Retention of an apparently functional plastome in an apparently mycoheterotrophic orchid, Dipodium roseum D.L.Jones & M.A.Clem. (Orchidaceae)
Todd G. B. McLay A B * , Michael J. Bayly A , Michael R. Whitehead A and Rachael M. Fowler A
A B * , Michael J. Bayly A , Michael R. Whitehead A and Rachael M. Fowler A
A School of Biosciences, The University of Melbourne, Parkville, Vic. 3010, Australia.
B National Herbarium of Victoria, Royal Botanic Gardens Victoria, South Yarra, Vic. 3141, Australia.
Australian Journal of Botany 71(6) 306-317 https://doi.org/10.1071/BT22075
Submitted: 13 July 2022 Accepted: 2 June 2023 Published: 23 June 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: Giving up photosynthetic function is a bold evolutionary step for a plant, yet the evolutionary transition from autotrophy to mycoheterotrophy has occurred dozens of times. Comparing plastome sequences of mycoheterotrophs and autotrophs has identified recurring patterns of gene loss throughout a range of land plants, though more comparisons are required to see how broadly these patterns apply across the diversity of terrestrial plants. Mycoheterotrophy is especially common in Orchidaceae, with more than 40 transitions from autotrophy to mycoheterotrophy in the family.
Aims: We sought to test generalised patterns of plastome degradation seen in other mycoheterotrophs by comparing two species in the genus Dipodium (Orchidaceae); one species is photosynthetic and the other appears to be a facultative mycoheterotroph species.
Methods: We sequenced and assembled the plastomes of two Dipodium species and compared the two genomes to identify gene degradation or loss.
Results: The two plastomes were nearly identical, with no degradation of photosynthesis genes in the putative mycoheterotroph, and both species have undergone loss or pseudogenisation of all plastid ndh (nicotinamide adenine dinucleotide + hydrogen specific dehydrogenase) genes.
Conclusions: These results contrast with most other comparisons between photosynthetic and likely mycoheterotrophic relatives, where rapid degradation in mycoheterotroph plastome genes is common, and may suggest the leafless Dipodium species are capable of photosynthesis and may be in the early stages of transitioning to a fully heterotrophic lifestyle.
Implications: Further investigation of trophic transitions in Dipodium, including sequencing more plastomes and measuring photosynthetic capability of the putative heterotrophs, will yield insights into the evolution of plant lineages that lose the ability to photosynthesise.
Keywords: Dipodium, gene loss/degradation, mixotrophy, mycoheterotrophy, ndh genes, Orchidaceae, plastome, symbiosis.
Introduction
Mycorrhizal symbiosis operates over a continuum of ecological syndromes. True mutualism lies at one extreme. In the middle are mixed strategies including both obligate and facultative mixotrophy, while obligate parasitism (mycoheterotrophy) lies on the other extreme (Selosse et al. 2006). Mycoheterotrophic plants act as obligate parasites of fungi, and having lost the capacity for photosynthesis, rely on mycorrhizal associations to obtain carbon. These plants feature similar evolutionary morphological trends as holoparasites: they are frequently not green, leaves are typically scale-like or entirely absent, vascularisation of the stems is often reduced, stomata are mostly absent from above-ground/vegetative parts, and the below-ground organs of mycoheterotrophic plants are typically highly modified (Merckx and Freudenstein 2010). Although losing the ability to photosynthesise seems like a significant evolutionary risk for plants, mycoheterotrophy has evolved approximately 47 times in land plants (Merckx 2013).
Photosynthetic ability in land plants is conferred by the chloroplast, a specialised plastid organelle obtained from a cyanobacteria in an ancient endosymbiosis event (Timmis et al. 2004). In plants that have evolved a degree of mixotrophy and no longer rely directly on autotrophic photosynthesis, the plastid genome (plastome) is typically reduced, featuring gene pseudogenisation, deletions or rearrangements (dePamphilis and Palmer 1990; Bungard 2004). This includes holoparasites (Epifagus Nutt., dePamphilis and Palmer 1990; Cytinus L., Roquet et al. 2016; Hydnora Thunb., Naumann et al. 2016; Cuscuta L., Braukmann et al. 2013; Cistanche Hoffmans. & Link, Li et al. 2013; Pilostyles Guill., Bellot and Renner 2016; Rhopalocnemis Jungh., Schelkunov et al. 2019; Parasitaxus de Laub., Qu et al. 2019), hemiparasites (Viscum L., Petersen et al. 2015), and mycoheterotrophs (Corrallorhiza Gagnebin, Barrett et al. 2019; Rhizanthella R.S.Rogers, Delannoy et al. 2011; Neottia Jacq., Logacheva et al. 2011; Epipogium R.Br., Schelkunov et al. 2015; Corsia dispar D.L.Jones & B.Gray, Bodin et al. 2016; Sciaphila Blume, Lam et al. 2015; Orobanchaceae Vent., Frailey et al. 2018; Burmannia L., Li et al. 2019; Epirixanthes Blume, Petersen et al. 2019; Monotropa L., Gruzdev et al. 2016). The ultimate state of degradation of plastome function in heterotrophic plants is exemplified by an apparent complete loss of the plastome in several holoparasitic taxa (Rafflesia R.Br., Molina et al. 2014; Schoepfia Schreb., Su and Hu 2016). Non-parasites have also been identified as having reduced plastomes and gene loss (i.e. Tahina spectabilis J.Dransf. & Rakotoarin, Barrett et al. 2016; Carnegiea gigantea Britton & Rose, Sanderson et al. 2015; Saniculiphyllum guangxiense C.Y.Wu & T.C.Ku, Folk et al. 2020). Sequencing heterotroph plastomes (including partial heterotrophs and mixotrophs) has led to inferences of general evolutionary trends of gene pseudogenisation or loss, including an increase in AT%, indels and gene rearrangements, and the degradation of photosynthetic ability (Graham et al. 2017).
Lineages that include both photosynthetic and fully mycoheterotrophic species have been studied to test whether there are common or repeated pathways of plastome degradation. The current consensus holds that losing genes in the NADH (nicotinamide adenine dinucleotide) dehydrogenase complex (ndh genes) can begin a heterotrophy cascade (Lam et al. 2016; Barrett et al. 2018), which leads to the degradation of a set of photosynthesis genes (including psa, psb, pet and rpo genes, and rbcL). These are lost soon after the transition to heterotrophy and lead to a loss of photosynthetic ability, though this does not always inhibit the plant from producing chlorophyll (Wickett et al. 2011; Barrett and Davis 2012; Cusimano and Wicke 2016; Feng et al. 2016). However, in many heterotrophic species the plastome is retained and patterns of reduction converge on a conserved minimal gene set (Delannoy et al. 2011; Logacheva et al. 2011). This core set includes ribosomal and transfer RNAs (rRNAs, tRNAs), ribosomal protein genes, clpP, accD, ycf1, and ycf2. For example, even the very reduced plastomes of Epipogeum (~11 kb) are thought to be functional in some way as they contain a high proportion of coding DNA, an absence of non-synonymous mutations in protein-coding genes, and apparently functional tRNAs (Schelkunov et al. 2015). Studies with dense sampling of mycoheterotrophs and related autotrophs have found significant plastome sequence variation at even very fine taxonomic scales (including within species variation) and rapid degradation of plastid genomes associated with a transition to heterotrophy (Barrett et al. 2018, 2019; Schneider et al. 2018). Conversely, examples where a photosynthetic species and an apparent (facultative) heterotroph have similar plastomes are rare in the literature (but see Kim et al. 2020). The extent of plastome gene degradation therefore appears to vary widely among transitions to heterotrophy. Identifying whether this variation is due to lineages being at different places on the same path, or whether different paths of degradation are taken by different lineages is key to better understanding the evolution of heterotrophy. Ideally, we need to accumulate more studies of phylogenetically independent autotrophic–heterotrophic contrasts.
Approximately half of all facultative mycoheterotrophs occur in the Orchidaceae. This represents at least 30 transitions to full mycoheterotrophy (Barrett et al. 2018) and many of the sequenced plastomes of mycoheterotrophs have been orchids. However, the plastomes of photosynthetic orchid species also commonly feature loss and degradation of ndh genes (Neyland and Urbatsch 1996; Kim et al. 2015). Sequencing across Orchidaceae has revealed that ndh gene loss is complex; within subfamilies it appears that loss of full-length copies of the gene family occurs multiple times (Kim and Chase 2017). Ndh genes have also been lost in several non-orchid mycoheterotrophs (Petrosavia Becc, Petrosaviaceae, Logacheva et al. 2014), parasitic plants (Cuscuta, Convolvulaceae, Haberhausen and Zetsche 1994), carnivorous plants (Utricularia L., Utriculariaceae, Silva et al. 2016; Genlisea Rchb., Lentibulariaceae, Silva et al. 2018), aquatic plants (Najas L., Hydrocharitaceae, Peredo et al. 2013), as well as other photosynthetic lineages (Gnetales and Pinaceae, Braukmann et al. 2009; Erodium L’Hér. (Chris Blazier et al. 2011) and Melianthus L. (Weng et al. 2014), Geraniaceae; Capparis L., Capparaceae, Maurya et al. 2020). Under optimal growth conditions these genes have been found to be dispensable (Burrows et al. 1998) as the NADH dehydrogenase complex functions to fine tune photosynthesis that may not be required in terrestrial habits (Ruhlman et al. 2015) or under low stress conditions (Wicke et al. 2011). In orchids, the complex patterns of ndh loss or retention irrespective of the mode of carbon acquisition may confuse generalisations about the loss of this gene family and mycoheterotrophy.
The orchid genus Dipodium R.Br. (Orchidaceae, subfamily Epidendroideae) provides a useful study system for understanding the transition from photosynthetic-capable (autotrophic) to non-photosynthetic (likely facultative mycoheterotrophs) as the genus includes both leafy, photosynthetic and leafless, likely mycoheterotrophic species. The photosynthetic species are typically epiphytic or climbers, have large leaves (up to 400 mm long), and occur in tropical regions of Australia and south-east Asia. The apparent mycoheterotrophic species are terrestrial and entirely subterranean when not flowering. The inflorescence stem is typically brown (Fig. 1a, b) with leaves reduced to scales (up to 30 mm long), and they occur in south-eastern Australia, New Caledonia and New Guinea (O’byrne 2014). One species, D. ensifolium is terrestrial but has leafy stems. The literature on whether the leafless terrestrial Dipodium species are fully or partially mycoheterotrophic is mixed (Dearnaley and Le Brocque 2006a; O’byrne 2014), and some populations of apparently mycoheterotrophic species can be found with green stems (Fig. 1c; M. Bayly, personal observation; Jeanes and Backhouse 2006; see images on Dipodium profile at VicFlora 2022). Mycorrhizal associations of the Australian terrestrial Dipodium are dominated by the genus Russula Pers. which is typically ectomycorrhizal with trees (Dearnaley and Le Brocque 2006a). This reflects associations of other mycoheterotrophic orchids with Russula (Cymbidium Swartz, Ogura-Tsujita et al. 2012; Limodorum Boehm., Girlanda et al. 2006; Goodyera R.Br., Suetsugu et al. 2019) and stands in contrast to the major diversity of Australian terrestrial orchids, which associate with saprophytic lineages Tulasnella J.Schröt., Serendipita P.Roberts, and Ceratobasidium D.P. Rogers (Warcup 1981; Dearnaley and Le Brocque 2006b; Whitehead et al. 2017; Freestone et al. 2021; Arifin et al. 2022). Here, we compare the plastomes of D. ensifolium, a likely photosynthetic species with green leaves, and D. roseum, an apparent facultative mycoheterotrophic species of Dipodium. This comparison aims to test generalised patterns of plastome degradation seen in other mycoheterotrophs.
Methods
Sampling, DNA extraction, library preparation and sequencing
To compare the plastome sequence of a mycoheterotroph and a photosynthetic species, we obtained a fresh sample of the leafless and putatively heterotrophic Dipodium roseum F.Muell. (MELUM155142a) and an herbarium specimen of the photosynthetic, green and leafy D. ensifolium F.Muell. (BRI AQ0839318). Genomic DNA was extracted using a modified CTAB (cetyltrimethylammonium bromide) protocol (Shepherd and McLay 2011). DNA extraction for each sample was performed at separate times to avoid contamination between samples. Libraries were prepared for Illumina sequencing following Schuster et al. (2018). The Dipodium samples were pooled equally and then pooled with two other libraries to be 45% of the final concentration. The final library was sequenced using a MiSeq V3 300PE kit, at the Walter and Eliza Hall Institute, Melbourne, Victoria, Australia.
Plastome assembly
Raw read data of total genomic DNA was de novo assembled using CLC genomics workbench ver. 8.5.1 (https://www.qiagenbioinformatics.com/) with default settings, minimum length 1000 bp. Resulting contigs were then mapped to Cymbidium aloifolium (NC021429, Yang et al. 2013) in Geneious ver. 9.1.8 (http://www.geneious.com, Kearse et al. 2012), using custom sensitivity settings; gaps allowed set to a maximum of 20% per read, maximum gap size of 3000 and two to three iterations. Four contigs spanned the entire reference genome in D. roseum and five contigs in D. ensifolium. The only gaps present between contigs represented the second inverted repeat (IR) region, otherwise contigs overlapped at each end. The IR region was duplicated and remapped to the genome allowing for draft assembly of a complete circular plastome. Raw reads were mapped back over the draft genome to check for any deletions and SNP (single nucleotide polymorphism) differences between de novo assembled contigs used to make the draft and the raw reads. Mean raw read coverage of the D. roseum plastome was 202× bp, and 74× bp for the D. ensifolium plastome. A consensus sequence was then made for each sample. Plastid genomes were aligned using the MAFFT pairwise alignment plug-in (MAFFT, ver. 7.222; Katoh and Standley 2013) in Geneious Prime ver. 2019.2.1 with default settings. Gene annotations were transferred from GenBank published reference genomes and manually checked for changes to length and reading frame.
Testing for relaxed selection in D. roseum plastome genes
To compare the ratio of substitution rates at non-synonymous and synonymous sites, we performed dN/dS tests in MEGA7 (Kumar et al. 2016) using the Nei-Gojobori method (Nei and Gojobori 1986); the variance of the difference was computed using the bootstrap method (1000 replicates). We compared 63 genes from the two Dipodium genomes to Cymbidium aloifolium. Ribosomal RNAs, translational RNAs, and ndh genes were not tested. We also tested for relaxed selection pressure on D. roseum genes using RELAX v.2.1 (Wertheim et al. 2015) in HyPhy (Pond et al. 2005) using the Datamonkey webserver (Pond and Frost 2005). RELAX extends dN/dS calculations by estimating an intensity parameter (k), and tests whether selection is relaxed or intensified along a branch. D. roseum was set as the test branch and D. ensifolium was set as the reference branch. Model acceptance or rejection was determined using a likelihood ratio test.
Phylogenetic position of Dipodium
Twenty-four complete plastid genomes from species in subfamily Epidendroideae were downloaded from GenBank. Gene regions were extracted from each genome (58 genes, excluding the ndh genes, ribosomal RNAs, translational RNAs), gene reading frames and annotations were checked by eye, and were aligned using the MUSCLE plug-in (Edgar 2004) in Geneious with default settings. All genes were concatenated together to produce an alignment 52 466 bp long. The alignment was analysed using RAxML (ver. 8, Stamatakis 2014) with five maximum likelihood searches under both the GTRCAT and GTRGAMMA model, with Cypripedium formosanum as an outgroup. Support for the RAxML topology was tested by performing 1000 ‘rapid’ bootstrap repetitions, the 10 independent runs were checked for topological convergence, and bootstrap values were mapped on to the best tree as defined by likelihood scores.
NADH dehydrogenase (ndh) gene evolution
The loss of the ndh complex in the plastome was characterised using the parameters outlined in Kim et al. (2015); genes are either present (1, i.e. full length and in frame), pseudogenised (2, i.e. stop codons reduce length of transcript), truncated (3, >10% gene length deleted), or completely absent (4). The loss of genes in the ndh complex was reconstructed onto a cladogram using a directional step matrix following Kim et al. (2015), where any transition towards loss (e.g. 1–>2, 3–>4, or 1–>4) required one step, and any reversal required five steps. The characters were mapped onto a cladogram featuring Phaelenopsis, Cymbidium, Dipodium, Erycina, Oncidium, with Calanthe as an outgroup as all ndh genes are fully functional in that genus. Character state reconstructions were performed in Mesquite v3.7 (Maddison and Maddison 2021) and only non-equivocal states were recorded.
Results
Comparative analyses of two Dipodium plastomes
The results of DNA sequencing and plastome assembly, including reads mapped to plastome, coverage, plastome size and GC (guanine-cytosine) content are shown in Table 1. Comparing the plastome structure and gene content between the two species shows that there is no loss in genes or change in structure in the apparent facultative mycoheterotrophic D. roseum compared to the photosynthetic D. ensifolium. Both species have the typical quadripartite structure and organisation of orchid plastomes (Fig. 2). The pairwise genetic similarity between the two species across the aligned plastomes was 97.77% and across protein-coding genes it was 99.25%.
| Species | Total reads | Reads mapped | Plastome length (bp) | LSC length (bp) | SSC length (bp) | IR length (bp) | Mean coverage | GC (%) | GenBank accession number | Voucher |
|---|---|---|---|---|---|---|---|---|---|---|
| Dipodium roseum (001) | 7 000 412 | 177 302 | 141 239 | 81 836 | 10 446 | 24 534 | 201 | 37.7 | OQ885084 | MELUM155142a |
| Dipodium ensifolium (004) | 2 085 448 | 44 603 | 142 016 | 82 176 | 10 998 | 24 430 | 73 | 36.9 | OQ885083 | BRI AQ0839318 |
LSC, large single copy; SSC, small single copy; IR, inverted repeat.
Plastome map of Dipodium roseum, made with OGDraw (using the chlorobox toolkit – https://chlorobox.mpimp-golm.mpg.de/OGDraw.html, accessed March 2020).
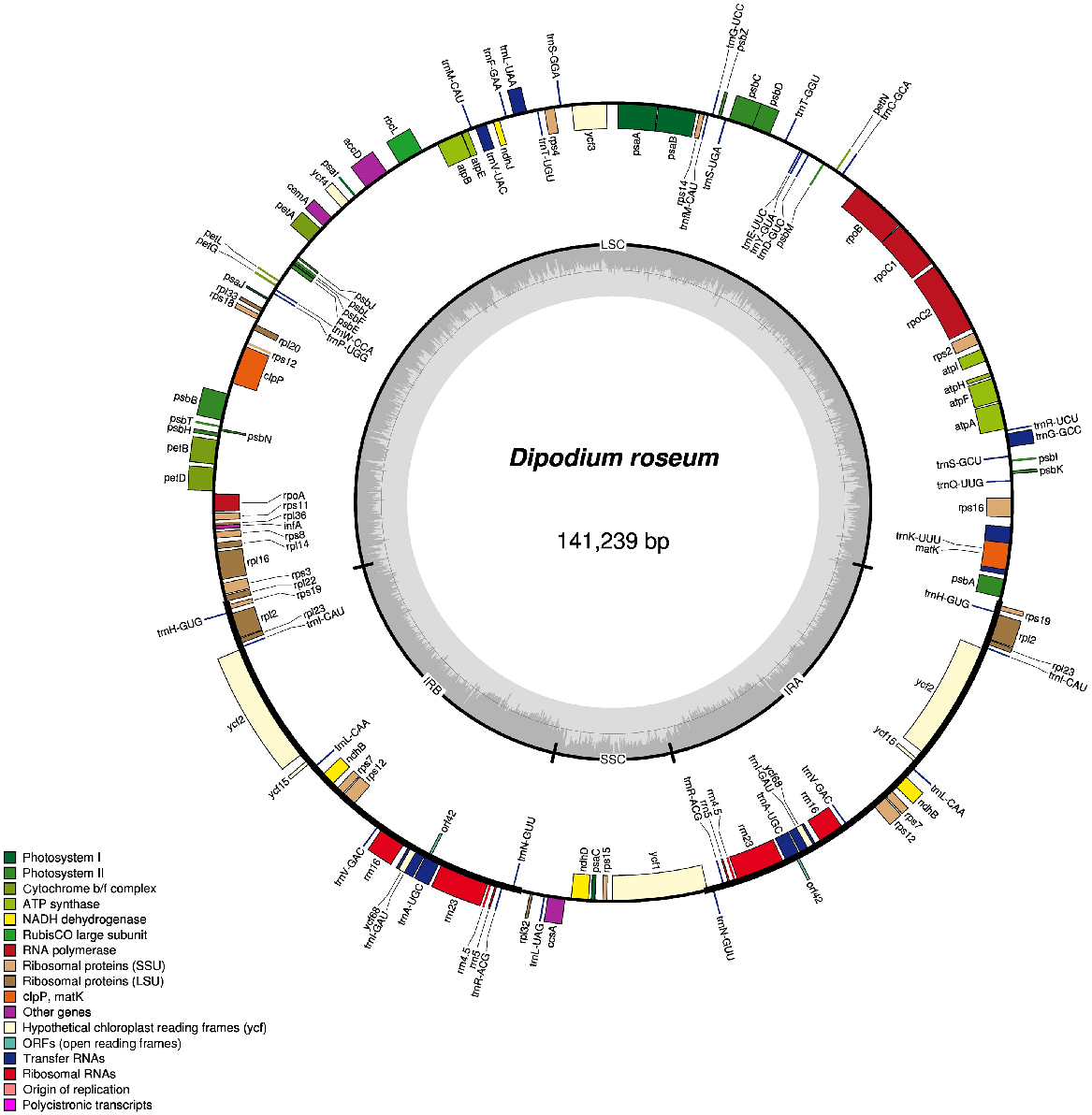
Structurally, the number and order of genes in both species of Dipodium remain consistent. However, relative to other members of Orchidaceae, there has been a small rearrangement in the placement/location and direction of petN and psbM genes compared to Cymbidium species. Both species of Dipodium exhibit an alternative initiation codon for matK relative to other closely related orchids with sequenced whole plastomes. Such alternative out-of-frame initiation codons are well documented, particularly in the Orchidaceae (Barthet et al. 2015).
No significant changes in selection between the Dipodium species
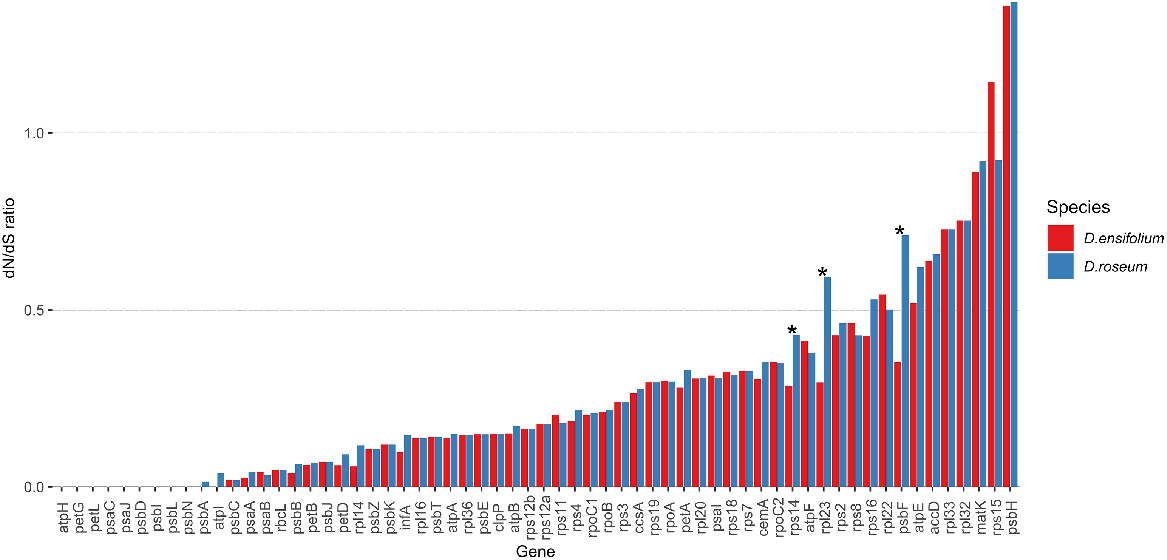
The Nei-Gojobori test indicated that three genes rps14 (300 bp), rpl23 (279 bp), and psbF (117 bp) had significantly different dN/dS ratios between the two species, indicating reduced selection in D. roseum in those genes (Fig. 3). There was no significant difference between the mycoheterotroph and the autotroph in the remaining 60 genes analysed; most genes in the D. roseum plastid genome are evolving under purifying selection (dN/dS < 1). For both species, the gene psbH (309 bp) appears to be under relaxed selection compared to Cymbidium. RELAX analyses of the same genes revealed no significant evidence for relaxation or intensification of selection on any genes between the two species.
Phylogenetic position of Dipodium
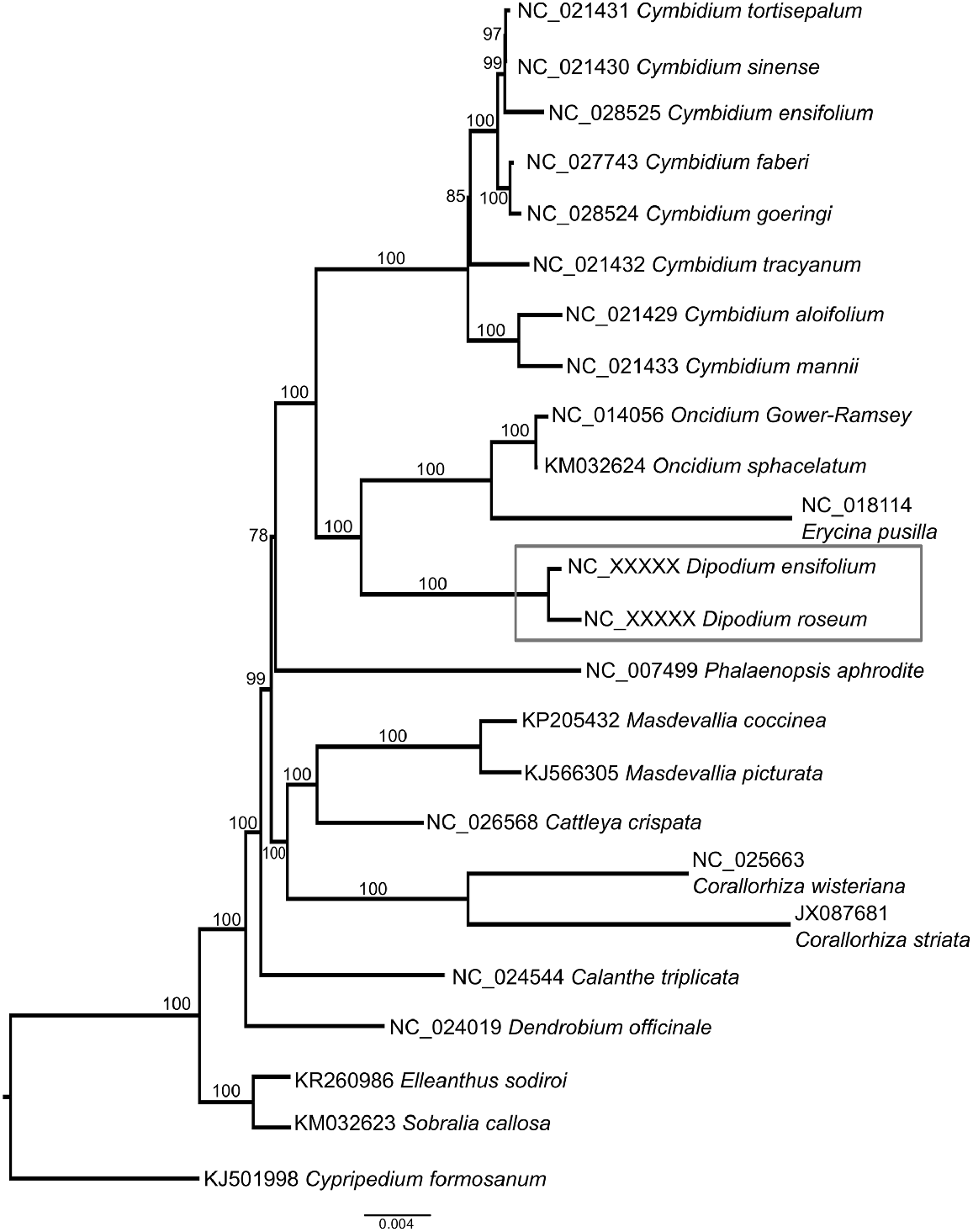
Phylogenetic analysis of 58 plastome genes resolved Dipodium in Orchidaceae subfamily Epidendroideae, sister to Oncidium and Erycina (Fig. 4). The 10 independent tree searches between the two models (GTRGAMMA and GTRCAT) produced identical topologies. Limited taxonomic sampling restricts the usefulness of this dataset to fully resolve the currently uncertain position of the Dipodium in the tribe Cymbidieae (Freudenstein and Chase 2015), or determine whether it belongs in its own subtribe, Dipodiinae (Li et al. 2016).
NADH dehydrogenase (ndh) gene evolution
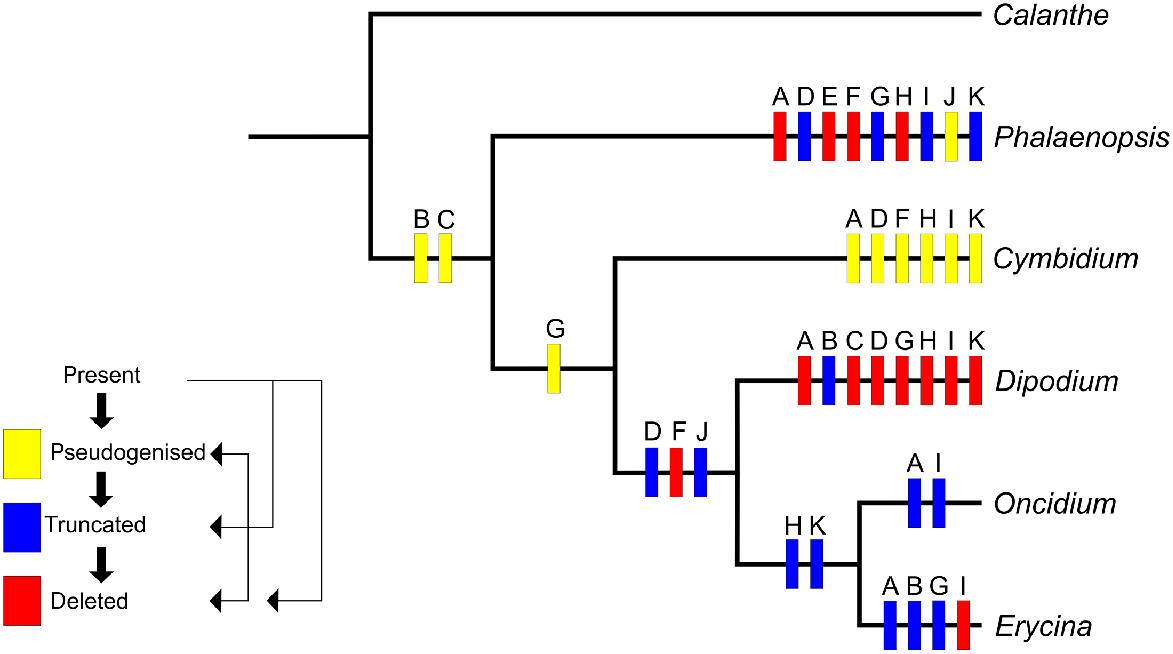
We found that all 11 ndh genes typically present in plant plastomes display identical changes in the plastomes of both Dipodium species. Using the parameters of ndh loss outlined by Kim et al. (2015), eight genes have been deleted (ndhA, ndhC, ndhE, ndhF, ndhG, ndhH, ndhI, ndhK) and three genes (ndhB, ndhD, ndhJ) have been truncated by greater than 10% of the original length (Table 2). Character state reconstruction of the ndh gene family on a simplified phylogeny shows that most of these losses or truncations have occurred within the Dipodium lineage, and that ndh gene degradation has independently occurred repeatedly within the genera compared here (Fig. 5, Supplementary material S1).
| Gene | Dipodium ensifolium OQ885083 (bp) | Dipodium roseum OQ885084 (bp) | Cymbidium aloifolium NC021429 (bp) |
|---|---|---|---|
| ndhA | Deleted (0) | Deleted (0) | Pseudogenised (2199) |
| ndhB | Truncated (811) | Truncated (800) | Pseudogenised (2224) |
| ndhC | Deleted (0) | Deleted (0) | Pseudogenised (363) |
| ndhD | Truncated (1179) | Truncated (1006) | Pseudogenised (1481) |
| ndhE | Deleted (0) | Deleted (0) | Functional (306) |
| ndhF | Deleted (0) | Deleted (0) | Pseudogenised (2211) |
| ndhG | Deleted (0) | Deleted (0) | Pseudogenised (531) |
| ndhH | Deleted (0) | Deleted (0) | Pseudogenised (1210) |
| ndhI | Deleted (0) | Deleted (0) | Pseudogenised (504) |
| ndhJ | Truncated (387) | Truncated (387) | Functional (477) |
| ndhK | Deleted (0) | Deleted (0) | Pseudogenised (886) |
| matK | Functional (1545) | Functional (1539) | Functional (1554) |
| psbG | Deleted (0) | Deleted (0) | Pseudogenised (746) |
| ycf15 | Functional (243) | Functional (243) | Pseudogenised (231) |
Inferred evolution of ndh genes in Epidendroideae, incorporating data and the approach to character state optimisation used by Kim et al. (2015). Letters A to K refer to the 11 genes, ndhA to ndhK. The data matrix used for this character mapping is presented in Supplementary material S1.
Discussion
We sequenced the plastomes of two Dipodium species; the photosynthetically capable D. ensifolium, and the putative mycoheterotroph, D. roseum. Although these species lie on opposite sides of a substantial evolutionary transition in metabolism, the genomic architecture of plastomes in both species was very similar. We found no evidence of genome size reduction, gene deletions or pseudogenisation in D. roseum relative to the autotrophic D. ensifolium. Three plastid genes showed a significant signal for reduced selection in D. roseum, but their sequences indicate they remain functional. These results contrast with the majority of other sequenced facultative mycoheterotroph plastomes, where genome size reduction and gene loss or pseudogenisation is common in the transition to mycoheterotrophy (Merckx and Freudenstein 2010; Barrett et al. 2018).
Our discovery that the likely-mycoheterotroph D. roseum has a near-identical plastome to a photosynthetic congener is unusual amongst studies of plastome evolution in mycoheterotrophs. The only other study to identify an apparent mycoheteroph and photosynthetic congener with a near-identical plastome was that of Kim and Chase (2017) in the leafless Cymbidium macrorhizon and relatives. That species lacks leaves and roots, but when flowering the inflorescence rachis is pale green. A complementary study found that species photosynthesises during flowering and fruiting, with the additional carbon resources likely contributing to seed production (Suetsugu et al. 2019). The more typical finding in similar studies is that substantial genomic changes are associated with the transition from autotrophy to mycoheterotrophy (Graham et al. 2017; Barrett et al. 2018).
Our finding of no substantial differences in plastome architecture is surprising in context with other contrasts at the sub-generic level. Perhaps the most well-studied transition to full mycoheterotrophy and loss of photosynthetic capability in orchids is in Corallorhiza. Extensive studies in the genus have found that fully mycoheterotrophic species have degradation in a suite of key photosynthetic genes (including pet, psa, rpo complexes and ccsA, cemA and rbcL, as well as ndh complex), and phylogenetic reconstruction of gene loss in the genus identified several independent losses of photosynthetic genes (Barrett et al. 2019). However, mycoheterotrophs did not necessarily have reduced plastome sizes (Barrett et al. 2018), leaflessness was not correlated with plastome degradation, and some mycoheterotrophs have detectable levels of chlorophyll (10-fold less than autotrophs; Barrett et al. 2014). Other examples include Hexalectris, an orchid genus of nine species with six independent losses of photosynthetic capability (Barrett and Kennedy 2018), and the tribe Neottieae (Feng et al. 2016; Zhou and Jin 2018), which has had three to four independent transitions to full mycoheterotrophy accompanied by a loss of important photosynthetic genes (Lallemand et al. 2019). These comparative analyses have shown that degradation can be rapid and frequently there are multiple transitions to full mycoheterotrophy amongst closely related species.
We found ndh gene loss in both species of Dipodium, which is consistent with generalisations that orchids appear to lose ndh genes readily, even in autotrophic species (Kim et al. 2020). The ndh gene family is important in environments with highly variable light intensities but functional ndh genes in the plastome are clearly dispensable within the Orchidaceae; loss of ndh genes is common and widespread in Orchidaceae and has been found in four of five subfamilies (Kim et al. 2020). Ndh gene loss appears to be randomly distributed throughout the family and within subfamilies (Kim et al. 2015), which makes any implication of ndh gene loss beginning a transition away from autotrophy difficult to tease out. Our data support this, with evidence of ndh loss in Dipodium further complicating the pattern in the Epidendroideae (Fig. 5) compared to the findings of Kim et al. 2015. There is some evidence that ndh subunits from the nuclear genome are lost alongside those from the plastome (Lin et al. 2017), but an alternative hypothesis of transferral of ndh genes to the nuclear or mitochondrial genome(s) is also plausible (Timmis et al. 2004). Further sequencing of both nuclear and mitochondrial genomes of orchids will allow testing of these hypotheses and determine the destination of ndh genes as they leave the plastome.
Relative to the currently understood general case of transition to mycoheterotrophy, Dipodium appears to be somewhat of an anomaly. D. roseum has many of the morphological characteristics of a mycoheterotroph but does not have any plastome degradation compared to the photosynthetic Dipodium ensifolium, illustrating that significant morphological change can be decoupled from functional loss of photosynthesis. Alternatively, D. roseum might represent a very recent transition away from autotrophy, and we happen to be observing that transition before the ensuing rapid gene loss. D. roseum is known to have fungal associations (Dearnaley and Le Brocque 2006a) with mycorrhizae commonly associated with mycoheterotrophy, and fungal peletons can be observed in the body of the plant (Gross 1991; M. Whitehead, personal observation). Based on the contents of its plastome this species is possibly still capable of photosynthesis. Some populations of D. roseum have green stems (Fig. 1c; M. Bayly, personal observation), which indicates the plants are still capable of producing chlorophyll, although the presence of chlorophyll does not necessarily confer photosynthetic capability (Julou et al. 2005). It is possible that D. roseum can still photosynthesise, but only does so during flowering, and is on an evolutionary pathway towards full mycoheterotrophy. Were this the case, D. roseum may be better described as a mixotrophic species, capable of obtaining carbon both through photosynthesis and fungal mutualism (Dearnaley and Le Brocque 2006a; Girlanda et al. 2006; Yagame et al. 2012; Bellino et al. 2014).
Further research is required to confirm whether other terrestrial Dipodium species are facultative mycoheterotrophs, how frequently mycoheterotrophy may have evolved in the genus, what the pattern of ndh loss is within autotrophic species, and an assessment of the associated fungal diversity. Ideally, this would include a dated genus-level phylogeny, sequencing plastomes of all species in the genus (ideally multiple samples within a species), measures of photosynthetic capability of populations, and characterisation of the fungal associations of leafy and terrestrial species. If the terrestrial species of Dipodium are in the early stages of a transition to mycoheterotrophy, the genus may prove to be important in understanding the bold evolutionary step of abandoning photosynthesis in favour of fungal symbioses.
Acknowledgements
Daniel Ohlsen (Royal Botanic Gardens Victoria) and Will Neal (Fire Rescue Victoria) assisted with collection of Dipodium roseum, Gill Brown (Queensland Herbarium) sent material of D. ensifolium, and Stephen Wilcox (Walter and Eliza Hall Institute of Medical Research) performed the sequencing run. Comments from two reviewers and the handling editor improved the manuscript.
References
Arifin AR, Reiter NH, May TW, Linde CC (2022) New species of Tulasnella associated with Australian terrestrial orchids in the subtribes Megastylidinae and Thelymitrinae. Mycologia 114, 388-412.
| Crossref | Google Scholar |
Barrett CF, Davis JI (2012) The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. American Journal of Botany 99, 1513-1523.
| Crossref | Google Scholar |
Barrett CF, Kennedy AH (2018) Plastid genome degradation in the endangered, mycoheterotrophic, North American orchid Hexalectris warnockii. Genome Biology and Evolution 10, 1657-1662.
| Crossref | Google Scholar |
Barrett CF, Freudenstein JV, Li J, Mayfield-Jones DR, Perez L, Pires JC, Santos C (2014) Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Molecular Biology and Evolution 31, 3095-3112.
| Crossref | Google Scholar |
Barrett CF, Baker WJ, Comer JR, Conran JG, Lahmeyer SC, Leebens-Mack JH, Li J, Lim GS, Mayfield-Jones DR, Perez L, Medina J, Pires JC, Santos C, Wm. Stevenson D, Zomlefer WB, Davis JI (2016) Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytologist 209, 855-870.
| Crossref | Google Scholar |
Barrett CF, Wicke S, Sass C (2018) Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytologist 218, 1192-1204.
| Crossref | Google Scholar |
Barrett CF, Sinn BT, Kennedy AH, Pupko T (2019) Unprecedented parallel photosynthetic losses in a heterotrophic orchid genus. Molecular Biology and Evolution 36, 1884-1901.
| Crossref | Google Scholar |
Barthet MM, Moukarzel K, Smith KN, Patel J, Hilu KW (2015) Alternative translation initiation codons for the plastid maturase MatK: unraveling the pseudogene misconception in the Orchidaceae. BMC Evolutionary Biology 15, 210.
| Crossref | Google Scholar |
Bellino A, Alfani A, Selosse M-A, Guerrieri R, Borghetti M, Baldantoni D (2014) Nutritional regulation in mixotrophic plants: new insights from Limodorum abortivum. Oecologia 175, 875-885.
| Crossref | Google Scholar |
Bellot S, Renner SS (2016) The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biology and Evolution 8, 189-201.
| Crossref | Google Scholar |
Bodin SS, Kim JS, Kim J-H (2016) Phylogenetic inferences and the evolution of plastid DNA in Campynemataceae and the mycoheterotrophic Corsia dispar D.L Jones & B. Gray (Corsiaceae). Plant Molecular Biology Reporter 34, 192-210.
| Crossref | Google Scholar |
Braukmann TWA, Kuzmina M, Stefanović S (2009) Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Current Genetics 55, 323-337.
| Crossref | Google Scholar |
Braukmann T, Kuzmina M, Stefanović S (2013) Plastid genome evolution across the genus Cuscuta (Convolvulaceae): two clades within subgenus Grammica exhibit extensive gene loss. Journal of Experimental Botany 64, 977-989.
| Crossref | Google Scholar |
Bungard RA (2004) Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. BioEssays 26, 235-247.
| Crossref | Google Scholar |
Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. The EMBO Journal 17, 868-876.
| Crossref | Google Scholar |
Chris Blazier J, Guisinger MM, Jansen RK (2011) Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Molecular Biology 76, 263-272.
| Crossref | Google Scholar |
Cusimano N, Wicke S (2016) Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytologist 210, 680-693.
| Crossref | Google Scholar |
Dearnaley JDW, Le Brocque AF (2006a) Molecular identification of the primary root fungal endophytes of Dipodium hamiltonianum (Orchidaceae). Australian Journal of Botany 54, 487-491.
| Crossref | Google Scholar |
Dearnaley JD, Le Brocque AF (2006b) Endophytic fungi associated with Australian orchids. Australasian Plant Conservation 15, 7-9.
| Google Scholar |
Delannoy E, Fujii S, Colas Des Francs-Small C, Brundrett M, Small I (2011) Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Molecular Biology and Evolution 28, 2077-2086.
| Crossref | Google Scholar |
dePamphilis CW, Palmer JD (1990) Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature 348, 337-339.
| Crossref | Google Scholar |
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792-1797.
| Crossref | Google Scholar |
Feng Y-L, Wicke S, Li J-W, Han Y, Lin C-S, Li D-Z, Zhou T-T, Huang W-C, Huang L-Q, Jin X-H (2016) Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biology and Evolution 8, 2164-2175.
| Crossref | Google Scholar |
Folk RA, Sewnath N, Xiang C-L, Sinn BT, Guralnick RP (2020) Degradation of key photosynthetic genes in the critically endangered semi-aquatic flowering plant Saniculiphyllum guangxiense (Saxifragaceae). BMC Plant Biology 20, 1-11.
| Crossref | Google Scholar |
Frailey DC, Chaluvadi SR, Vaughn JN, Coatney CG, Bennetzen JL (2018) Gene loss and genome rearrangement in the plastids of five Hemiparasites in the family Orobanchaceae. BMC Plant Biology 18, 30.
| Crossref | Google Scholar |
Freestone MW, Swarts ND, Reiter N, Tomlinson S, Sussmilch FC, Wright MM, Holmes GD, Phillips RD, Linde CC (2021) Continental-scale distribution and diversity of Ceratobasidium orchid mycorrhizal fungi in Australia. Annals of Botany 128, 329-343.
| Crossref | Google Scholar |
Freudenstein JV, Chase MW (2015) Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Annals of Botany 115, 665-681.
| Crossref | Google Scholar |
Girlanda M, Selosse MA, Cafasso D, Brilli F, Delfine S, Fabbian R, Ghignone S, Pinelli P, Segreto R, Loreto F, Cozzolino S, Perotto S (2006) Inefficient photosynthesis in the Mediterranean orchid Limodorum abortivum is mirrored by specific association to ectomycorrhizal Russulaceae. Molecular Ecology 15, 491-504.
| Crossref | Google Scholar |
Graham SW, Lam VKY, Merckx VSFT (2017) Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes. New Phytologist 214, 48-55.
| Crossref | Google Scholar |
Gruzdev EV, Mardanov AV, Beletsky AV, Kochieva EZ, Ravin NV, Skryabin KG (2016) The complete chloroplast genome of parasitic flowering plant Monotropa hypopitys: extensive gene losses and size reduction. Mitochondrial DNA Part B 1, 212-213.
| Crossref | Google Scholar |
Haberhausen G, Zetsche K (1994) Functional loss of all ndh genes in an otherwise relatively unaltered plastid genome of the holoparasitic flowering plant Cuscuta reflexa. Plant Molecular Biology 24, 217-222.
| Crossref | Google Scholar |
Julou T, Burghardt B, Gebauer G, Berveiller D, Damesin C, Selosse M-A (2005) Mixotrophy in orchids: insights from a comparative study of green individuals and nonphotosynthetic individuals of Cephalanthera damasonium. New Phytologist 166, 639-653.
| Crossref | Google Scholar |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772-780.
| Crossref | Google Scholar |
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647-1649.
| Crossref | Google Scholar |
Kim HT, Chase MW (2017) Independent degradation in genes of the plastid ndh gene family in species of the orchid genus Cymbidium (Orchidaceae; Epidendroideae). PLoS ONE 12, e0187318.
| Crossref | Google Scholar |
Kim HT, Kim JS, Moore MJ, Neubig KM, Williams NH, Whitten WM, Kim J-H (2015) Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS ONE 10, e0142215.
| Crossref | Google Scholar |
Kim Y-K, Jo S, Cheon S-H, Joo M-J, Hong J-R, Kwak M, Kim K-J (2020) Plastome evolution and phylogeny of Orchidaceae, with 24 new sequences. Frontiers in Plant Science 11, 22.
| Crossref | Google Scholar |
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870-1874.
| Crossref | Google Scholar |
Lallemand F, Logacheva M, Le Clainche I, Bérard A, Zheleznaia E, May M, Jakalski M, Delannoy É, Le Paslier M-C, Selosse M-A (2019) Thirteen new plastid genomes from mixotrophic and autotrophic species provide insights into heterotrophy evolution in Neottieae orchids. Genome Biology and Evolution 11, 2457-2467.
| Crossref | Google Scholar |
Lam VKY, Soto Gomez M, Graham SW (2015) The highly reduced plastome of mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection. Genome Biology and Evolution 7, 2220-2236.
| Crossref | Google Scholar |
Lam VKY, Merckx VSFT, Graham SW (2016) A few-gene plastid phylogenetic framework for mycoheterotrophic monocots. American Journal of Botany 103, 692-708.
| Crossref | Google Scholar |
Li X, Zhang T-C, Qiao Q, Ren Z, Zhao J, Yonezawa T, Hasegawa M, Crabbe MJC, Li J, Zhong Y (2013) Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS ONE 8, e58747.
| Crossref | Google Scholar |
Li M-H, Zhang G-Q, Liu Z-J, Lan S-R (2016) Subtribal relationships in Cymbidieae (Epidendroideae, Orchidaceae) reveal a new subtribe, Dipodiinae, based on plastid and nuclear coding DNA. Phytotaxa 246, 037-048.
| Crossref | Google Scholar |
Li X, Qian X, Yao G, Zhao Z, Zhang D (2019) Plastome of mycoheterotrophic Burmannia itoana Mak. (Burmanniaceae) exhibits extensive degradation and distinct rearrangements. PeerJ 7, e7787.
| Crossref | Google Scholar |
Lin C-S, Chen JJW, Chiu C-C, Hsiao HCW, Yang C-J, Jin X-H, Leebens-Mack J, de Pamphilis CW, Huang Y-T, Yang L-H, Chang W-J, Kui L, Wong GK-S, Hu J-M, Wang W, Shih M-C (2017) Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. The Plant Journal 90, 994-1006.
| Crossref | Google Scholar |
Logacheva MD, Schelkunov MI, Penin AA (2011) Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biology and Evolution 3, 1296-1303.
| Crossref | Google Scholar |
Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA (2014) The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biology and Evolution 6, 238-246.
| Crossref | Google Scholar |
Maddison WP, Maddison DR (2021) Mesquite: a modular system for evolutionary analysis. Version 3.70. Available at http://www.mesquiteproject.org
Maurya S, Darshetkar AM, Datar MN, Tamhankar S, Li P, Choudhary RK (2020) Plastome data provide insights into intra and interspecific diversity and ndh gene loss in Capparis (Capparaceae). Phytotaxa 432, 206-220.
| Crossref | Google Scholar |
Merckx VSFT (2013) Mycoheterotrophy: an introduction. In ‘Mycoheterotrophy: the biology of plants living on fungi’. (Ed. VSFT Merckx) pp. 1–17. (Springer: New York, NY, USA) doi:10.1007/978-1-4614-5209-6
Merckx VSFT, Freudenstein JV (2010) Evolution of mycoheterotrophy in plants: a phylogenetic perspective. New Phytologist 185, 605-609.
| Crossref | Google Scholar |
Molina J, Hazzouri KM, Nickrent D, Geisler M, Meyer RS, Pentony MM, Flowers JM, Pelser P, Barcelona J, Inovejas SA, Uy I, Yuan W, Wilkins O, Michel C-I, LockLear S, Concepcion GP, Purugganan MD (2014) Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Molecular Biology and Evolution 31, 793-803.
| Crossref | Google Scholar |
Naumann J, Der JP, Wafula EK, Jones SS, Wagner ST, Honaas LA, Ralph PE, Bolin JF, Maass E, Neinhuis C, Wanke S, dePamphilis CW (2016) Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biology and Evolution 8, 345-363.
| Crossref | Google Scholar |
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular Biology and Evolution 3, 418-426.
| Crossref | Google Scholar |
Neyland R, Urbatsch LE (1996) Phylogeny of subfamily Epidendroideae (Orchidaceae) inferred from ndh F chloroplast gene sequences. American Journal of Botany 83, 1195-1206.
| Crossref | Google Scholar |
Ogura-Tsujita Y, Yokoyama J, Miyoshi K, Yukawa T (2012) Shifts in mycorrhizal fungi during the evolution of autotrophy to mycoheterotrophy in Cymbidium (Orchidaceae). American Journal of Botany 99, 1158-1176.
| Crossref | Google Scholar |
O’byrne P (2014) On the evolution of Dipodium R. BR. Reinwardtia 14, 123.
| Crossref | Google Scholar |
Peredo EL, King UM, Les DH (2013) The plastid genome of Najas flexilis: adaptation to submersed environments is accompanied by the complete loss of the NDH Complex in an aquatic angiosperm. PLoS ONE 8, e68591.
| Crossref | Google Scholar |
Petersen G, Cuenca A, Seberg O (2015) Plastome evolution in hemiparasitic mistletoes. Genome Biology and Evolution 7, 2520-2532.
| Crossref | Google Scholar |
Petersen G, Darby H, Lam VKY, Pedersen HAE, Merckx VSFT, Zervas A, Seberg O, Graham SW (2019) Mycoheterotrophic Epirixanthes (Polygalaceae) has a typical angiosperm mitogenome but unorthodox plastid genomes. Annals of Botany 124, 791-807.
| Crossref | Google Scholar |
Pond SLK, Frost SDW (2005) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531-2533.
| Crossref | Google Scholar |
Pond SLK, Frost SDW, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21(5), 676-679.
| Crossref | Google Scholar |
Qu X-J, Fan S-J, Wicke S, Yi T-S, Sloan D (2019) Plastome reduction in the only parasitic gymnosperm Parasitaxus is due to losses of photosynthesis but not housekeeping genes and apparently involves the secondary gain of a large inverted repeat. Genome Biology and Evolution 11, 2789-2796.
| Crossref | Google Scholar |
Roquet C, Coissac É, Cruaud C, Boleda M, Boyer F, Alberti A, Gielly L, Taberlet P, Thuiller W, Van Es J, Lavergne S (2016) Understanding the evolution of holoparasitic plants: the complete plastid genome of the holoparasite Cytinus hypocistis (Cytinaceae). Annals of Botany 118, 885-896.
| Crossref | Google Scholar |
Ruhlman TA, Chang W-J, Chen JJW, Huang Y-T, Chan M-T, Zhang J, Liao D-C, Blazier JC, Jin X, Shih M-C, Jansen RK, Lin C-S (2015) NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biology 15, 100.
| Crossref | Google Scholar |
Sanderson MJ, Copetti D, Búrquez A, Bustamante E, Charboneau JLM, Eguiarte LE, Kumar S, Lee HO, Lee J, McMahon M, Steele K, Wing R, Yang T-J, Zwickl D, Wojciechowski MF (2015) Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat. American Journal of Botany 102, 1115-1127.
| Crossref | Google Scholar |
Schelkunov MI, Shtratnikova VY, Nuraliev MS, Selosse M-A, Penin AA, Logacheva MD (2015) Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biology and Evolution 7, 1179-1191.
| Crossref | Google Scholar |
Schelkunov MI, Nuraliev MS, Logacheva MD (2019) Rhopalocnemis phalloides has one of the most reduced and mutated plastid genomes known. PeerJ 7, e7500.
| Crossref | Google Scholar |
Schneider AC, Chun H, Stefanović S, Baldwin BG (2018) Punctuated plastome reduction and host–parasite horizontal gene transfer in the holoparasitic plant genus Aphyllon. Proceedings of the Royal Society B: Biological Sciences 285, 20181535.
| Crossref | Google Scholar |
Schuster TM, Setaro SD, Tibbits JFG, Batty EL, Fowler RM, McLay TGB, Wilcox S, Ades PK, Bayly MJ (2018) Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae). PLoS ONE 13, e0195034.
| Crossref | Google Scholar |
Selosse M-A, Richard F, He X, Simard SW (2006) Mycorrhizal networks: des liaisons dangereuses? Trends in Ecology & Evolution 21, 621-628.
| Crossref | Google Scholar |
Shepherd LD, McLay TGB (2011) Two micro-scale protocols for the isolation of DNA from polysaccharide-rich plant tissue. Journal of Plant Research 124, 311-314.
| Crossref | Google Scholar |
Silva SR, Diaz YCA, Penha HA, Pinheiro DG, Fernandes CC, Miranda VFO, Michael TP, Varani AM (2016) The chloroplast genome of Utricularia reniformis sheds light on the evolution of the ndh gene complex of terrestrial carnivorous plants from the Lentibulariaceae family. PLoS ONE 11, e0165176.
| Crossref | Google Scholar |
Silva SR, Michael TP, Meer EJ, Pinheiro DG, Varani AM, Miranda VFO (2018) Comparative genomic analysis of Genlisea (corkscrew plants—Lentibulariaceae) chloroplast genomes reveals an increasing loss of the ndh genes. PLoS ONE 13, e0190321.
| Crossref | Google Scholar |
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312-1313.
| Crossref | Google Scholar |
Su H-J, Hu J-M (2016) The complete chloroplast genome of hemiparasitic flowering plant Schoepfia jasminodora. Mitochondrial DNA Part B 1, 767-769.
| Crossref | Google Scholar |
Suetsugu K, Yamato M, Matsubayashi J, Tayasu I (2019) Comparative study of nutritional mode and mycorrhizal fungi in green and albino variants of Goodyera velutina, an orchid mainly utilizing saprotrophic rhizoctonia. Molecular Ecology 28, 4290-4299.
| Crossref | Google Scholar |
Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature Reviews Genetics 5, 123-135.
| Crossref | Google Scholar |
VicFlora (2022) Flora of Victoria. Royal Botanic Gardens Victoria. Available at https://vicflora.rbg.vic.gov.au [Last accessed 07 January 2022]
Warcup JH (1981) The mycorrhizal relationships of Australian orchids. New Phytologist 87, 371-381.
| Crossref | Google Scholar |
Weng M-L, Blazier JC, Govindu M, Jansen RK (2014) Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Molecular Biology and Evolution 31, 645-659.
| Crossref | Google Scholar |
Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K (2015) RELAX: detecting relaxed selection in a phylogenetic framework. Molecular Biology and Evolution 32(3), 820-832.
| Crossref | Google Scholar |
Whitehead MR, Catullo RA, Ruibal M, Dixon KW, Peakall R, Linde CC (2017) Evaluating multilocus Bayesian species delimitation for discovery of cryptic mycorrhizal diversity. Fungal Ecology 26, 74-84.
| Crossref | Google Scholar |
Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Molecular Biology 76, 273-297.
| Crossref | Google Scholar |
Wickett NJ, Honaas LA, Wafula EK, Das M, Huang K, Wu B, Landherr L, Timko MP, Yoder J, Westwood JH, dePamphilis CW (2011) Transcriptomes of the parasitic plant family Orobanchaceae reveal surprising conservation of chlorophyll synthesis. Current Biology 21, 2098-2104.
| Crossref | Google Scholar |
Yagame T, Orihara T, Selosse MA, Yamato M, Iwase K (2012) Mixotrophy of Platanthera minor, an orchid associated with ectomycorrhiza-forming Ceratobasidiaceae fungi. New Phytologist 193, 178-187.
| Crossref | Google Scholar |
Yang J-B, Tang M, Li H-T, Zhang Z-R, Li D-Z (2013) Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evolutionary Biology 13, 84.
| Crossref | Google Scholar |
Zhou T, Jin X-H (2018) Molecular systematics and the evolution of mycoheterotrophy of tribe Neottieae (Orchidaceae, Epidendroideae). PhytoKeys 94, 39-49.
| Crossref | Google Scholar |