Chloroplast DNA phylogeography reveals the island colonisation route of Eucalyptus urophylla (Myrtaceae)
Kitt G. Payn A B C , William S. Dvorak B and Alexander A. Myburg AA Forest Molecular Genetics Programme, Department of Genetics, Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria 0002, South Africa.
B Camcore, North Carolina State University, PO Box 7626, Raleigh, NC 27695, USA.
C Corresponding author. Email: kitt.payn@fabi.up.ac.za
Australian Journal of Botany 55(7) 673-683 https://doi.org/10.1071/BT07056
Submitted: 29 March 2007 Accepted: 5 June 2007 Published: 15 November 2007
Abstract
We present a study of the colonisation patterns of a tropical tree species among an island archipelago. Eucalyptus urophylla (S.T.Blake) is an economically important plantation species endemic to the volcanic slopes of seven islands in eastern Indonesia. In the present study, we investigated the geographical distribution of chloroplast DNA sequence variation in E. urophylla to gain insight into its historical seed-migration routes. DNA sequence data were obtained from 198 plants from which 20 haplotypes were identified. A moderate to high level of chloroplast genetic differentiation (GST = 0.581, NST = 0.724) and significant phylogeographic structure (NST > GST; P < 0.01) were observed, suggesting low levels of recurrent seed-mediated gene flow among the islands. The highest levels of haplotype diversity were observed on the eastern islands of Wetar and Timor. The two most westerly islands, Flores and Lomblen, were fixed for what appeared to be the ancestral haplotype. Chloroplast haplotype diversity therefore exhibited a decreasing trend from east to west in the species’ range, consistent with an east-to-west colonisation route across the seven islands. Environmental factors that may have contributed to the contemporary spatial distribution of chloroplast DNA haplotypes include island paleogeology, ocean currents, fluctuations in sea levels and possible hybridisation events.
Introduction
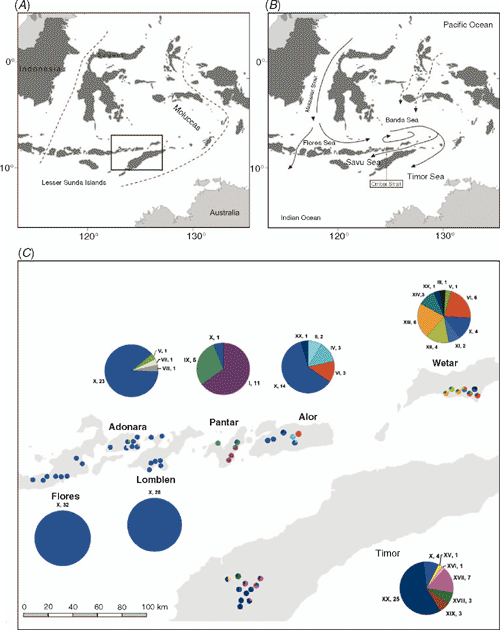
Situated at the interface of the Asian and Australian biotic realms are the islands of Indonesia. These islands contain some of the most diverse collections of flora and fauna on earth and have long been a region of major biogeographical interest (Wallace 1860; Myers et al. 2000; Brown et al. 2004). The division between Asian and Australian biota in Indonesia, first described by Alfred Wallace in the 19th century, is now recognised as a biogeographic region of transition, named Wallacea (Fig. 1A). Wallacea encompasses Sulawesi, the Lesser Sunda Islands and the Moluccas. This area is recognised not only for its rich biodiversity but also exhibits a high level of species endemicity (Myers et al. 2000). Several geological and environmental factors have influenced the contemporary distribution of Indo-Malay and Australasian biota on the islands of Indonesia. They include the continuing northward drift of the Indo-Australian plate into the region (Michaux 1991), the associated volcanic activity on many of the islands and sea-level fluctuations during the Pleistocene (Voris 2000) creating land bridges that facilitate migration among islands.
|
The genus Eucalyptus, comprising more than 600 species, is primarily endemic to the Australian continent (Ladiges et al. 2003). E. urophylla is one of only two Eucalyptus species that occurs exclusively outside of Australia, the other being E. deglupta Blume. Its natural distribution is limited to a series of disjunct populations located on seven of the Lesser Sunda Islands in eastern Indonesia (Fig. 1C). It occurs from almost sea level to high volcanic mountain slopes (3000-m elevation), with the largest stands found on the islands of Timor and Wetar, whereas more scattered stands occur on the islands of Adonara, Alor, Flores, Lomblen (Lembata) and Pantar (Eldridge et al. 1993). On the lower slopes of these islands it forms a mosaic distribution pattern with E. alba Reinw. ex Blume, which, unlike E. urophylla, is also indigenous to northern Australia and Papua (Pryor et al. 1995). In tropical and subtropical regions of Africa, South America and Asia, E. urophylla is commercially planted to produce wood that is used for a diverse array of products including pulp, sawn timber and fuel wood. The species is often crossed with E. grandis to produce hybrid progeny displaying rapid growth and superior disease resistance compared with the E. grandis parent (Pepe et al. 2004).
The Lesser Sunda Islands form part of the Banda arc, which comprises the inner volcanic arc and the outer non-volcanic arc (Norvick 1979). The eastern region of the inner arc includes the islands of Flores through Wetar, whereas the island of Timor forms part of the outer arc. These arcs were formed by the collision and subduction of the Australian plate beneath the Asian plate during the Pliocene. The colonisation and historical migration patterns of E. urophylla among the islands are unclear. An earlier isozyme study supported the hypothesis that E. urophylla existed on Timor before colonising the remaining islands because populations on Timor and nearby Alor contained the highest number alleles observed in the species (House and Bell 1994). The putative initial colonisation of Timor from an Australian source would have been aided by a period of low sea level during a glacial maximum, bringing emergent lands into close proximity (Ladiges et al. 2003). However, it is thought that there has never been a continuous land link between the Sunda Islands and Australia (Hall 2001), which suggests that historical seed migration would have required the successful crossing of ocean water.
Phylogeography is a field of study that analyses the geographical distribution of genealogical lineages, especially those at the intraspecific level (Avise 1998). The resolved phylogeographic structure of E. urophylla would provide insight into its evolutionary history and may be used to infer historical migration routes, and previous occurrences of genetic bottlenecks or population expansions. Plant phylogeographic studies predominantly make use of genetic variation in the chloroplast genome. The genome is inherited from a single parent, is effectively haploid and does not recombine (Birky 1995), thereby eliminating analytical complications involving interallelic recombination and heterozygosity. Notably, both the reduced ploidy and the uniparental mode of inheritance of organelle DNA decrease the effective population size, consequently increasing genetic drift and resulting in greater phylogeographic structure (McCauley 1995).
The chloroplast genome appears to be maternally inherited in most angiosperms (Corriveau and Coleman 1988), including Eucalyptus (Byrne et al. 1993; McKinnon et al. 2001a), thus, the distribution of chloroplast DNA (cpDNA) haplotypes may be used to infer seed-mediated migration routes. For example, two studies based on cpDNA variation of Quercus spp. supported the existence of refugia located in southern Iberia, Italy and the Balkans and proposed several postglacial migration routes into northern Europe from each refugium (Dumolin-Lapegue et al. 1997; Ferris et al. 1998). Similar studies have been carried out on Liriodendron tulipifera in North America (Sewell et al. 1996) and Cedrela odorata in Measoamerica (Cavers et al. 2003). More recently, a large-scale project called CYTOFOR (http://www.pierroton.inra.fr/Cytofor), comprising nine research groups representing six European countries, investigated the patterns of chloroplast genetic diversity and the postglacial recolonisation history of 22 widespread European trees and shrubs (Petit et al. 2003). Together these studies have shown that cpDNA markers are very useful in providing insight into the evolutionary history of a species.
Chloroplast DNA variation has also been extensively examined in Eucalyptus (Byrne and Moran 1994; Steane et al. 1998; Jackson et al. 1999; McKinnon et al. 1999). These studies were based on restriction fragment length polymorphism (RFLP) variation in cpDNA. More recently, Vaillancourt and Jackson (2000) found the JLA region (an intergenic spacer on either side of the junction between the large single-copy region and inverted repeat A of the chloroplast genome; Goulding et al. 1996) to be hypervariable in Eucalyptus. The DNA sequence data were shown to accurately identify haplotypes from divergent E. globulus cpDNA lineages previously identified by RFLP analysis (Jackson et al. 1999).
A subsequent study by Freeman et al. (2001) expanded the sampling of E. globulus to a finer geographic resolution and extended the JLA region in the 3′ direction to cover the complete trnH gene and the trnH–psbA intergenic spacer. Analysis of the extended hypervariable sequence, termed JLA+ (Freeman et al. 2001), found the distribution of major haplotype clades to be broadly consistent with that in the former study of E. globulus (Jackson et al. 1999), but allowed for a greater resolution of the phylogenetic relationships between and within haplotype clades. A continental Australian origin of E. globulus was supported by the widespread distribution of the basal JCg haplotypes on continental Australia. There was also evidence of glacial refugia in the coastal areas of eastern and south-eastern Tasmania, with the most recent seed migration of E. globulus between Tasmania and continental Australia occurring along a western island migration route during a glacial maximum and accompanying reduced sea level (Freeman et al. 2001).
More recently, the JLA region was used to investigate chloroplast variation and population structure in E. grandis (Jones et al. 2006), which predominantly occurs in subtropical eastern Australia, with smaller populations located in the tropical north. According to Jones et al. (2006), there was a low level of chloroplast differentiation among populations (GST = 0.30) that was possibly due to a relatively recent geographical isolation of the northern populations of E. grandis. It was further suggested that the northern populations might have been colonised from the southern populations, because of the greater number of haplotypes in the latter populations (Jones et al. 2006).
In contrast with the Eucalyptus species endemic to the Australian mainland, volcanic peaks and seawater geographically isolate the distributions of E. urophylla populations. Therefore, a high degree of population structure at the chloroplast level is expected. Furthermore, the hypothesis of an initial colonisation of Timor and/or Wetar, followed by a westerly migration route, would be supported by a gradient of decreasing chloroplast genetic diversity along the chain of islands towards Flores. With this view, the aim of the present study was to investigate and describe the contemporary phylogeographic structure of E. urophylla among the seven islands of the Sunda archipelago. We report the first estimates of chloroplast sequence variation in the species and infer historical seed-migration routes among the islands on the basis of the relationship between haplotypes, as defined by polymorphisms in the hypervariable JLA+ region and their present geographical distribution. The results are interpreted in light of the known geological and oceanographic patterns of the region.
Materials and methods
Plant material and DNA isolation
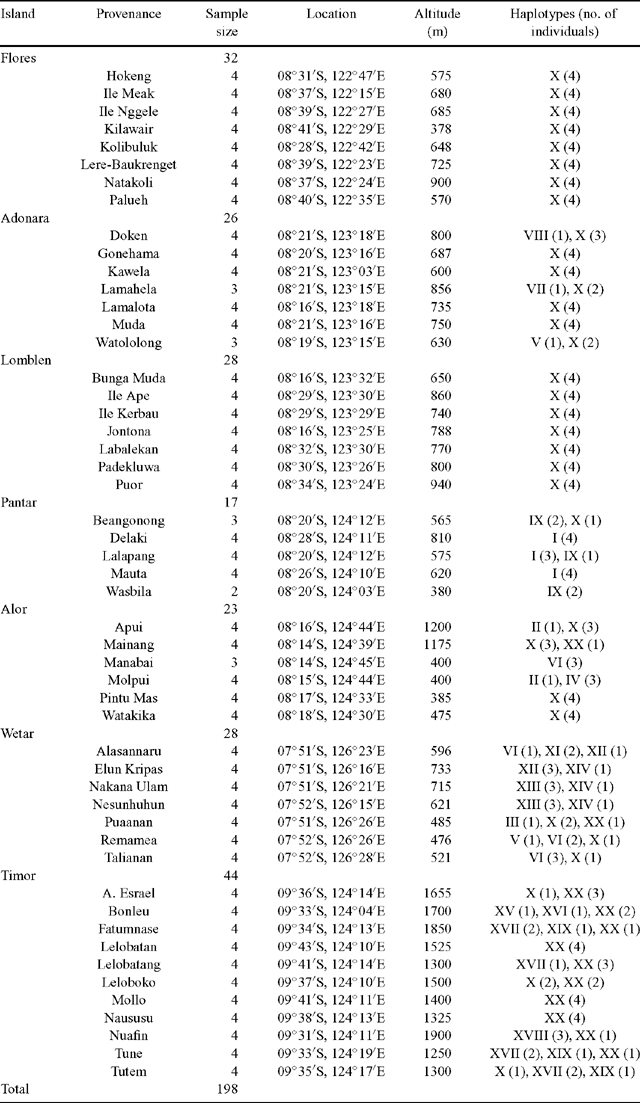
Seed collections were conducted by the research staff of PT Sumalindo Lestari Jaya, a private Indonesian forestry company, and Camcore, North Carolina State University, USA, an international tree conservation and domestication program (Pepe et al. 2004). This series of collections comprised seed from 1104 mother trees distributed across 62 provenances (geographic locations) representing the natural distribution of E. urophylla, barring the region of East Timor, which was experiencing political unrest at the time of seed collection. A subset of 51 provenances was included in the present study. Seeds were sown in a commercial nursery in South Africa (Mondi Business Paper South Africa). Leaf tissue was sampled from 198 plants of known family and provenance origin (Table 1).
|
Total genomic DNA was extracted from 50 mg of fresh leaf tissue with the DNeasy plant mini kit (Qiagen, Valencia, CA). Samples were homogenised for 30–60 s in a FastPrep FP120 instrument (QBiogene, Carlsbad, CA) set at 4.0 m/s. In order to improve efficiency, cell lysis was performed at 65°C for 30 min. Thereafter, all steps were performed as described in the DNeasy plant mini kit manual. DNA quality and quantity were determined by agarose gel electrophoresis and spectrophotometry (Nanodrop Technologies, Wilmington, DE).
Chloroplast DNA amplification and sequencing
The extended JLA+ region used previously by Freeman et al. (2001) was further extended in the 5′ direction in the present study. A forward primer (euro_rpl2; GCGTCCTGTAGTAAGAGGAG) was designed to anneal to a conserved region 151 bp upstream of the forward primer rpl2 previously developed by Goulding et al. (1996) and used in Freeman et al. (2001). We used this primer, together with the reverse primer eucpsbA (eucpsbA; GGAGCAATAACCAACACTCTTG) developed by Freeman et al. (2001). The reverse primer anneals to a conserved region found in eucalypt species 45–66 bp downstream of the stop codon of the psbA gene (Freeman et al. 2001). PCR amplification reactions were performed in 20-μL volumes containing 5 ng of genomic DNA, 0.8 U of Exsel polymerase (Southern Cross), 1 × PCR Exsel buffer, 0.2 mm dNTPs and 0.4 μm of each primer. PCR amplifications were performed with an iCycler (Bio-Rad Laboratories, Hercules, CA), with the following cycling conditions: an initial denaturation step of 94°C for 1 min, followed by 25 cycles of 94°C for 20 s, 64°C for 30 s and 72°C for 40 s with a 1-s increase per cycle; and a final extension step of 68°C for 10 min. The total PCR product length was ~780 bp.
PCR products were cleaned with the QIAquick PCR Product purification kit (Qiagen) and sequenced in both directions, by using primers euro_rpl2 and eucpsbA, with the Big Dye terminator kit (v3.1, Applied Biosystems, Foster City, CA) on an ABI 3100 Automated DNA sequencer (Applied Biosystems).
Genetic-diversity analysis
Sequence data were assembled and aligned with the software package SeqScape (v2.1, Applied Biosystems). The sequence alignment length was reduced to 576 bp to ensure no missing data across 198 samples. Indel (insertion/deletion) mutations were further removed from the analysis as it was unknown whether the indels were produced by a single mutational event or several events. Consequently, the exclusion of indel mutations provided a more conservative estimate of sequence divergence.
Haplotype diversity (h) (Nei 1987) was calculated by using DnaSP version 4.0 (Rozas et al. 2003). Two estimates of population differentiation, GST and NST, were determined by the Hapstep program (version 2001, Pons and Petit 1996). The GST estimate depends only on the frequencies of the haplotypes, but both haplotype frequencies and the genetic distances between haplotypes influence NST. Provenances were treated as populations for the differentiation analysis. The Hapstep program requires populations with sample sizes smaller than three individuals to be excluded from the analysis. Therefore, differentiation parameter estimates (GST and NST) were based on 50 populations (n < 3 for Wasbila provenance from the island of Pantar, Table 1).
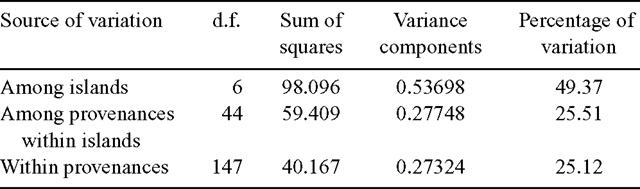
Hierarchical analysis of molecular variance (AMOVA, Excoffier et al. 1992) was implemented by using Arelequin software (version 2.000; Schneider et al. 2000) to apportion variance within provenances, between provenances within islands, and among islands. The significance of variance components was tested by a non-parametric permutation procedure with 1000 permutations.
Haplotype network and nested clade analysis
The program TCS (version 1.2.1, Clement et al. 2000) was used to construct a network of haplotypes by using statistical parsimony (Templeton et al. 1992). Closed haplotype loops were removed by the procedures described by Crandall and Templeton (1993). The haplotype network was converted manually into a nested design according to the procedures defined by Templeton et al. (1987) and Templeton and Sing (1993).
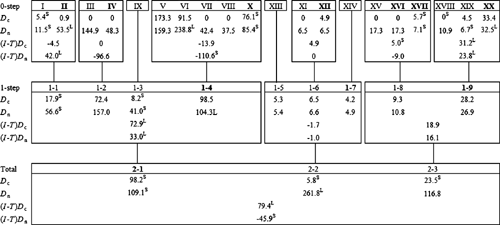
A nested clade analysis (NCA) was performed by the program Geodis (version 2.4; Posada et al. 2000) to assess geographical associations of haplotypes and infer historical patterns of colonisation and dispersal. Clades without geographical or genetic variation were not included in the following analyses. The geographical coordinates of each provenance were used to calculate two statistics, the clade distance (Dc), which measures the geographical spread of a clade, and the nested clade distance (Dn), which measures how a clade is geographically distributed relative to other clades in the same higher-level nesting category (Posada et al. 2000). In addition, for each nesting clade, the average differences in Dc and Dn values between older (interior) and more recent (tip) clades were calculated, abbreviated (I – T)Dc and (I – T)Dn, respectively. To determine whether any of these distance parameters were significantly small or large, all clades within a nesting clade were permuted randomly across localities (1000 times) to generate a null distribution against which the observed values were tested. The output of significant parameters (Dc, Dn, (I – T)Dc, (I – T)Dn) was entered into a program called Autoinfer (version 1.0; Zhang et al. 2006), which inferred biological events by implementing the algorithm by Templeton (2004).
Results
DNA sequence data were obtained from 198 seedlings, representing seven islands and 51 provenances, with a harmonic mean sample size of 26.3 and 3.8, respectively. Each seedling was derived from a unique maternal parent tree because in angiosperms the progeny from the same maternal parent typically share the same chloroplast genome (Corriveau and Coleman 1988).
Haplotype polymorphism and geographic distribution
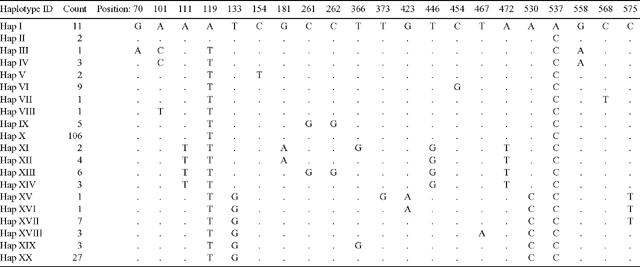
In the present study, the forward primer was moved 151 bp in the 5′ direction of the JLA+ region in an effort to identify additional polymorphic sites. However, all observed nucleotide substitutions were found to occur within the JLA+ region defined by Freeman et al. (2001). In total, 21 polymorphic sites comprising 18 parsimony informative sites and three singleton sites were detected. Twenty haplotypes (Haplotypes I–XX) were identified from the polymorphic sites (Table 2). Haplotype frequencies ranged from 0.005 to 0.535 (Table 2). Haplotype X was the most prevalent, with 106 observations. Five haplotypes were observed in a single individual.
|
The geographic distribution of the haplotypes is listed in Table 1 and illustrated in Fig. 1C. Haplotype X was the most geographically widespread haplotype, occurring in provenances on all seven islands. Haplotype XX was observed in provenances on three adjacent islands, namely Timor, Wetar and Alor. Haplotypes V and VI were observed in provenances on Wetar and were also present on the islands of Adonara and Alor, respectively. The remaining haplotypes were geographically restricted to single islands.
Haplotype diversity and population differentiation
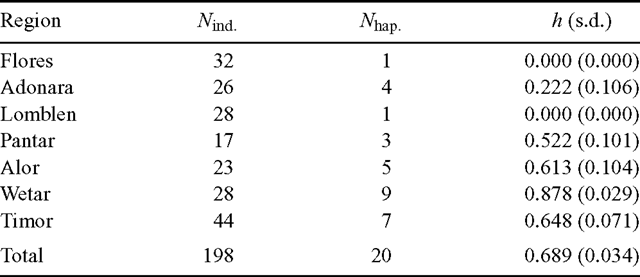
Haplotype diversity (h) across the entire region was 0.689 (Table 3). At the island level, Wetar exhibited the greatest haplotype diversity of 0.878, whereas the populations on the western islands of Flores and Lomblen were fixed for Haplotype X.
|
As defined in ‘Materials and methods’, provenances were treated as populations for the population differentiation analysis. A moderate to high proportion of variation resulted from differences among populations, GST = 0.581. The parameter NST was used to investigate whether related haplotypes were clustered according to geographical location. The NST estimate of 0.724 was larger than the GST estimate and the difference was significant (P < 0.01). According to Pons and Petit (1996), a higher NST than GST usually indicates the presence of phylogeographic structure, with closely related haplotypes being found more often in the same area than less closely related haplotypes.
Hierarchical AMOVA revealed that cpDNA variation within provenances accounted for 25.1% of the total molecular variance (Table 4). A further 25.5% of the total variation was distributed among provenances within islands, whereas 49.4% of the total molecular variance occurred among islands.
|
Population history inferred from NCA
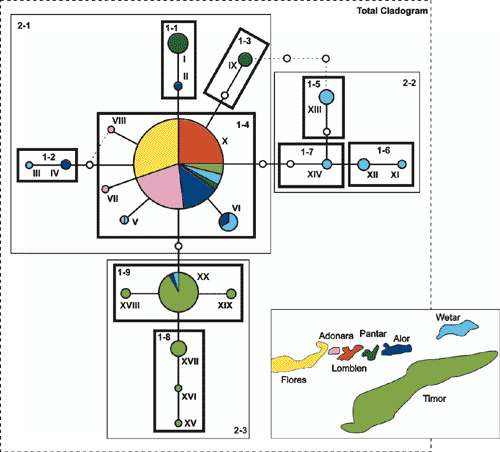
Chloroplast haplotypes were connected in a single most parsimonious network with 95% probability (Fig. 2). Two closed loops, each a consequence of more than one parsimonious connection of a haplotype to the rest of the network, were resolved following the criteria suggested by Crandall and Templeton (1993). Accordingly, a haplotype connection was maintained to high-frequency haplotypes with an interior position in the network rather than to low-frequency haplotypes located in tip clades. In addition, connections between haplotypes occurring in the same geographical area were preferentially maintained.
|
Ancestral haplotypes are identifiable by their internal position in the network, by the number of lineages that arise from them, and by their commonness (Castelloe and Templeton 1994). Statistical parsimony implemented in the TCS program identified Haplotype X as the ancestral haplotype (Fig. 2). Haplotype X was connected to multiple lower-frequency haplotypes, which is consistent with the expectation that older haplotypes have a higher probability of producing mutational derivatives than do younger haplotypes, thereby becoming interior haplotypes (Crandall and Templeton 1993). Related haplotypes derived from Haplotype X were mostly clustered according to geographical location (Fig. 2).
The nested clade analysis was performed manually on the resolved haplotype network according to the algorithm by Templeton et al. (1987) (Fig. 2). Haplotype XVII was the only observed haplotype symmetrically stranded and was grouped with the nesting category that had the smallest sample size, in accordance with Templeton and Sing (1993). Stranded haplotypes that were missing intermediates were left unnested (Templeton and Sing 1993). The nesting design resulted in a three-step hierarchy, with a total of 13 clades. Of these, 10 clades (Table 5) contained both geographical and genetic variation and could therefore be tested for geographical association.
|
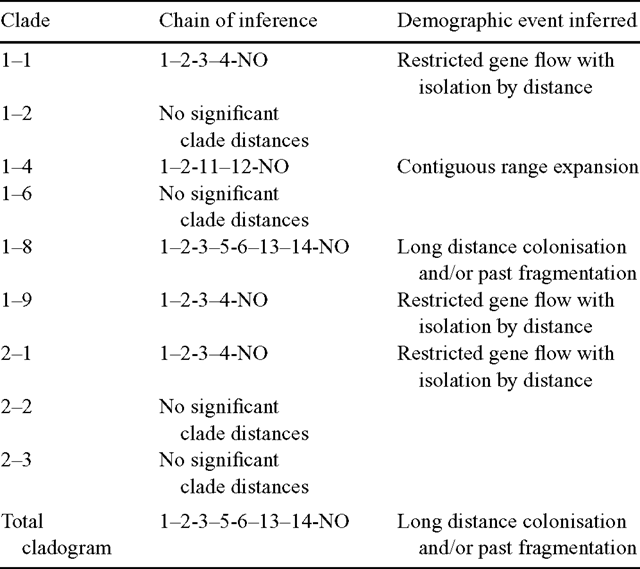
The results of the nested clade analysis are presented in Fig. 3. Nested clades that showed significant spatial genetic structuring were used to infer biological events according to the algorithm by Templeton (2004). Restricted gene flow with isolation by distance was the inferred biological process leading to significant geographical–genetic associations for the haplotypes nested in Clades 1–1 and 1–9, and for the one-step level clades nested in Clade 2–1 (Table 5). The biological processes inferred for the remaining clades and for the total cladogram included contiguous range expansion (Clade 1–4) and long-distance colonisation and/or past fragmentation (Clade 1–8 and total cladogram). In the present study, there was insufficient evidence to discriminate between long-distance colonisation and past fragmentation. It is feasible that both may have played a role leading to the present-day distribution of E. urophylla cpDNA haplotypes.
|
Discussion
In the present paper, we report the first investigation of cpDNA variation in E. urophylla, a tropical forest species that occurs in a series of disjunct populations distributed on seven islands of the Sunda archipelago in eastern Indonesia. Our results allow us to infer possible seed-dispersal routes that may explain the observed patterns in chloroplast and nuclear (House and Bell 1994) genetic diversity within E. urophylla. A moderate to high level of chloroplast genetic differentiation was found. The observed chloroplast genetic structure supports the hypothesis of an east-to-west historical seed-migration route among the seven islands.
Limited population differentiation was observed for E. urophylla when biparentally inherited nuclear DNA markers were used (GST = 0.12; House and Bell 1994), indicating that most of the nuclear genetic diversity in the species is contained within, rather than among, populations. However, the level of chloroplast differentiation among populations observed in the present study was substantially higher (GST = 0.581, NST = 0.724) and was close to the average cytoplasmic differentiation for angiosperm species (GST = 0.64, Petit et al. 2005). There was also a significant phylogeographic structure in the chloroplast variation (NST > GST; P < 0.01), with an estimated 49.37% of the total variance explained by differences among islands and 25.51% by differences among provenances within islands (Table 4). Similar results were reported for Santalum austrocaledonicum, an economically important forest tree species endemic to the New Caledonia and Vanuatu archipelagos, whereby populations were highly differentiated on the basis of chloroplast microsatellite markers (FST = 0.66; Bottin et al. 2007) and the majority of the total variance was explained by differences among islands. The higher level of differentiation observed with chloroplast markers than with nuclear markers in E. urophylla was expected since Eucalyptus seeds are mainly dispersed by gravity, whereas pollen is typically dispersed by insect or even bird vectors (House 1997). The gravitational seed dispersal is relatively limited, particularly where high volcanic mountains and seawater form formidable dispersal barriers at the provenance and island level, respectively.
The islands of Wetar in the east and Timor in the south contained most of the chloroplast genetic diversity of E. urophylla (Table 3). Notably, there was a high amount of morphological variation observed on these two islands that resulted in a proposed separation of two new species from E. urophylla sensu lato, namely E. wetarensis and E. orophila on the islands of Wetar and Timor, respectively (Pryor et al. 1995). However, a subsequent isozyme study did not fully support the proposal, although there was a large degree of allelic diversity on the two islands (House and Bell 1994). Numerous studies of forest trees have described the trend of comparatively high cpDNA diversity in glacial refugia and less diversity in regions colonised more recently following deglaciation (Demesure et al. 1996; Petit et al. 1997; Ferris et al. 1998; King and Ferris 1998; Marchelli et al. 1998). Our data are, therefore, consistent with the original colonisation by E. urophylla of its present natural range occurring on the eastern and/or southern islands, followed by a more recent colonisation of the western region.
The hypothesis of a historical east-to-west migration pattern is further supported by the haplotype network (Fig. 2). Both Wetar and Timor had clades of haplotypes that were private to each island and exhibited considerable divergence from the ancestral Haplotype X. This suggests that E. urophylla was present on both islands for a relatively long time before colonising the other islands to the west. The island of Alor appeared most similar to Wetar and Timor in terms of shared and related haplotypes but the haplotypes were not highly diverged, suggesting a shorter period of occupation on Alor. The observed haplotype distribution on Pantar was quite different from its neighbouring islands in that it had a high frequency of private Haplotypes I and IX. Notably, Haplotype I was closely related to Haplotye II, which was observed on the neighbouring island of Alor (Fig. 2). One suggestion is that Pantar was colonised by individuals with Haplotype II from Alor, which subsequently gave rise to Haplotype I. Other colonisation events may have included individuals with Haplotype X from which Haplotype IX was likely derived. Flores, Lomblen and Adonara appeared to be the most recently colonised islands. A likely source would have been from the island of Alor, which had a high frequency of Haplotype X. The absence of observed cpDNA variation for the islands of Flores and Lomblen (Table 3) is the signature of a recent founder event. Several environmental factors may have contributed to the historical seed-migration patterns that lead to the contemporary distribution of cpDNA variation. These include, among others, island paleogeology and subsequent proximity to the mainland, ocean currents, fluctuations in sea level and possible hybridisation events.
The Lesser Sunda Islands form part of the Banda arc that represents the convergence zone between the still northward-drifting Australian continental margin and the inner arc. The inner arc, which started to appear ~12 million years ago (Audley-Charles 2004), was built up before the collision with the age of inception decreasing eastward (van der Werff 1995). The outer arc arose in the front part of the convergence zone where low-density sedimentary rocks were uplifted by their buoyancy. The emergence of Timor island occurred after the arc-continent collision at ~3.5–2 million years ago (Audley-Charles 2004). On the basis of the proposed order of geological events, one might assume that E. urophylla may have colonised several of the inner arc islands, possibly starting with the older islands in the west, before the emergence and subsequent colonisation of the outer arc island of Timor. However, Ladiges et al. (2003) proposed that E. urophylla diverged from Australian taxa in the subgenus Symphyomyrtus relatively recently, ~5–2 million years ago during the compression of Timor between the inner Banda arc and the north-west region of the Australian continental crust. The putative initial colonisation of Timor from an Australian or New Guinea source would have been assisted by a period of lower sea levels occurring during the Quaternary, bringing emergent lands closer together. The greater number of nuclear DNA alleles on Timor and nearby Alor (House and Bell 1994), together with the greater chloroplast haplotype diversity on the islands of Wetar, Timor and Alor (Table 3), further support the hypothesis of an initial southern or eastern colonisation, followed by a more recent east-to-west colonisation process.
Sea-surface currents are another environmental factor that may be important for predicting ecological and genetic connections among island populations. The currents proximal to the Lesser Sunda Islands primarily comprise North Pacific water flowing from the Makassar Strait into the Flores and Banda Seas, before curling southwards into the Timor Sea and Indian Ocean (Fig. 1B; Gordon and Fine 1996). In addition, there are currents that are channelled into the Ombai Strait between Alor and Timor islands and are generally directed towards the Savu (Sawu) Sea, but there is an occasional reversal of flow in a north-eastern direction, entering the Savu Sea from the Indian Ocean (Molcard et al. 2001). The highly structured distribution of cpDNA at the island level suggests that seed migration across bodies of water is an uncommon occurrence (Fig. 1C). However, if plant material drifted among the islands at the mercy of prevailing currents, it could be assumed that long-distance colonisation (Table 5) would more likely occur in a westerly direction.
During the Pleistocene, glaciation and deglaciation led to fluctuating sea levels that greatly affected landmass configurations in South-east Asia (Voris 2000). At the peak of the glacial maxima, sea levels were at a minimum and many of the present islands, currently separated by shallow seas, merged to form composite islands. Of the seven islands on which E. urophylla naturally occurs, Flores, Adonara and Lomblen were connected when sea levels were 60–120 m below the present level, whereas the other four islands remained separated (Heaney 1991; Voris 2000). According to How et al. (1996), a land bridge between Flores and Lomblen during glacial maxima was considered to be a major factor explaining why populations of several species of snake on Flores and Lomblen were more similar to one another than they were to conspecific populations on adjacent islands to the west (Lombok, Sumba) and east (Alor). Furthermore, the most pronounced morphological differentiation occurred among snake populations existing on different islands that remained separate throughout the Pleistocene. Notably, all of the E. urophylla samples obtained from the islands of Flores and Lomblen, and the majority of samples from Adonara, were fixed for Haplotype X (Fig. 1C). These data suggest a recent colonisation of the western region followed by a founder effect. The likelihood of Haplotype X being fixed in samples from both Flores and Lomblen would have been increased if long-distance seed colonisation occurred during a relatively recent era when these western islands were joined.
Chloroplast DNA variation in Eucalyptus generally appears to be geographically structured, but does not always conform to species boundaries as a result of hybridisation (Steane et al. 1998; Jackson et al. 1999; McKinnon et al. 1999, 2001b). For example, intraspecific cpDNA polymorphism in 14 of 17 species sampled in Tasmania was coupled with extensive sharing of identical haplotypes across populations of different species in the same geographic area (McKinnon et al. 2001b). They concluded that sharing of cpDNA haplotypes among Tasmanian species of Eucalyptus subgenus Symphyomyrtus is the rule rather than the exception. E. urophylla, which occupies a wide altitudinal range on volcanic slopes (180–3000 m, Pepe et al. 2004), forms a mosaic distribution pattern with E. alba at low-elevation sites. Here, natural E. urophylla × E. alba hybrids do exist but they are considered rare as mature trees (Martin and Cossalter 1976).
Putative E. urophylla × E. alba hybrids have been observed in both the first-generation E. urophylla and E. alba provenance trials established in South Africa, suggesting that hybridisation is bi-directional (K. G. Payn, unpubl. data). We obtained three E. alba samples from each of the islands of Flores, Wetar and Timor, and a single sample from New Guinea. All the samples from Flores, Wetar and Timor possessed Haplotype X (K. G. Payn, unpubl. data), the putative ancestral haplotype of E. urophylla. The sample from New Guinea had a highly related haplotype, with only two additional substitutions. These findings suggest that haplotype sharing does occur between E. urophylla and E. alba. Hence, it raises the question whether natural hybridisation events at lower elevation have influenced the distribution of chloroplast haplotypes observed in E. urophylla, particularly Haplotype X (Fig. 1C). However, it is important to note that we presently do not have enough information on the cpDNA haplotype diversity within E. alba to determine whether Haplotype X is ancestral to both species, which also could explain the high prevalence of this haplotype in the small number of E. alba samples that we have analysed.
Conclusions
The present study demonstrates the capacity of cpDNA variation to reveal the phylogeographic history of island-dispersed plant species such as E. urophylla and to draw inferences regarding past migratory routes and possible interactions with other species. The geographical distribution of chloroplast haplotype diversity suggests an east-to-west colonisation pattern. Timor was likely the first island to be colonised, on the basis of its high haplotype diversity and proximity to Australia or New Guinea. The haplotype diversity observed on the islands of Wetar and Alor suggests that they too could be islands of early colonisation, whereas the lack of chloroplast haplotype diversity on the islands of Flores and Lomblen suggest a more recent colonisation event. Restricted gene flow with isolation by distance and long-distance colonisation events, possibly assisted by sea currents, are considered largely responsible for the spatial distribution of cpDNA haplotypes within extant populations of the species.
Pollen flow among provenances and even among islands is likely to be largely responsible for the low estimate of population differentiation with nuclear markers (House and Bell 1994). However, a gradient of decreasing nuclear genetic diversity from east to west was also observed, with the exception being the populations on the island of Flores. On the basis of our chloroplast data, we propose that the high nuclear genetic diversity reported for Flores may be a result of hybridisation with E. alba. This hypothesis is supported by the observation that provenances from Flores appear to have a higher frequency of putative hybrids in the first-generation E. urophylla provenance trials established in South Africa (K. G. Payn, unpubl. data).
Proficient management of this valuable genetic resource, with respect to conservation and breeding strategies, will benefit from the knowledge of the nature and distribution of the chloroplast and nuclear genetic variation across the native range of E. urophylla. In addition, an understanding of the spatial distribution of cpDNA variability in E. urophylla may be used for practical applications such as seed-source certification and the determination of geographic origin of unknown samples.
Acknowledgements
The authors thank G. E. McKinnon for valuable comments on the manuscript and D. Posada for the review of the nested clade design. We are grateful to PT Sumalindo Lestari Jaya for the seed collections, Camcore for locality and reference information, Mondi Business Paper South Africa for maintenance of the plant materials, Sappi Forestry South Africa for providing the E. alba plant material, and F. Maleka for technical assistance. Financial support for this work was provided by Mondi Business Paper South Africa through the Wood and Fibre Molecular Genetics Programme and by Camcore, Raleigh, NC, USA. Additional funding was provided by the Technology and Human Resources for Industry Programme (THRIP) in South Africa.
Audley-Charles MG
(2004) Ocean trench blocked and obliterated by Banda forearc collision with Australian proximal continental slope. Tectonophysics 389, 65–79.
| Crossref | GoogleScholarGoogle Scholar |

Avise J
(1998) The history and purview of phylogeography: a personal reflection. Molecular Ecology 7, 371–379.
| Crossref | GoogleScholarGoogle Scholar |

Birky CW
(1995) Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proceedings of the National Academy of Sciences USA 92, 11331–11338.
| Crossref | GoogleScholarGoogle Scholar |

Bottin L,
Tassin J,
Nasi R, Bouvet JM
(2007) Molecular, quantitative and abiotic variables for the delineation of evolutionary significant units: case of sandalwood (Santalum austrocaledonicum Vieillard) in New Caledonia. Conservation Genetics 8, 99–109.
| Crossref | GoogleScholarGoogle Scholar |

Brown P,
Sutikna T,
Morwood MJ,
Soejono RP,
Jatmiko
,
Saptomo EW, Due RA
(2004) A new small-bodied hominin from the late Pleistocene of Flores, Indonesia. Nature 431, 1055–1061.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Byrne M,
Moran GF, Tibbits WN
(1993) Restriction map and maternal inheritance of chloroplast DNA in Eucalyptus nitens. Journal of Heredity 84, 218–220.

Byrne M, Moran GF
(1994) Population divergence in the chloroplast genome of Eucalyptus nitens. Heredity 73, 18–28.

Castelloe J, Templeton AR
(1994) Root probabilities for intraspecific gene trees under neutral coalescent theory. Molecular Phylogenetics and Evolution 3, 102–113.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Cavers S,
Navarro C, Lowe AJ
(2003) Chloroplast DNA phylogeography reveals colonization history of Neotropical tree, Cedrela odorata L., in Mesoamerica. Molecular Ecology 12, 1451–1460.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Clement M,
Posada D, Crandall KA
(2000) tcs: a computer program to estimate gene genealogies. Molecular Ecology 9, 1657–1659.
| Crossref | tcs: a computer program to estimate gene genealogies.&journal=Molecular Ecology&volume=9&pages=1657-1659&publication_year=2000&author=KA%20Crandall&hl=en&doi=10.1046/j.1365-294x.2000.01020.x" target="_blank" rel="nofollow noopener noreferrer" class="reftools">GoogleScholarGoogle Scholar | PubMed |

Corriveau JL, Coleman AW
(1988) Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. American Journal of Botany 75, 1443–1458.
| Crossref | GoogleScholarGoogle Scholar |

Crandall KA, Templeton AR
(1993) Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134, 959–969.
| PubMed |

Demesure B,
Comps B, Petit RJ
(1996) Chloroplast DNA phylogeography of the common beech (Fagus sylvatica L.) in Europe. Evolution 50, 2515–2520.
| Crossref | GoogleScholarGoogle Scholar |

Dumolin-Lapegue S,
Demesure B,
Fineschi S,
Le Corre V, Petit RJ
(1997) Phylogeographic structure of white oaks throughout the European continent. Genetics 146, 1475–1487.
| PubMed |

Excoffier L,
Smouse P, Quattro JM
(1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
| PubMed |

Ferris C,
King RA,
Vainola R, Hewitt GM
(1998) Chloroplast DNA recognizes three refugial sources of European oaks and suggests independent eastern and western immigrations to Finland. Heredity 80, 584–594.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Freeman JS,
Jackson HD,
Steane DA,
McKinnon GE,
Dutkowski GW,
Potts BM, Vaillancourt RE
(2001) Chloroplast phylogeography of Eucalyptus globulus. Australian Journal of Botany 49, 585–596.
| Crossref | GoogleScholarGoogle Scholar |

Gordon AL, Fine RA
(1996) Pathways of water between the Pacific and Indian oceans in the Indonesian seas. Nature 379, 146–149.
| Crossref | GoogleScholarGoogle Scholar |

Goulding SE,
Olmstead RG,
Morden CW, Wolfe KH
(1996) Ebb and flow of the chloroplast inverted repeat. Molecular & General Genetics 252, 195–206.

Heaney LR
(1991) A synopsis of climatic and vegetational change in southeast Asia. Climatic Change 19, 53–61.
| Crossref | GoogleScholarGoogle Scholar |

House APN, Bell JC
(1994) Isozyme variation and mating system in Eucalyptus urophylla S.T.Blake. Silvae Genetica 43, 167–176.

How RA,
Schmitt LH, Suyanto A
(1996) Geographical variation in the morphology of four snake species from the Lesser Sunda Islands, eastern Indonesia. Biological Journal of the Linnean Society 59, 439–456.
| Crossref | GoogleScholarGoogle Scholar |

Jackson HD,
Steane DA,
Potts BM, Vaillancourt RE
(1999) Chloroplast DNA evidence for reticulate evolution in Eucalyptus (Myrtaceae). Molecular Ecology 8, 739–751.
| Crossref | GoogleScholarGoogle Scholar |

Jones ME,
Shepherd M,
Henry RJ, Delves A
(2006) Chloroplast DNA variation and population structure in the widespread forest tree, Eucalyptus grandis. Conservation Genetics 7, 691–703.
| Crossref | GoogleScholarGoogle Scholar |

King RA, Ferris C
(1998) Chloroplast DNA phylogeography of Alnus glutinosa (L.) Gaertn. Molecular Ecology 7, 1151–1161.
| Crossref | GoogleScholarGoogle Scholar |

Ladiges PY,
Udovicic F, Nelson G
(2003) Australian biogeographical connections and the phylogeny of large genera in the plant family Myrtaceae. Journal of Biogeography 30, 989–998.

Marchelli P,
Gallo L,
Scholz F, Ziegenhagen B
(1998) Chloroplast DNA markers reveal a geographical divide across Argentinean southern beech Nothofagus nervosa (Phil.) Dim. et Mil. distribution area. Theoretical and Applied Genetics 97, 642–646.
| Crossref | GoogleScholarGoogle Scholar |

Martin B, Cossalter C
(1976) Les eucalyptus des iles de la Sonde. Bois et Foréts des Tropiques 164, 3–14.

McCauley DE
(1995) The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends in Ecology & Evolution 10, 198–202.
| Crossref | GoogleScholarGoogle Scholar |

McKinnon GE,
Steane DA,
Potts BM, Vaillancourt RE
(1999) Incongruence between chloroplast and species phylogenies in Eucalyptus subgenus Monocalyptus (Myrtaceae). American Journal of Botany 86, 1038–1046.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

McKinnon GE,
Vaillancourt RE,
Tilyard PA, Potts BM
(2001a) Maternal inheritance of the chloroplast genome in Eucalyptus globulus and interspecific hybrids. Genome 44, 831–835.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

McKinnon GE,
Vaillancourt RE,
Jackson HD, Potts BM
(2001b) Chloroplast sharing in the Tasmanian eucalypts. Evolution 55, 703–711.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Michaux B
(1991) Distributional patterns and tectonic development in Indonesia. Australian Systematic Botany 4, 25–36.
| Crossref | GoogleScholarGoogle Scholar |

Molcard R,
Fieux M, Syamsudin F
(2001) The throughflow within Ombai Strait. Deep-sea Research. Part I. Oceanographic Research Papers 48, 1237–1253.
| Crossref | GoogleScholarGoogle Scholar |

Myers N,
Mittermeier RA,
Mittermeier CG,
da Fonseca GAB, Kent J
(2000) Biodiversity hotspots for conservation priorities. Nature 403, 853–858.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Norvick MS
(1979) The tectonic history of the Banda Arcs, eastern Indonesia: a review. Journal of the Geological Society London 136, 519–527.

Pepe B,
Surata K,
Suhartono F,
Sipayung M,
Purwanto A, Dvorak WS
(2004) Conservation status of natural populations of Eucalyptus urophylla in Indonesia and international efforts to protect dwindling gene pools. Forest Genetic Resources [FAO: Rome] 31, 62–64.

Petit RJ,
Pineau E,
Demesure B,
Bacilierir R,
Ducousso A, Kremer A
(1997) Chloroplast DNA footprints of postglacial recolonization by oaks. Proceedings of the National Academy of Sciences USA 94, 9996–10001.
| Crossref | GoogleScholarGoogle Scholar |

Petit RJ,
Aguinagalde I,
de Beaulieu JL,
Bittkau C,
Brewer S,
Cheddadi R,
Ennos R,
Fineschi S,
Grivet D,
Lascoux M,
Mohanty A,
Müller-Starck G,
Demesure-Musch B,
Palmé A,
Martin JP,
Rendell S, Vendramin GG
(2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300, 1563–1565.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Petit RJ,
Duminil J,
Fineschi S,
Hampe A,
Salvini D, Vendramin GG
(2005) Comparative organisation of chloroplast, mitochondrial and nuclear diversity in plant populations. Molecular Ecology 14, 689–701.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pons O, Petit RJ
(1996) Measuring and testing genetic differentiation with ordered vs. unodered alleles. Genetics 144, 1237–1245.
| PubMed |

Posada D,
Crandall KA, Templeton AR
(2000) Geodis: a program for cladistic nested analysis of the geographical distribution of genetic haplotypes. Molecular Ecology 9, 487–488.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pryor LD,
Williams ER, Gunn BV
(1995) A morphometric analysis of Eucalyptus urophylla and some related taxa with descriptions of two new species. Australian Systematic Botany 8, 57–70.
| Crossref | GoogleScholarGoogle Scholar |

Rozas J,
Sánchez-DelBarrio JC,
Messeguer X, Rozas R
(2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Sewell MM,
Parks CR, Chase MW
(1996) Intraspecific chloroplast DNA variation and biogeography of North American Liriodendron L. (Magnoliaceae). Evolution 50, 1147–1154.
| Crossref | GoogleScholarGoogle Scholar |

Steane DA,
Byrne M,
Vaillancourt RE, Potts BM
(1998) Chloroplast DNA polymorphism signals complex interspecific interactions in Eucalyptus (Myrtaceae). Australian Systematic Botany 11, 25–40.
| Crossref | GoogleScholarGoogle Scholar |

Templeton AR
(2004) Statistical phylogeography: methods of evaluating and minimizing inference errors. Molecular Ecology 13, 789–809.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Templeton AR, Sing CF
(1993) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analysis with cladogram uncertainty and recombination. Genetics 134, 659–669.
| PubMed |

Templeton AR,
Boerwinkle E, Sing CF
(1987) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in Drosophila. Genetics 117, 343–352.
| PubMed |

Templeton AR,
Crandall KA, Sing CF
(1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132, 619–633.
| PubMed |

Vaillancourt RE, Jackson HD
(2000) A chloroplast DNA hypervariable region in eucalypts. Theory of Applied Genetics 101, 473–477.
| Crossref | GoogleScholarGoogle Scholar |

Voris HK
(2000) Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. Journal of Biogeography 27, 1153–1167.
| Crossref | GoogleScholarGoogle Scholar |

Wallace AR
(1860) On the zoological geography of the Malay archipelago. Journal of the Linnean Society of London 4, 172–184.

van der Werff W
(1995) Cenozoic evolution of the Savu Basin, Indonesia: forearc basin response to arc-continent collision. Marine and Petroleum Geology 12, 247–262.
| Crossref | GoogleScholarGoogle Scholar |

Zhang AB,
Tan S, Sota T
(2006) Autoinfer 1.0: a computer program to infer biogeographical events automatically. Molecular Ecology Notes 6, 597–599.
| Crossref | GoogleScholarGoogle Scholar |



