underpinning ‘APCalign’ is available on the project’s GitHub repository, https://github.com/traitecoevo/APCalign. Articles on the GitHub repository’s website provide installation instructions and offer additional details on the functions, including example usage; see https://traitecoevo.github.io/APCalign. The APC and APNI, the Australia’s National Species List resources pertaining to vascular plants, can be accessed at https://biodiversity.org.au/nsl/services/export/index. A preprint version of this article is available at https://www.biorxiv.org/content/10.1101/2024.02.02.578715v1.
Declaration of funding
The AusTraits project received investment (https://doi.org/10.47486/DP720) from the Australian Research Data Commons (ARDC). The ARDC is funded by the National Collaborative Research Infrastructure Strategy (NCRIS). F. K. was funded by a UNSW Research Infrastructure Grant to Falster.
Author contributions
E. W. and D. F. led the development of the algorithms to align and update taxon names. E. W., F. K., W. C. and D. F. wrote the software. A. F., A. M. and H. S. offered expertise to ensure the algorithms correctly aligned and updated all categories of correct and erroneous taxon names. All authors contributed to the writing of the paper.
Acknowledgements
We thank the ARDC for their commitment to funding projects that increase community access to and use of informatics resources. We thank Rachael Gallagher for many conversations about how to build the best tool for Australia’s research community. We thank David Coleman and Julian Schrader for testing the APCalign functions on two large datasets, offering feedback on needed algorithm refinements.
References
Barker WL (2005) Standardising informal names in Australian publications. Australian Systematic Botany Society Newsletter 122, 11-12.
| Google Scholar |
Borsch T, Berendsohn W, Dalcin E, Delmas M, Demissew S, Elliott A, Fritsch P, Fuchs A, Geltman D, Güner A, Haevermans T, Knapp S, le Roux MM, Loizeau P-A, Miller C, Miller J, Miller JT, Palese R, Paton A, Parnell J, Pendry C, Qin H-N, Sosa V, Sosef M, von Raab-Straube E, Ranwashe F, Raz L, Salimov R, Smets E, Thiers B, Thomas W, Tulig M, Ulate W, Ung V, Watson M, Jackson PW, Zamora N (2020) World Flora Online: placing taxonomists at the heart of a definitive and comprehensive global resource on the world’s plants. TAXON 69, 1311-1341.
| Crossref | Google Scholar |
Boyle B, Hopkins N, Lu Z, Raygoza Garay JA, Mozzherin D, Rees T, Matasci N, Narro ML, Piel WH, Mckay SJ, Lowry S, Freeland C, Peet RK, Enquist BJ (2013) The taxonomic name resolution service: an online tool for automated standardization of plant names. BMC Bioinformatics 14, 16.
| Crossref | Google Scholar | PubMed |
Chamberlain SA, Szöcs E (2013) taxize: taxonomic search and retrieval in R. F1000Research 2, 191.
| Crossref | Google Scholar |
Chamberlain S, Szoecs E, Foster Z, Arendsee Z, Boettiger C, Ram K, Bartomeus I, Baumgartner J, O’Donnell J, Oksanen J, Tzovaras BG, Marchand P, Tran V, Salmon M, Li G, Grenié M, rOpenSci (https://ropensci.org/) (2022) taxize: taxonomic information from around the web. Available at https://github.com/ropensci/taxize
Falster D, Gallagher R, Wenk EH, Wright IJ, Indiarto D, Andrew SC, Baxter C, Lawson J, Allen S, Fuchs A, Monro A, Kar F, Adams MA, Ahrens CW, Alfonzetti M, Angevin T, Apgaua DMG, Arndt S, Atkin OK, Atkinson J, Auld T, Baker A, von Balthazar M, Bean A, Blackman CJ, Bloomfield K, Bowman DMJS, Bragg J, Brodribb TJ, Buckton G, Burrows G, Caldwell E, Camac J, Carpenter R, Catford JA, Cawthray GR, Cernusak LA, Chandler G, Chapman AR, Cheal D, Cheesman AW, Chen S-C, Choat B, Clinton B, Clode PL, Coleman H, Cornwell WK, Cosgrove M, Crisp M, Cross E, Crous KY, Cunningham S, Curran T, Curtis E, Daws MI, DeGabriel JL, Denton MD, Dong N, Du P, Duan H, Duncan DH, Duncan RP, Duretto M, Dwyer JM, Edwards C, Esperon-Rodriguez M, Evans JR, Everingham SE, Farrell C, Firn J, Fonseca CR, French BJ, Frood D, Funk JL, Geange SR, Ghannoum O, Gleason SM, Gosper CR, Gray E, Groom PK, Grootemaat S, Gross C, Guerin G, Guja L, Hahs AK, Harrison MT, Hayes PE, Henery M, Hochuli D, Howell J, Huang G, Hughes L, Huisman J, Ilic J, Jagdish A, Jin D, Jordan G, Jurado E, Kanowski J, Kasel S, Kellermann J, Kenny B, Kohout M, Kooyman RM, Kotowska MM, Lai HR, Laliberté E, Lambers H, Lamont BB, Lanfear R, van Langevelde F, Laughlin DC, Laugier-Kitchener B-A, Laurance S, Lehmann CER, Leigh A, Leishman MR, Lenz T, Lepschi B, Lewis JD, Lim F, Liu U, Lord J, Lusk CH, Macinnis-Ng C, McPherson H, Magallón S, Manea A, López-Martinez A, Mayfield M, McCarthy JK, Meers T, van der Merwe M, Metcalfe DJ, Milberg P, Mokany K, Moles AT, Moore BD, Moore N, Morgan JW, Morris W, Muir A, Munroe S, Nicholson Á, Nicolle D, Nicotra AB, Niinemets Ü, North T, O’Reilly-Nugent A, O’Sullivan OS, Oberle B, Onoda Y, Ooi MKJ, Osborne CP, Paczkowska G, Pekin B, Guilherme Pereira C, Pickering C, Pickup M, Pollock LJ, Poot P, Powell JR, Power SA, Prentice IC, Prior L, Prober SM, Read J, Reynolds V, Richards AE, Richardson B, Roderick ML, Rosell JA, Rossetto M, Rye B, Rymer PD, Sams MA, Sanson G, Sauquet H, Schmidt S, Schönenberger J, Schulze E-D, Sendall K, Sinclair S, Smith B, Smith R, Soper F, Sparrow B, Standish RJ, Staples TL, Stephens R, Szota C, Taseski G, Tasker E, Thomas F, Tissue DT, Tjoelker MG, Tng DYP, de Tombeur F, Tomlinson K, Turner NC, Veneklaas EJ, Venn S, Vesk P, Vlasveld C, Vorontsova MS, Warren CA, Warwick N, Weerasinghe LK, Wells J, Westoby M, White M, Williams NSG, Wills J, Wilson PG, Yates C, Zanne AE, Zemunik G, Ziemińska K (2021) AusTraits, a curated plant trait database for the Australian flora. Scientific Data 8, 254.
| Crossref | Google Scholar | PubMed |
Franz NM, Peet RK (2009) Perspectives: towards a language for mapping relationships among taxonomic concepts. Systematics and Biodiversity 7, 5-20.
| Crossref | Google Scholar |
Gagolewski M (2022) stringi: fast and portable character string processing in R. Journal of Statistical Software 103, 1-59.
| Crossref | Google Scholar |
Garnett ST, Christidis L, Conix S, Costello MJ, Zachos FE, Bánki OS, Bao Y, Barik SK, Buckeridge JS, Hobern D, Lien A, Montgomery N, Nikolaeva S, Pyle RL, Thomson SA, van Dijk PP, Whalen A, Zhang Z-Q, Thiele KR (2020) Principles for creating a single authoritative list of the world’s species. PLOS Biology 18, e3000736.
| Crossref | Google Scholar | PubMed |
Govaerts R, Nic Lughadha E, Black N, Turner R, Paton A (2021) The World Checklist of Vascular Plants, a continuously updated resource for exploring global plant diversity. Scientific Data 8, 215.
| Crossref | Google Scholar | PubMed |
Grenié M, Berti E, Carvajal-Quintero J, Dädlow GML, Sagouis A, Winter M (2023) Harmonizing taxon names in biodiversity data: a review of tools, databases and best practices. Methods in Ecology and Evolution 14, 12-25.
| Crossref | Google Scholar |
Martín-Forés I, Guerin GR, Lewis D, Gallagher RV, Vilà M, Catford JA, Pauchard A, Sparrow B (2023) The Alien Flora of Australia (AFA), a unified Australian national dataset on plant invasion. Scientific Data 10, 834.
| Crossref | Google Scholar | PubMed |
Ooms J (2014) The jsonlite package: a practical and consistent mapping between JSON data and R objects. arXiv:14032805 [StatCO]. Available at https://arxiv.org/abs/1403.2805
Ooms J, Wickham H, R Studio (2024) curl: a modern and flexible web client for R. Available at https://jeroen.r-universe.dev/curl, https://curl.se/libcurl/
R Core Team (2024) ‘R: a language and environment for statistical computing.’ (R Foundation for Statistical Computing: Vienna, Austria) Available at https://www.R-project.org/
Richardson N, Cook I, Crane N, Dunnington D, François R, Keane J, Moldovan-Grünfeld D, Ooms J, Wujciak-Jens J, Apache Arrow (2024) arrow: integration to ‘Apache’ ‘Arrow’. Available at https://github.com/apache/arrow/
Sandall EL, Maureaud AA, Guralnick R, McGeoch MA, Sica YV, Rogan MS, Booher DB, Edwards R, Franz N, Ingenloff K, Lucas M, Marsh CJ, McGowan J, Pinkert S, Ranipeta A, Uetz P, Wieczorek J, Jetz W (2023) A globally integrated structure of taxonomy to support biodiversity science and conservation. Trends in Ecology & Evolution 38, 1143-1153.
| Crossref | Google Scholar | PubMed |
Schellenberger Costa D, Boehnisch G, Freiberg M, Govaerts R, Grenié M, Hassler M, Kattge J, Muellner-Riehl AN, Rojas Andrés BM, Winter M, Watson M, Zizka A, Wirth C (2023) The big four of plant taxonomy – a comparison of global checklists of vascular plant names. New Phytologist 240, 1687-1702.
| Crossref | Google Scholar | PubMed |
Toelken HR, Miller RT (2012) Notes on Hibbertia (Dilleniaceae) 8. Seven new species, a new combination and four new subspecies from subgen. Hemistemma, mainly from the central coast of New South Wales. Journal of the Adelaide Botanic Garden 25, 71-96.
| Google Scholar |
van der Loo MPJ (2014) The stringdist package for approximate string matching. The R Journal 6, 111-122.
| Crossref | Google Scholar |
Walker B (2021) kewr: R package to access kew data APIs. Available at https://barnabywalker.github.io/kewr
Whitbread G (2018) Taxon, taxon concept and taxon name usage: definitions and relationships (GitHub issue). Available at https://github.com/tdwg/tnc/issues/1
Wickham H (2011) testthat: get started with testing. The R Journal 3, 5-10.
| Crossref | Google Scholar |
Wickham H (2023) httr: tools for working with URLs and HTTP. Available at https://github.com/r-lib/httr
Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H (2019) Welcome to the tidyverse. Journal of Open Source Software 4, 1686.
| Crossref | Google Scholar |
Wieczorek J, Bloom D, Guralnick R, Blum S, Döring M, Giovanni R, Robertson T, Vieglais D (2012) Darwin core: an evolving community-developed biodiversity data standard. PLoS ONE 7, e29715.
| Crossref | Google Scholar | PubMed |


