Origins of the rare Australian daisy Erigeron conyzoides and its implications for biological control research and conservation management
Stephanie H. Chen A , Alicia Grealy A , Michelle A. Rafter B , Ben Gooden C and Alexander N. Schmidt-Lebuhn A *
A , Alicia Grealy A , Michelle A. Rafter B , Ben Gooden C and Alexander N. Schmidt-Lebuhn A *
A
B
C
Abstract
Confidence in risk analyses for weed biological control (biocontrol) agents is underpinned by knowledge of the phylogenetic associations between the target weed and off-target plant species, with an emphasis on native taxa that co-occur with the weed in its introduced range. The origins of off-target plant species are also considered when assessing potential adverse effects of releasing weed biocontrol agents into the environment. Erigeron bonariensis L. (flaxleaf fleabane), native to South America, is a major cropping weed in North America, Europe, and Australia. Phylogenomic analysis of the weed’s tribe, Astereae, for a biocontrol program has put into question the existence of native Australian fleabanes.
We aimed to resolve the establishment means of a supposed native species to Australia by testing its phylogenetic and morphological associations with other Erigeron taxa at a global scale.
Target-sequence capture data were combined with traditional taxonomy.
We rediscovered the closest presumed native relative of flaxleaf fleabane, the rare and declining E. conyzoides F.Muell. (daisy fleabane), during field work in Victoria, Australia. Molecular data and morphology indicated that E. conyzoides and E. acer L. from the northern hemisphere are not distinct.
Erigeron conyzoides is very likely not a distinct species endemic to Australia but rather a disjunct population of E. acer.
This finding improves confidence in the host-specificity of candidate biocontrol agents for fleabane in Australia, because we argue that the closest related truly native species is much more distantly related to fleabane than previously thought.
Keywords: Asteraceae, Astereae, Erigeron conyzoides, host-specificity test, phylogenetics, risk analyses, target capture, weed biological control.
Introduction
Invasive plants (hereafter weeds) have wide-ranging adverse ecological and socio-economic impacts (Pyšek et al. 2012). Weed biological control (biocontrol) involves the use of natural enemies (usually fungal pathogens and herbivorous invertebrates) to reduce the risk of spread and impacts of weeds on recipient ecosystems (Schwarzländer et al. 2018; Hinz et al. 2020; Cullen et al. 2022). Biocontrol offers a sustainable and cost-effective approach to weed control (Cullen et al. 2022; Fowler et al. 2024; Paterson et al. 2024). The risks of off-target impacts of candidate weed biocontrol agents are assessed through host-specificity testing that is informed by research to understand evolutionary (phylogenetic) relationships between the target weed and related plants. Off-target plant species more closely related to the target weed tend to be at a greater risk of off-target damage by a candidate biocontrol agent and are thus prioritised for host-specificity testing (Wapshere 1974; Sheppard et al. 2002; Briese 2005; Chen et al. 2024).
Priority in host-specificity testing tends to be given to closely related taxa that are either native or species of high economic importance (e.g. crops, ornamental garden plants) (Barton 2004; Gilbert et al. 2012). As such, resolving the phylogenetic associations between a target weed and co-occurring plant species as well as the establishment means of resident taxa are critical factors underpinning confidence in weed-biocontrol risk assessments. For example, Gildenhuys et al. (2013, 2015) clarified that several balloon vine species in the genus Cardiospermum are native to southern Africa. Conversely, Senecio madagascariensis Poir. was in Australia for several decades considered to be part of a native species complex, until consultation with a South African taxonomist suggested its true identity (Michael 1994). This was subsequently confirmed with genetic data (Scott et al. 1998) and was the basis for initiating exploratory research in Madagascar and South Africa for invertebrates and pathogens as suitable biocontrol for S. madagascariensis management in Australia. Uncertain phylogenetic associations with indigenous flora can hamper progress on biocontrol research and, subsequently, effective invasive-plant management.
In this study, we sought to use target-sequence capture-data phylogenies along with traditional taxonomy to resolve the origin and introduction status of off-target plant species and improve confidence in weed-biocontrol risk assessments, by using the case study of Erigeron bonariensis L. (flaxleaf fleabane) invasion in Australia. There are ~490 species of Erigeron L. (Bánki et al. 2023), with seven species found in Australia, including E. bilbaoanus (J.Rémy) Cabrera, E. bonariensis, E. canadensis L., E. conyzoides F.Muell., E. karvinskianus DC., E. primulifolius (Lam.) Greuter, and E. sumatrensis Retz (Council of Heads of Australasian Herbaria 2024). Erigeron bonariensis is a widespread and common invasive species in Australia; the species is native to South America, and the first botanical collections in Australia date back to the 1940s (Wu 2007). Erigeron bonariensis is an approved candidate for biocontrol in Australia (Rafter and Morin 2017) by using the rust agent Puccinia cnici-oleracei Pers. (ex. Conyza) (Morin et al. 2020; Gooden 2023).
As the closest native relative of E. bonariensis and the only native Australian species of the genus Erigeron, E. conyzoides (daisy fleabane) was identified as at highest risk of off-target damage from a candidate biocontrol agent. It was therefore included in the proposed test list for host-specificity testing (Hunter et al. 2018). However, E. conyzoides is very rare and apparently declining (Fig. 1a). Previously found across much of the Australian Alps, it has not been collected in New South Wales since 1978 (Fig. 1b). It could, therefore, not be located for previous host-specificity testing (Morin et al. 2020), neither during field work nor in seedbanks, and an attempt to raise plants from seeds on herbarium specimens failed.
(a) Collections of Erigeron conyzoides (purple) have declined in recent decades in proportion to overall Asteraceae collections in its area of occurrence (grey). (b) Historical distribution of Erigeron conyzoides collections in south-eastern continental Australia with geocoded localities in 1854–1899 (light purple circles), 1900–1979 (mid-purple triangles), and 1980–2024 (dark purple squares).
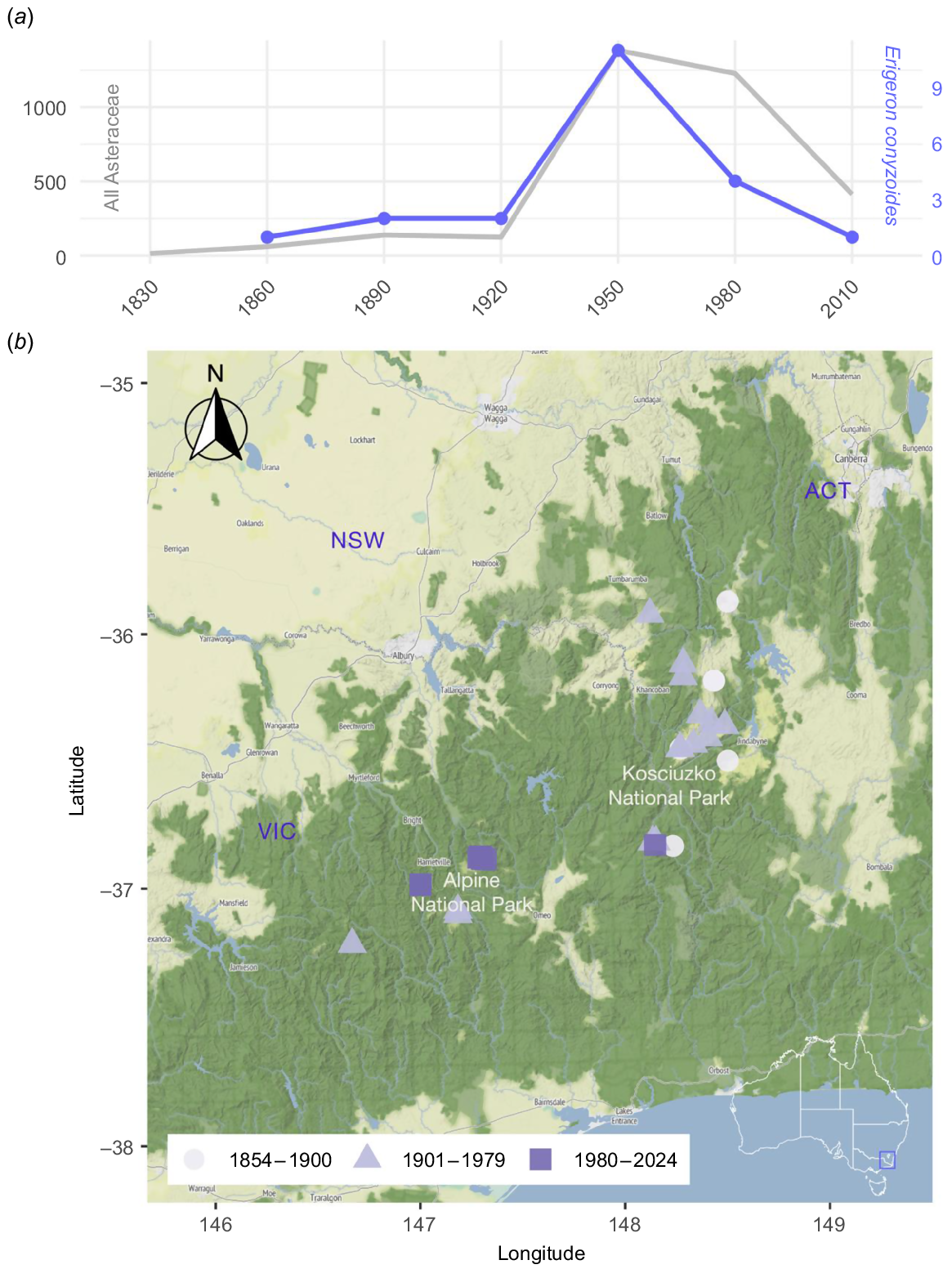
Erigeron conyzoides is listed as Endangered under the Victorian Government’s Flora and Fauna Guarantee Act 1988 Threatened List (Department of Energy, Environment and Climate Action 2024). It was first listed as Vulnerable on the Advisory List of Rare or Threatened Plants in Victoria 2014 (Department of Environment and Primary Industries 2014) and subsequently re-assessed and officially listed as Endangered in 2021 (Department of Environment, Land, Water and Planning 2021). However, a recent target sequence capture phylogeny of Australian Astereae casts doubt on E. conyzoides being native (Chen et al. 2024). The species is phylogenetically isolated from other native species of tribe Astereae in Australia and biogeographically isolated from its close relatives in the northern hemisphere. Apart from its original description by Mueller (1855) and flora treatments, Erigeron conyzoides appears to have been the subject of only one scientific paper, a study of its chemical compounds (Sorensen and Sorensen 1969).
Prompted by the requirement for additional biocontrol options for Erigeron bonariensis, the aim of this study was to find an extant population of Erigeron conyzoides in Australia, collect seeds to take the species into cultivation for use in host-specificity testing, and increase our understanding of its biology and native or introduced status by using a combination of phylogenomics, morphology, and historical observations.
Materials and methods
Field work
Field work was conducted in January 2024 in Kosciuszko National Park, New South Wales, and Alpine National Park, Victoria, in Australia. After discovering an extant population of Erigeron conyzoides at Falls Creek, Victoria (36.8746°S, 147.3001°E), young leaf tissue was collected from 15 individuals spatially distributed across the population and dried on silica gel.
Herbarium specimens and floras
To explore the possibility that Erigeron conyzoides is in fact an introduced species in Australia, we first attempted to identify specimens held at the Australian National Herbarium with keys published in the following floras from the northern hemisphere: Flora Europaea (Tutin 1976), the Flora of China (Chen and Brouillet 2011), the Flora of North America (Nesom 2006), and the Flora of the USSR (Schischkin 1999). In addition, we consulted a taxonomic treatment of South American Erigeron (Solbrig 1962).
To visualise historical collections, records of preserved specimens of Asteraceae in circles of 5 km radius around 16 historical localities of E. conyzoides in Australia were queried from the Atlas of Living Australia (https://ala.org.au, accessed 29 May 2024) by using the R package galah (v.2.0.2; Westgate et al. 2024). To allow automatic removal of duplicates with R (v.4.4.0; R Core Team 2024), collector names and localities were harmonised with OpenRefine (v.3.8.1; Ham 2013) by using the key collision method and both fingerprint and n-Gram fingerprint keying functions. Duplicates of collector name and collection number were removed except for ‘s.n.’ (specimens without collection numbers), in which case duplicates of collector name and exact locality description were removed. The data were binned into 30-year intervals and plotted with R. We also mapped specimens of Erigeron conyzoides in Australia, by using records from the Australasian Virtual Herbarium (https://avh.ala.org.au, accessed 14 January 2024).
DNA extraction and library preparation
We used existing data from our recently published phylogeny of Asteraceae tribe Astereae in Australia (Chen et al. 2024) and the Plant and Fungal Tree of Life (PAFTOL), supplementing these with newly generated sequences of Erigeron conyzoides and the morphologically most similar northern hemisphere species, E. acer (Supplementary Table S1).
We followed DNA-extraction and library-preparation methods described in Chen et al. (2024). Briefly, genomic DNA was extracted from 5–15 mg of silica dried leaf tissue or herbarium material by using the QIAGEN DNeasy Plant 96 Kit. Libraries were built using the QIAGEN Ultralow Input Library Kit, by using a protocol adapted for automation by using an Echo 525 Acoustic Liquid Handler (Beckman-Coulter). Sequence capture was conducted on pools of 16 libraries by using the myBaits Expert Angiosperms353 (v.1; Johnson et al. 2019) kit (Daicel Arbor Biosciences), following the manufacturer’s standard protocol for plants (v5 September 2020). Enriched libraries were sequenced on Illumina NovaSeq 6000 SP with v1.5 paired-end 2 × 150 cycle chemistry with 8-bp dual indexing.
Demultiplexed reads were quality filtered and paired using Trimmomatic (v.0.38; Bolger et al. 2014), with illuminaclip:adapters, fa:4:20:10, minimum length of 30, and average quality of 25. Sequences were further filtered with BBDuk (Bushnell 2022) with entropy of 0.8, entropy window of 20, and low-entropy parts of sequences were masked. The target capture data were assembled with an Asteraceae-specific target file (McLay et al. 2021) by using HybPiper (Johnson et al. 2016), implemented through the Nextflow pipeline hybpiper-nf (Jackson et al. 2023).
ITS phylogeny from GenBank Sanger data
To complement our genomic-scale capture dataset, which included a limited number of samples per species, we inferred an Erigeron phylogeny by using nuclear ribosomal internal transcribed spacer (ITS) sequences, the most widely used Sanger-sequence marker. We queried all ITS sequences of Erigeron and its synonym Conyza from GenBank (accessed 24 January 2024), aligned them by using MAFFT (v.7.453; Katoh and Standley 2013), and excluded a small number of poor-quality sequences after visual inspection of the alignment. We assembled filtered and paired reads of seven samples of E. conyzoides and two of E. acer from target capture data by using hybpiper-nf and an ITS sequence of E. acer (Genbank accession OK447875) as the reference. We obtained six high-quality ITS sequences for E. conyzoides and one for E. acer. It is likely that the target capture reaction was too efficient for the other samples, leaving insufficient off-target reads for ITS assembly. To test whether the use of E. acer as the reference introduced reference bias, we repeated the assembly by using distantly related E. varius Webb. (KR059038). The resulting sequences were identical, indicating no reference bias.
The newly assembled ITS sequences were added to the alignment of sequences from GenBank by using MAFFT (v.7.453). The resulting alignment had 867 sequences with 984 characters, of which 363 were parsimony informative, 162 variable but uninformative, and 459 constant. Phylogenetic analysis was conducted with IQ-TREE (v.1.6.12; Nguyen et al. 2015) without partitioning, under automatic model testing, and by using the approximate likelihood-ratio test (aLRT) (Anisimova and Gascuel 2006) as support values with 1000 replicates. SYM+R4 was chosen as the preferred model. The phylogeny was outgroup rooted on E. rhizomatus Cronquist (AF046992).
Target-sequence capture (Angiosperm353) phylogeny
We took three different approaches to generate the phylogeny. First, two-step coalescent analysis was performed directly from the paralog sequences retrieved by hybpiper-nf. Sequences were aligned with MAFFT (v.7.490; Katoh and Standley 2013) and gene trees were generated with IQ-TREE 2 (v.2.2.0.5; Nguyen et al. 2015; Minh et al. 2020). The best substitution model was estimated with ModelFinder (Kalyaanamoorthy et al. 2017) and support values were estimated with SH-aLRT (Guindon et al. 2010) and 10,000 ultrafast bootstrap replicates (Hoang et al. 2018). The gene trees were combined into a species tree with ASTRAL-Pro (v.1.15.1; Zhang et al. 2020), which allows for multi-copy genes and therefore deals with paralogs. Branch supports were measured as local posterior probabilities, and branch lengths for non-terminal branches were measured in coalescent units.
Second, the paralog sequences retrieved by hybpiper-nf were processed with paragone-nf (v.1.0.1; Jackson et al. 2023) to resolve paralogy (Yang and Smith 2014). The monophyletic-outgroups (MO) algorithm was used in favour of both the rooted-subtrees (RT) and maximum-inclusion (MI) algorithms, as MO produces a more complete matrix. The multiple gene alignments were cleaned and checked to be in-frame by using a custom Python script (https://bitbucket.csiro.au/projects/NRCA/repos/bioinformatics-and-phylogenetics/browse/check_odd_indels_and_frames2.py). For the concatenated analysis, which produced the main results discussed in this paper, concatenation was performed with a custom Python script (https://bitbucket.csiro.au/projects/NRCA/repos/bioinformatics-and-phylogenetics/browse/concat7.py), with a minimum of 20 genes and five taxa per alignment. One sample (Erigeron_conyzoides_SL2138H) was removed because it had fewer than 20 genes, whereas four genes (6064_2, 6448_1, 6705_1, and 6958_1) were removed. With an input of 33 taxa and 260,349 total sites (15.5% missing data), a phylogeny was constructed with IQ-TREE 2 (v.2.2.0.5; Nguyen et al. 2015; Minh et al. 2020) with partition models (Chernomor et al. 2016). There were 8004 parsimony-informative sites, 11,199 singleton sites, and 161,151 constant sites. The data were partitioned by gene, with a total of 332 partitions. The best substitution model for each partition was estimated with ModelFinder (Kalyaanamoorthy et al. 2017), and support was estimated with SH-aLRT (Guindon et al. 2010) and 10,000 ultrafast bootstrap replicates (Hoang et al. 2018).
Third, for other two-step coalescent analyses after resolution of paralogy, the MO outputs were aligned with MAFFT (v.7.490: Katoh and Standley 2013) and gene trees were constructed with IQ-TREE 2 (v.2.2.0.5; Nguyen et al. 2015; Minh et al. 2020) by using ModelFinder (Kalyaanamoorthy et al. 2017), SH-aLRT (Guindon et al. 2010), and 10,000 ultrafast bootstraps (Hoang et al. 2018) with partition models (Chernomor et al. 2016). The gene trees were combined into a species tree with ASTRAL (v.5.7.8; Zhang et al. 2018).
Variant calling and population genomics
The trimmed and quality- filtered target-sequence capture reads for Erigeron species were aligned to the nine chromosomes of the Erigeron canadensis reference genome (Laforest et al. 2020) by using BWA v.0.7.17 (Li 2013), and variants were called with freebayes (v.1.3.7; Garrison and Marth 2012).
The variant call format (VCF) file was filtered using vcftools (v.0.1.16; Danecek et al. 2011) and vcflib (v.1.0.1; Garrison et al. 2022). Low-quality sites were removed (QUAL/DP > 0.25 and --minQ 20 --min-meanDP 4 --max-meanDP 40 --mac 2 --max-missing-count 32 –remove-indels). Filtering was then performed for allelic balance (AB > 0.25 and AB < 0.75| AB < 0.01) and discrepancy in mapping quality between reference and alternative alleles (MQM/MQMR < 2 and MQMR/MQM < 2). The adegenet package (Jombart 2008) was used to perform principal-component analysis (PCA) in (R v.4.0.4; R Core Team 2024).
Results
Field observations
During field surveys by the New South Wales National Parks and Wildlife Service (NPWS) and its contractors in 2019, Erigeron conyzoides was recorded from several sites in Kosciuszko National Park (bionet.nsw.gov.au, accessed 22 December 2023). We visited one site north of Thredbo, just east of Kosciuszko Walk. At and around the exact coordinates recorded, we were unable to find the species. The local habitat was predominantly alpine heath. No species were present that could plausibly have been misidentified as E. conyzoides, except perhaps two species of Pappochroma Raf. (formerly classified as part of Erigeron), with very different inflorescence structure.
There has been a historical decline in collections of Erigeron conyzoides (Fig. 1). The most recent herbarium collection of Erigeron conyzoides was made in 2011 (P.A. French s.n., MELUD106780a, MELUD106792a), along a road directly south of Falls Creek, Victoria. The population was described as ‘growing in gravel up to 60 cm [tall]. Five specimens growing closely together but not occurring anywhere else in surroundings.’. We were unable to find this population. Given its small size and the time since it was last observed, it may be extinct in that location.
The second most recent herbarium collection with precise location data was made in 1998, also near Falls Creek, but at Rocky Valley Dam (N.G. Walsh 4720, MEL 2044671A). This was the only population we were able to find in January 2024. Erigeron conyzoides grew in large numbers on a terraced rock slope of the northern side of the dam in a weed-dominated community (Fig. 2a) with Euchiton japonicus (Thunb.) Holub, Hypochaeris radicata L., and a Verbascum species. It also grew in smaller numbers along the creek under the dam, but not in undisturbed, native vegetation further away from the dam. We did not count individuals, but the population is likely to have onstituted at least a few hundred individuals, including sterile, young rosettes, mid-sized plants producing one flowering stem, and old plants producing multiple flowering stems each year, indicating a stable, reproducing population and a perennial habit.
(a) Erigeron conyzoides in Victoria, Australia, Schmidt-Lebuhn 2138 (CANB 997869), photographer Alexander Schmidt-Lebuhn. (b) Erigeron acer subsp. politus in Russia, iNaturalist observation 38471468, reproduced with permission from Igor Pospelov (taimyr).

The largest specimens were ~50 cm tall, smaller than the largest herbarium specimens deposited at CANB, perhaps because of the exposed, often windy location. Stems were reddish-purple, and flowering capitula showed a striking contrast between reddish-purple phyllaries and white ray florets. We revisited the population on 5 February 2024 and found that older ray florets had turned light purple (Fig. 2a).
During the second visit in February, fertilised capitula were bagged. They were harvested by the Victorian Conservation Seedbank on 20 February 2024, resulting in the retrieval of ~17,000 seeds, of which an estimated 75% were filled, as estimated from X-ray imaging of a sample (Rebecca Miller, pers. comm.). Seeds were divided between the Victorian Conservation Seedbank and the National Seed Bank, and some were set aside to raise plants for host-specificity testing in weed biocontrol research.
Morphological affinities of Erigeron conyzoides
In the Flora Europaea (Tutin 1976), Erigeron conyzoides somewhat doubtfully keyed out to Erigeron acer subsp. politus (Fries) H.Lindberg. Couplet 11 in the key to the species of Erigeron and Couplet 2 in the key to subspecies of E. acer are problematic. The first lead of the former requires the plant to be an ‘annual or short-lived perennial’, with basal leaf rosette ‘usually absent at anthesis’, which is not the case for E. conyzoides, but the lead matches its growth form and capitulescence. The latter requires phyllaries to be ‘glabrous or almost so’ to lead to E. acer subsp. politus, whereas E. conyzoides has glandular and/or hispid phyllaries. However, the implied morphology of E. acer subsp. politus in the lead is contradicted by that described in the Flora of China.
In the Flora of China, Erigeron conyzoides keyed out to Erigeron acer subsp. politus. The only difference between the two taxa is that E. acer subsp. politus is described as having pink to lilac rays, whereas those of E. conyzoides are pure-white when young. However, as observed during field work, the rays of E. conyzoides turn purplish-pink as the capitula age, and the two taxa share deep purple stems and phyllaries, sparsely hispid stem indumentum, leaves that are more or less glabrous on the surfaces and ciliate on the margins, and phyllaries short-stipitate glandular to hispid.
In the Flora of the USSR, which applied an extremely narrow (splitting) species concept to Erigeron, E. conyzoides keyed out to E. elongatus Ledeb., a synonym of E. acer subsp. politus. In the Flora of North America, Erigeron conyzoides keyed out to E. acer. No match could be made to South American species in Solbrig’s taxonomic treatment (Solbrig 1962). The morphologically closest species were arguably E. pazensis Sch.Bip. ex Rusby and E. pratensis Phil. However, the former does not have a leaf rosette, and the latter has ray florets that are too few and too long, and no glands on the phyllaries.
Morphological comparison of herbarium specimens of Erigeron conyzoides and E. acer (some of which were collected by co-author A. N. Schmidt-Lebuhn in Germany) confirmed their affinity. Erigeron acer is a highly variable species complex, as reflected in the number of subspecies and segregate species variously recognised by taxonomists. Plants varied in overall size from ~7 to ~70 cm, annual to perennial life cycle, colouration of stem and phyllaries from green across brown to deep purple, and indumentum type and density from subglabrous to glandular and/or hispid. Crucially, however, the morphology of Australian E. conyzoides fell into the variation exhibited by E. acer. Except for the relatively small size of the plants, a specimen of E. acer subsp. politus that was available to us (G. Samuelson s.n., CANB 130739) was most similar to specimens of E. conyzoides in phyllary and stem colouration and indumentum (Fig. 3).
(a) Phyllaries of Erigeron conyzoides. (b) Phyllaries of E. acer subsp. politus. (c) Stem of E. conyzoides. (d) Stem of E. acer subsp. politus. A.N. Schmidt-Lebuhn 2138 (CANB 997869) (a, c). G. Samuelson s.n. (CANB 130739) (b, d). Scale bars indicate 1 mm. Note matching indumentum and purple colouration of phyllaries and stem.

Images of living specimens of E. acer subsp. politus present a match both to stem and phyllary colour of E. conyzoides and to its frequent occurrence on rocky substrate (iNaturalist taxon ID 235867 and iNaturalist observations 35498956, 38471468, and 28487382; Fig. 2b). However, they differ from E. conyzoides as observed in the field in having considerably fewer cauline leaves.
ITS phylogeny
ITS sequence data did not provide sufficient resolution to place Erigeron conyzoides confidently either within or outside of E. acer. The samples of E. conyzoides were placed in a clade containing all samples of E. acer but also those of several related species (Supplementary Fig. S1). Within this clade, all except one were placed in the basal polytomy of a subclade comprising most sequences of E. acer, E. altaicus Popov, E. atticus Vill., E. borealis (Vierh.) Simmons, E. elongatus (a synonym of E. acer subsp. politus), most sequences of E. thunbergii A.Gray, and one sequence of E. uniflorus L. The sixth sample of E. conyzoides was placed in a subclade of sequences mostly of E. acer and E. elongatus.
Target-sequence capture phylogeny
Mapped genes from HybPiper averaged 312 of 353 in the bait kit (n = 34, s.d. = 64), with on-target reads ranging from 1.4% to 37.4%, with an average of 15.4% (s.d. = 12.4%) (Table S2). This translates to the percentage of on-target base-pairs recovered ranging from 0.13% (sample excluded) to 80.7%, with an average of 36.3% (s.d. = 25.7%) (Table S2).
The phylogeny of available target-sequence data for species of Erigeron showed E. conyzoides and E. acer forming one clade with maximum support (100 UFBoot and 100 SH-aLRT; Fig. 4a). Within that clade, samples of both species were intermingled without any discernible geographic or population structure, implying that the two taxa cannot be genetically distinguished with these data. The same pattern was shown by coalescent analysis with both ASTRAL-Pro (Fig. S2) and ASTRAL (Fig. S3), despite several samples with <10% on-target base pairs (Erigeron_acer_CANB340701, Erigeron_acris_CANB361138, Erigeron_conyzoides_SL2138B, Erigeron_conyzoides_SL2138G, Erigeron_conyzoides_SL2138H, and Erigeron_conyzoides_SL2138M).
(a) Phylogenetic tree based on concatenated analysis (260,349 total sites with 15.5% missing data and 332 partitions) of target-sequence capture data (Angiosperms353) provides evidence that Erigeron conyzoides (purple) is not distinct from E. acer (teal). Branch support values are from 10,000 replicates for SH-aLRT and UFboot. Branch lengths represent estimated nucleotide substitutions per site. (b) Principal-component analysis (PCA; 19,508 single-nucleotide polymorphisms) also demonstrated the lack of distinction between E. conyzoides and E. acer.
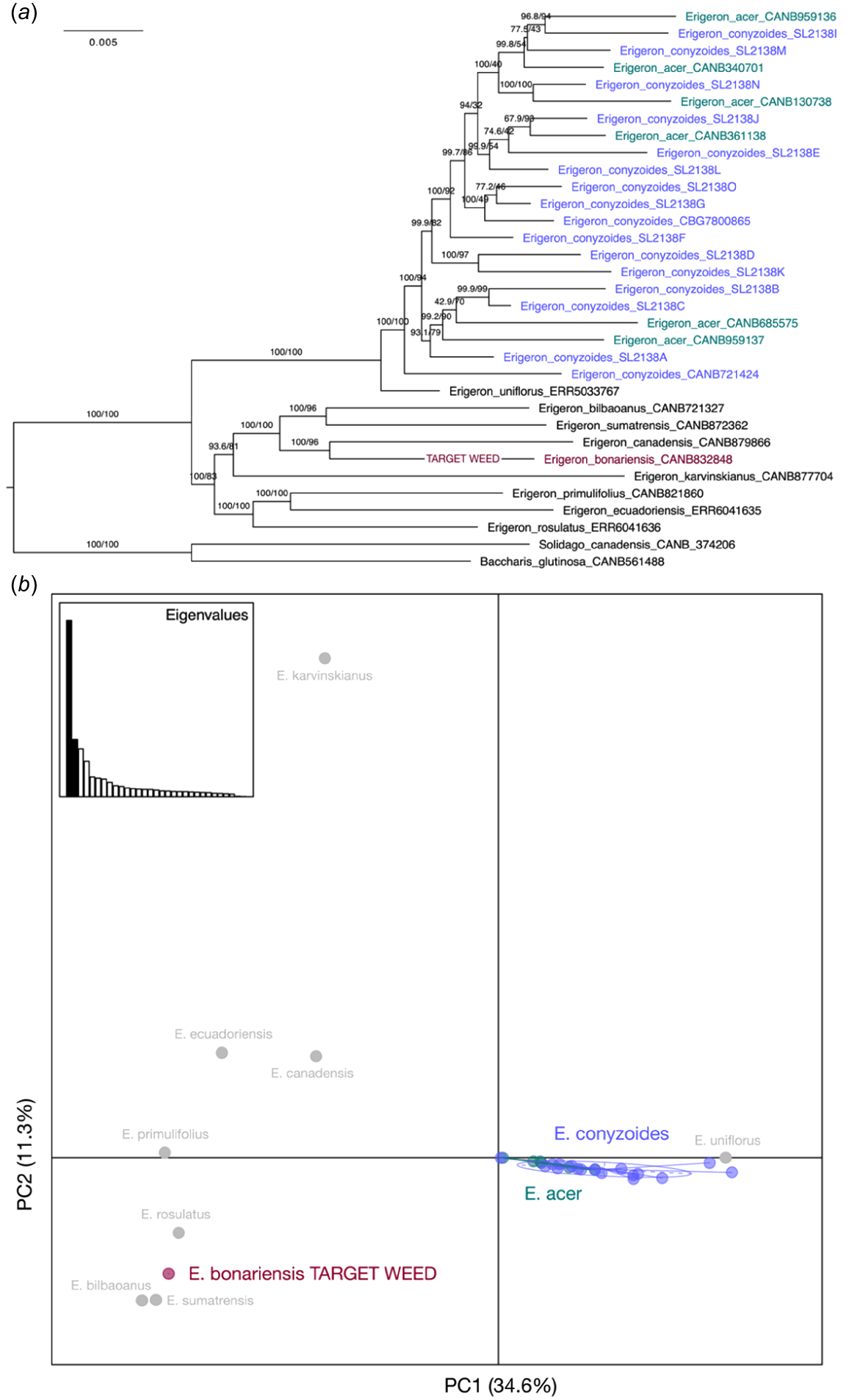
Mapping the sequence data for 32 individuals to the E. canadensis reference genome (Laforest et al. 2020) yielded 7,243,408 sites in the raw VCF file, and the mean depth of coverage for each site was 3.98×, and the mean depth for individuals was 8.31× (s.d. = 4.11). Following filtering, 19,508 sites remained. The result from the phylogeny is corroborated by the PCA where E. conyzoides and E. acer are indistinct (Fig. 4b). The first two PCs explained 34.6% and 11.6% of the variance respectively.
We did not undertake further population analyses nor calculate population statistics due to the poor quality of the data. The average missingness of the filtered VCF across 32 samples was 52% (s.d. = 30%). Further, data quality was particularly poor for the ingroup with 79% (s.d. = 13%) missingness for E. acer (n = 6) and 63% (s.d. = 20%) missingness for E. conyzoides (n = 17).
Discussion
Is Erigeron conyzoides a distinct species?
Several independent lines of evidence suggest that Erigeron conyzoides is conspecific with E. acer in its broader circumscription, i.e. unless adopting a splitting approach to the E. acer complex as in the Flora of the USSR (Schischkin 1999). Although ITS sequences do not provide sufficient phylogenetic resolution and branch support to place E. conyzoides with high confidence (aLRT < 50), its samples were placed in the same clade as those of E. acer and some of its synonyms such as E. elongatus. Target capture data from hundreds of low-copy nuclear genes showed that E. conyzoides formed a clade with E. acer samples from various locations in the northern hemisphere with maximum (100%) branch support, and both species were intermingled in that clade.
The intermingling of geographically dispersed specimens in the E. acer clade suggests that these protein-coding, slowly evolving genes are likely to not provide sufficient phylogenetic support within this species complex to resolve subspecies or geographic provenance, and, indeed, support within the E. acer–E. conyzoides clade is often low for a dataset of tens of thousands of nucleotide characters. Placement at the shallowest taxonomic levels will therefore have to depend on morphological characters as discussed below, or a future combination of broader sampling of E. acer across the northern hemisphere and a more finely resolving genetic approach, for example, genotyping-by-sequencing approaches such as restriction site-associated DNA sequencing (RAD-seq) (Narum et al. 2013). The data obtained from mapping raw capture sequencing to the E. canadensis reference genome (Laforest et al. 2020) and a method specifically devised for Angiosperm353 data (Slimp et al. 2021) were insufficient for understanding genetic diversity and divergence between lineages because of the high missingness, sequencing bias, and the limited number of single-nucleotide polymorphisms (SNPs) produced. For now, our conclusions regarding infraspecific taxonomy are based on morphology.
Erigeron conyzoides shows strong morphological affinity with E. acer. The two species share overall growth form, habit, and inflorescence structure, leaf arrangement, shape, and size, phyllary shape and number of phyllary rows, trimorphic florets, and ray florets narrow and in many rows. Although E. conyzoides keys out to E. acer subsp. politus in older floras, its subspecific placement within the E. acer complex is less clear. Two recent, regional taxonomic revisions, one for Fennoscandia (Olander and Tyler 2017) and one for Murmansk, Russia (Sennikov and Kozhin 2023), circumscribed the subspecies or, in Russia, species, more narrowly, with E. acer subsp. politus being characterised by fewer cauline leaves and a more corymbose capitulescence than for E. conyzoides. However, none of the taxonomic entities recognised in the complex by these two treatments matches the character combination exhibited by E. conyzoides, i.e. numerous cauline leaves, more or less racemose appearance of the capitulescence, phyllaries hairy on their entire length, stem and phyllaries intensely purple, and ray florets pure-white when young (Table 1).
| Taxon | Stem colour | Cauline leaves under capitulescence | Appearance of paniculate capitulescence | Phyllary indumentum | Phyllary colour | Phyllary length (mm) | Ray colour | |
|---|---|---|---|---|---|---|---|---|
| Erigeron acer L. A | Green to intensely purple | 5–10! | ±Corymbose! | Dense, on whole length | Green or purple at tips! | 6.0–7.5 | Pale pink! | |
| E. acer L. subsp. acer B | ? | (1–)2–8(−17) | ? | Glandular and eglandular, dense, on whole length | Medium-green to dark purplish-brown | (3.5–)4.3–5.5(−6.0) | Purplish! | |
| E. brachycephalus H.Lindb. A | Purple | 8–14 | ±Racemose | Moderate | Slightly to moderately purple | 5.5–6.0 | Pink! | |
| E. conyzoides F. Muell. | Purple | 8–18 | ±Racemose | Glandular and eglandular, dense, on whole length | Purple | 4–8 | Pure white (light purple when senescing) | |
| E. acer subsp. decoloratus (H.Lindb.) Hiitonen B | ? | 3–5(−6)! | ? | Glandular, dense, on whole length | Pale green! | 4.5–5.5 | Pure white | |
| E. droebachiensis O.F.Müll. A | Green or slightly purple! | 12–20 | ±Racemose | Sparse, only basal part! | Slightly or moderately purple | 5.5–6.0 | Pink! | |
| E. acer subsp. droebachiensis (O.F.Müll.) Arcang B | ? | (2–)4–11(−15) | ? | Glandular and eglandular, dense, on whole length | Pale to medium green or rarely brownish or pinkish! | (3.5–)3.8–4.6(−5.0) | Whitish to purplish | |
| E. politus Fr. A | Intensely purple to green | 3–8! | ±Corymbose! | Sparse, only basal part, or subglabrous! | Purple | 6.0–7.5 | Dark lilac to pale pinkish! | |
| E. acer subsp. politus (Fr.) H.Lindb. B | ? | (2–)3–7(−11)! | ? | Glandular and eglandular, dense, on whole length | Medium green to dark purplish-brownish | (4.5–)4.9–5.9(−6.5) | Purplish, rarely pure white | |
| E. rigidus Fr. A | Purple | 4–8(−12)! | ±Corymbose! | Dense, on whole length | Purple | 6.0–7.5 | Lilac! | |
| E. uralensis Less. A | Purple | 8–12 | ±Racemose | Sparse | Slightly to moderately purple | 5.5–6.0 | Pink! |
Therefore, we conclude that Erigeron conyzoides is not very likely to be a distinct species endemic to Australia, but a disjunct population of E. acer, and also that its placement at subspecies level must await a comprehensive revision of the E. acer complex across its entire range.
Is Erigeron conyzoides native to Australia?
The lack of morphological and genetic divergence of Australian Erigeron conyzoides from E. acer of the northern hemisphere suggests very recent dispersal to Australia. However, to decide on its native or introduced status, the question arises whether the dispersal occurred during European colonisation of Australia in the past ~200 years, or naturally, for example, via seeds attached to a migratory bird, in the last tens or at most hundreds of thousands of years.
Under the hypothesis of European introduction, the potentially most surprising observation is the presence of Erigeron conyzoides as early as 1854 in the two different locations where Ferdinand Mueller found it according to his publication notes (‘sources of the Murray and Snowy Rivers’) (Mueller 1855). Although this would require earlier introduction and spread than has been documented in many other introduced species (Kloot 1987), it is plausible in the light of how quickly European graziers reached, and thus likely affected, the mountains of Australia.
Graziers brought livestock into the mountain areas by the early 1820s (The Australian Alps National Parks 2013), and colonists visited alpine areas in the 1830s (Costin et al. 2000). A well-referenced study of the history of human impacts in the Monaro Region, which it defines to include the alpine areas of New South Wales, provides more detail (Hancock 1972). ‘Squatting runs’ had been well-established across most of the region by 1848–1850, including in montane areas such as Jindabyne, Adaminaby, and Cooma. Alpine areas were not permanently occupied but primarily used when lower elevations experienced droughts, but as early as 1834 a grazier lost livestock to a surprise blizzard at what is now Kiandra in Kosciuszko National Park. Another grazier established outstations at ‘the headwaters of the snow-fed rivers’ west of the Goodradigbee River from 1839. All of this implies that by the time Mueller found Erigeron conyzoides, the alpine landscape had experienced at least 20 years of grazing impacts and the possibility of introduction of weeds through contaminated equipment or produce.
Most early colonists were British or Irish, and a complicating factor is that subspecies showing ecological and morphological affinities with Erigeron conyzoides, such as E. acer subsp. droebachiensis and E. acer subsp. politus, do not occur in the British Isles. However, they occur in Scandinavia, the European Alps, and Russia, so that immigrants from these areas could be potential vectors of introduction, as could be products imported from them. A precedent for a weed introduction early in British colonisation but from a source region other than the British Empire is Gypsophila tubulosa (Jaub. & Spach) Boiss. (Caryophyllaceae). The species is native to western Turkey, but was first collected in Australia in 1843 and was found by Mueller at Mount Timbertop (Victoria, 1259 masl) by 1853, a year before he encountered E. conyzoides.
It may also be asked why Mueller, a European immigrant to Australia, would not recognise a northern hemisphere species and instead described it as an Australian species. Even though the subspecific identity of the Australian populations remains unresolved, their ecology suggests that it would likely have to be an alpine and/or subarctic subspecies, and Mueller grew up in temperate, lowland areas of northern Germany in what are now the states of Mecklenburg-Vorpommern and Schleswig-Holstein. It is thus plausible that he would not have been familiar with the northern hemisphere source populations of Erigeron conyzoides.
The decline of Erigeron conyzoides in recent decades is another important piece of the puzzle. The species has not been collected in New South Wales since 1978, but has persisted in Victoria (Fig. 1b). This parallels the removal of livestock from the Alps in both states, which happened much earlier in New South Wales. Grazing was removed from the alpine areas of Kosciuszko National Park in 1958 (Costin et al. 2000), but only much later from many parts of the Victorian Alps, such as from the Howitt Plains in 1988 and from the Bogong High Plains in 1991 (The Australian Alps National Parks 2013).
This pattern of decline suggests that in Australia, Erigeron conyzoides may be an opportunistic, disturbance-dependent weed. Further evidence for this proposition is some collectors’ notes on its habitat; the species has often been collected in uncompetitive habitats such as rocky slopes or boulder fields (B.G. Briggs 2547, NSW 569129; G.W. Carr s.n., MEL 0670729A; N.G. Walsh 2430, CANB 721424, HO 562220, MEL 2339440) but also on a weedy road verge (N.G. Walsh 4720, MEL 2044671A) and even a playground (Rogers s.n., MEL 600723). The population we were able to study in the field occupied an engineered structure, a dam, and edges of a creek as part of a community of other weeds, but was absent from undisturbed native vegetation in the surrounding area.
In summary, Erigeron conyzoides appears dependent on low-competition and/or anthropogenic habitats and has been declining in parallel with the recovery of native vegetation since the removal of grazing from alpine environments in Australia. This behaviour is consistent with the hypothesis that it is a European introduction.
Conclusions
These findings have several practical implications. An understanding of the origins of E. conyzoides means that the risk of off-target damage for a flaxleaf fleabane biocontrol program in Australia is reduced. The next-closest native species, namely the clades of Brachyscominae and Lagenophorinae and Celmisia with part of Olearia within the subtribe Celmisiinae, are much more distantly related to flaxleaf fleabane than is E. conyzoides (Chen et al. 2024).
Previous efforts to source E. conyzoides for host-specificity testing had been unsuccessful. It is now available to the wider research community because seeds were deposited at the Victorian Conservation Seedbank and the National Seed Bank. On the basis of the molecular and morphological findings presented in this paper, we consider Erigeron conyzoides to be a synonym of Erigeron acer and are preparing a separate paper to address taxonomic and nomenclatural issues.
Data availability
The raw sequence data newly generated in this study are available in Sequence Read Archive (SRA) under BioProject ID PRJNA1111666. Phylogenetic trees, the filtered VCF file, and historical records used for plotting are available on the CSIRO Data Access Portal (https://doi.org/10.25919/cxd2–3902).
Conflicts of interest
Ben Gooden is an Associate Editor of Australian Journal of Botany. To mitigate this potential conflict of interest they had no editor-level access to this manuscript during peer review. The authors have no further conflicts of interest to declare.
Declaration of funding
Sequencing was supported by Bioplatforms Australia through the Environomics Future Science Platform (CSIRO).
Author contributions
Stephanie Chen: conceptualisation, data curation, formal analysis, software, writing – original draft, writing – review and editing. Alicia Grealy: data curation, investigation, writing – review and editing. Michelle Rafter: writing – review and editing. Ben Gooden: conceptualisation, writing – review and editing. Alexander Schmidt-Lebuhn: conceptualisation, data curation, formal analysis, software, writing – original draft, writing – review and editing.
Acknowledgements
We are grateful to the Victorian government Department of Energy, Environment, and Climate Action for authorisation to sample protected flora, the New South Wales Government Department of Planning, Industry and Environment for a Scientific Licence (SL100750), Parks Victoria for an access agreement, to Juanna Durrant (Victoria), Philip Gleeson (NSW), Henri Maxwell (NSW), and Elaine Thomas (Victoria) for helpful communications and advice regarding field work, to Laura Swiderski (National Seed Bank) and Rebecca Miller (Royal Botanic Gardens Victoria) for seed bagging and collection, and to Igor Pospelov (Institute of Ecology and Evolution of the Russian Academy of Sciences) for permission to reproduce an image. Sequencing was performed by the Biomolecular Resource Facility (BRF) at the John Curtin School of Medical Research, Australian National University, Canberra, Australia.
References
Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Systematic Biology 55, 539-552.
| Crossref | Google Scholar | PubMed |
Bánki O, Roskov Y, Döring M, Ower G, Hernández Robles DR, Plata Corredor CA, Stjernegaard Jeppesen T, Örn A, Vandepitte L, Hobern D, Schalk P, DeWalt RE, Keping M, Miller J, Orrell T, Aalbu R, Abbott J, Adlard R, Adriaenssens EM, et al. (2023) Catalogue of Life Checklist (Annual Checklist 2023). Catalogue of Life, Leiden, Netherlands. Available at https://doi.org/10.48580/dfsr
Barton J (2004) How good are we at predicting the field host-range of fungal pathogens used for classical biological control of weeds? Biological Control 31, 99-122.
| Crossref | Google Scholar |
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120.
| Crossref | Google Scholar | PubMed |
Briese DT (2005) Translating host-specificity test results into the real world: the need to harmonize the yin and yang of current testing procedures. Biological Control 35, 208-214.
| Crossref | Google Scholar |
Bushnell B (2022) BBtools. Available at sourceforge.net/projects/bbmap/
Chen SH, Gooden B, Rafter MA, Hunter GC, Grealy A, Knerr N, Schmidt-Lebuhn AN (2024) Phylogenomics-driven host test list selection for weed biological control. Biological Control 193, 105529.
| Crossref | Google Scholar |
Chernomor O, von Haeseler A, Minh BQ (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65, 997-1008.
| Crossref | Google Scholar | PubMed |
Costin AB, Gray M, Totterdell CJ, Wimbush DJ (2000) ‘Kosciuszko Alpine Flora.’ (CSIRO Publishing) Available at https://books.google.com.au/books?id=dqvyql9N54cC
Council of Heads of Australasian Herbaria (2024) Australian Plant Census IBIS database. Available at https://biodiversity.org.au/nsl/services/search/taxonomy [Accessed 5 July 2024]
Cullen JM, Sheppard AW, Raghu S (2022) Effectiveness of classical weed biological control agents released in Australia. Biological Control 166, 104835.
| Crossref | Google Scholar |
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27, 2156-2158.
| Crossref | Google Scholar | PubMed |
Department of Energy, Environment and Climate Action (2024) Flora and Fauna Guarantee Act 1988 - Threatened List. Victoria State Government: Melbourne, Vic, Australia. Available at https://www.environment.vic.gov.au/__data/assets/pdf_file/0036/698571/FFG_Threatened_List_February_2024.pdf
Fowler SV, Groenteman R, Paynter Q (2024) The highs and the lows: a cost benefit analysis of classical weed biocontrol in New Zealand. BioControl 69, 253-267.
| Crossref | Google Scholar |
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. arXiv Preprint arXiv:12073907. 10.48550/arXiv.1207.3907
Garrison E, Kronenberg ZN, Dawson ET, Pedersen BS, Prins P (2022) A spectrum of free software tools for processing the VCF variant call format: vcflib, bio-vcf, cyvcf2, hts-nim and slivar. PLoS Computational Biology 18, e1009123.
| Crossref | Google Scholar | PubMed |
Gilbert GS, Magarey R, Suiter K, Webb CO (2012) Evolutionary tools for phytosanitary risk analysis: phylogenetic signal as a predictor of host range of plant pests and pathogens. Evolutionary Applications 5, 869-878.
| Crossref | Google Scholar | PubMed |
Gildenhuys E, Ellis A, Carroll S, Roux JL (2013) The ecology, biogeography, history and future of two globally important weeds: Cardiospermum halicacabum Linn. and C. grandiflorum Sw. NeoBiota 19, 45-65.
| Crossref | Google Scholar |
Gildenhuys E, Ellis AG, Carroll SP, Le Roux JJ (2015) Combining natal range distributions and phylogeny to resolve biogeographic uncertainties in balloon vines (Cardiospermum, Sapindaceae). Diversity and Distributions 21, 163-174.
| Crossref | Google Scholar |
Gooden B (2023) Advances in the biological control of flaxleaf fleabane with a novel rust fungus. GRDC Update Paper RDC1607-003OPX. Grains Research and Development Corporation. Available at https://grdc.com.au/resources-and-publications/grdc-update-papers/tab-content/grdc-update-papers/2023/02/advances-in-the-biological-control-of-flaxleaf-fleabane-with-a-novel-rust-fungus
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59, 307-321.
| Crossref | Google Scholar | PubMed |
Ham K (2013) OpenRefine (version 2.5). http://openrefine.org. Free, open-source tool for cleaning and transforming data. Journal of the Medical Library Association 101, 233-234.
| Crossref | Google Scholar |
Hancock WK (1972) ‘Discovering Monaro: a study of man’s impact on his environment.’ (Cambridge University Press) Available at https://books.google.com.au/books?id=pPu_UPKoUkYC
Hinz HL, Winston RL, Schwarzländer M (2020) A global review of target impact and direct nontarget effects of classical weed biological control. Current Opinion in Insect Science 38, 48-54.
| Crossref | Google Scholar | PubMed |
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35, 518-522.
| Crossref | Google Scholar | PubMed |
Jackson C, McLay T, Schmidt-Lebuhn AN (2023) hybpiper-nf and paragone-nf: containerization and additional options for target capture assembly and paralog resolution. Applications in Plant Sciences 11, e11532.
| Crossref | Google Scholar | PubMed |
Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, Zerega NJC, Wickett NJ (2016) HybPiper: Extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Applications in Plant Sciences 4, apps.1600016.
| Crossref | Google Scholar |
Johnson MG, Pokorny L, Dodsworth S, Botigué LR, Cowan RS, Devault A, Eiserhardt WL, Epitawalage N, Forest F, Kim JT, Leebens-Mack JH, Leitch IJ, Maurin O, Soltis DE, Soltis PS, Wong GK, Baker WJ, Wickett NJ (2019) A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Systematic Biology 68, 594-606.
| Crossref | Google Scholar | PubMed |
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403-1405.
| Crossref | Google Scholar | PubMed |
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587-589.
| Crossref | Google Scholar | PubMed |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772-780.
| Crossref | Google Scholar | PubMed |
Kloot PM (1987) The naturalised flora of South Australia 2. Its development through time. Journal of the Adelaide Botanic Garden 10, 91-98.
| Google Scholar |
Laforest M, Martin SL, Bisaillon K, Soufiane B, Meloche S, Page E (2020) A chromosome-scale draft sequence of the Canada fleabane genome. Pest Management Science 76, 2158-2169.
| Crossref | Google Scholar | PubMed |
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv:13033997. 10.48550/arXiv.1303.3997
McLay TGB, Birch JL, Gunn BF, Ning W, Tate JA, Nauheimer L, Joyce EM, Simpson L, Schmidt-Lebuhn AN, Baker WJ, Forest F, Jackson CJ (2021) New targets acquired: Improving locus recovery from the Angiosperms353 probe set. Applications in Plant Sciences 9, e11420.
| Crossref | Google Scholar |
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37, 1530-1534.
| Crossref | Google Scholar | PubMed |
Mueller F (1855) Descriptive characters of new alpine plants, from continental Australia. Transactions of the Philosophical Society of Victoria 1, 96-111.
| Google Scholar |
Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA (2013) Genotyping-by-sequencing in ecological and conservation genomics. Molecular Ecology 22, 2841-2847.
| Crossref | Google Scholar | PubMed |
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32, 268-274.
| Crossref | Google Scholar | PubMed |
Olander S, Tyler T (2017) Morphometrics and taxonomy of Erigeron acris sensu lato (Asteraceae) in Fennoscandia. New Journal of Botany 7, 39-50.
| Crossref | Google Scholar |
Paterson ID, Motitsoe SN, Coetzee JA, Hill MP (2024) Recent post-release evaluations of weed biocontrol programmes in South Africa: a summary of what has been achieved and what can be improved. BioControl 69, 279-291.
| Crossref | Google Scholar |
Pyšek P, Jarošík V, Hulme PE, Pergl J, Hejda M, Schaffner U, Vilà M (2012) A global assessment of invasive plant impacts on resident species, communities and ecosystems: the interaction of impact measures, invading species’ traits and environment. Global Change Biology 18, 1725-1737.
| Crossref | Google Scholar |
R Core Team (2024) R: A Language and Environment for Statistical Computing. Available at https://www.R-project.org/
Schwarzländer M, Hinz HL, Winston RL, Day MD (2018) Biological control of weeds: an analysis of introductions, rates of establishment and estimates of success, worldwide. BioControl 63, 319-331.
| Crossref | Google Scholar |
Scott LJ, Congdon BC, Playford J (1998) Molecular evidence that fireweed (Senecio madagascariensis, Asteraceae) is of South African origin. Plant Systematics and Evolution 213, 251-257.
| Crossref | Google Scholar |
Sennikov AN, Kozhin MN (2023) Taxonomic revision of the Erigeron acris group (Asteraceae) in Murmansk Region, Russia, reveals a complex pattern of native and alien taxa. PhytoKeys 235, 83-128.
| Crossref | Google Scholar | PubMed |
Sheppard AW, Heard TA, Briese DT (2002) What is needed to improve the selection, testing and evaluation of weed biological control agents: workshop synthesis and recommendations. In ‘Proceedings of the CRC Australian Weed Management Biological Control Weeds Symposium Workshop’, Perth, WA, Australia. pp. 87–100. (Weeds CRC: Perth, WA, Australia)
Slimp M, Williams LD, Hale H, Johnson MG (2021) On the potential of Angiosperms353 for population genomic studies. Applications in Plant Sciences 9, e11419.
| Crossref | Google Scholar |
Solbrig OT (1962) The South American Species of Erigeron. Contributions from the Gray Herbarium of Harvard University 191, 3-79.
| Crossref | Google Scholar |
Sorensen JS, Sorensen NA (1969) Studies related to naturally occurring acetylene compounds. XXXV. Investigation of Erigeron spp. from the Australian mountains and Tasmania. Australian Journal of Chemistry 22, 751-760.
| Crossref | Google Scholar |
The Australian Alps National Parks (2013) Grazing in the Australian Alps. Available at https://theaustralianalpsnationalparks.org/wp-content/uploads/2013/11/grazing.pdf
Tutin TG (1976) ‘Flora Europaea: Vol. 4. Plantaginaceae to Compositae (and Rubiaceae).’ (Cambridge University Press) Available at https://books.google.com.au/books?id=QXRooltqAVMC
Wapshere AJ (1974) A strategy for evaluating the safety of organisms for biological weed control. Annals of Applied Biology 77, 201-211.
| Crossref | Google Scholar |
Westgate M, Stevenson M, Kellie D, Newman P (2024) galah: Biodiversity Data from the GBIF Node Network. R package version 2.1.0. Available at https://CRAN.R-project.org/package=galah
Wu H (2007) The biology of Australian weeds 49. Conyza bonariensis (L.) Cronquist. Plant Protection Quarterly 22, 122-131.
| Google Scholar |
Yang Y, Smith SA (2014) Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Molecular Biology and Evolution 31, 3081-3092.
| Crossref | Google Scholar | PubMed |
Zhang C, Rabiee M, Sayyari E, Mirarab S (2018) ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19, 153.
| Crossref | Google Scholar | PubMed |
Zhang C, Scornavacca C, Molloy EK, Mirarab S (2020) ASTRAL-Pro: quartet-based species-tree inference despite paralogy. Molecular Biology and Evolution 37, 3292-3307.
| Crossref | Google Scholar | PubMed |


