Maintaining separate maternal lines increases the value and applications of seed collections
Marlien M. van der Merwe A * , Jason G. Bragg A B , Richard Dimon
A * , Jason G. Bragg A B , Richard Dimon  A C , Patrick S. Fahey
A C , Patrick S. Fahey  A C , Patricia M. Hogbin A , Patricia Lu-Irving A , Allison A. Mertin A D , Maurizio Rossetto A C , Trevor C. Wilson A E and Jia-Yee Samantha Yap A
A C , Patricia M. Hogbin A , Patricia Lu-Irving A , Allison A. Mertin A D , Maurizio Rossetto A C , Trevor C. Wilson A E and Jia-Yee Samantha Yap A
A Research Centre for Ecosystem Resilience, Botanic Gardens of Sydney, Sydney, NSW 2000, Australia.
B School of Biological, Earth and Environmental Sciences, University of New South Wales, Sydney, NSW 2033, Australia.
C Queensland Alliance of Agriculture and Food Innovation, University of Queensland, St Lucia, Qld 4067, Australia.
D School of BioSciences, University of Melbourne, Parkville, Vic. 3010, Australia.
E Plant Discovery and Evolution, Botanic Gardens of Sydney, Sydney, NSW 2000, Australia.
Handling Editor: Catherine Offord
Australian Journal of Botany - https://doi.org/10.1071/BT22136
Submitted: 9 December 2022 Accepted: 6 July 2023 Published online: 3 August 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: Given the effort and resources that go into collecting and maintaining seed collections, it is crucial that we maximise their usefulness. Conservation, restoration and research rely heavily on good quality collections in order to establish new populations, create habitat, minimise extinction and address scientific questions.
Aims: Although seed viability, excellent metadata and genetic representativeness make for good quality collections, we provide 10 detailed reasons why the maintenance of separate maternal lines further increases the quality and usefulness of seed collections.
Key results: Maternal line seed collections can accommodate new information, this is especially important given the increasing longevity of seed collections. For example, maintaining separate maternal lines facilitates accommodation of taxonomic changes, minimises the impact of erroneous plant identifications, and facilitates separation of polyploid races, hybrids and inappropriate lineages. Separate maternal line collections also facilitate better estimates of the genetic diversity captured, and consequently better inform conservation translocations and the establishment of conservation gardens and seed orchards. Separate maternal line collections can also expedite breeding for specific traits, such as disease resistance or other selective challenges that impact on biodiversity conservation. New seed microbiome data show how only some maternal lines contain pathogenic fungi, reminding seed collectors and collections managers that contamination can be better contained by keeping each maternal line separate.
Conclusions and implications: Maintaining separate maternal lines is a simple and effective way to increase the value of seed collections for multiple applications.
Keywords: biodiversity, breeding, conservation collections, disease management, germplasm collections, hybridisation, inbreeding, kinship, restoration ecology, seed research, population genetics, taxonomy, translocations.
Introduction
With increasing environmental pressures on many native plant ecosystems and the looming extinction crisis, ex situ collections of plant material are becoming increasingly valuable. Seed banking is one of the most cost-effective means of ex situ storage (Engels and Engelmann 1998; Li and Pritchard 2009) and apart from providing some level of long-term security, good quality seed collections have multiple applications, including plant propagation for restoration (León-Lobos et al. 2012) and conservation (O’Donnell and Sharrock 2017; Chapman et al. 2019; White et al. 2023), breeding programs (Byrne et al. 2018), horticulture (Li et al. 2016), and research (Sommerville et al. 2013; Everingham et al. 2021; Rauschkolb et al. 2022). Sourcing seed from wild populations can be time consuming and often require multiple visits to remote areas for each accession (Griffith et al. 2015). Seed harvesting itself can also have a detrimental effect on wild populations, especially for rare species (Bucharova et al. 2023). As long-term storage is one of the principal attractions of seed banking, particularly for state-of-the-art facilities, collections should be of the highest possible quality, fit for multiple purposes, and retain potential for future, perhaps even unanticipated, applications.
So, what attributes maximise long-term value and utility of a seed collection? Seed viability is of utmost importance and there are many guidelines available on how to collect, process and store seed to maintain viability (such as Martyn Yenson et al. 2021). However, it can be argued (Ellis et al. 1985) that even seed collections with 100% viability lose much of their value if they are not backed by good quality metadata (see Crawford et al. (2021) for details on quality metadata). It is increasingly accepted that collections that capture a good representation of the genetic diversity in a species best facilitate long-term conservation and restoration objectives through increased adaptive capacity (Reed and Frankham 2003; Hoffmann and Sgrò 2011; Raven et al. 2013; Prober et al. 2019). Here, we argue that in addition to genetically representative, viable seed with good metadata, seed collections that maintain separate maternal lines are better suited to multiple downstream applications, are more amenable to taxonomic changes and enable a more accurate understanding of the level of genetic diversity in a collection.
We present 10 reasons for the maintenance of separate maternal lines in seed collections. Supporting examples, from our original research and from the published literature, are provided to illustrate the argument behind the reasons.
Reason 1: Taxonomic changes can be accommodated
Taxonomy is the science of classifying, describing and naming organisms based on all available information (Godfray 2002). The naming of a species (nomenclature) is a hypothesis based on the interpretation of the available information used by a taxonomist to describe the species. Ideally, names represent distinct evolutionary lineages (Nelson 1971; Mayr and Bock 2002). The available information can differ vastly between groups of organisms and across time. Some groups of organisms have been studied intensely, leading to a large amount of information available across their geographic distribution, including in-depth morphological characterisations, biochemical studies, phylogenetic data, population genetic data, and whole genome sequences (Hao et al. 2022). There are still many other groups that lack this level of information and consequently have species that are described from only a handful of characters that a taxonomist deems adequate to delineate diverged taxonomic units. As new information becomes available the taxonomic status of a lineage can change, leading to either the naming of new lineages (such as species) or the synonymisation of taxa. If seed collections identify separate maternal lines, their integrity can be spared from depreciation as a result of taxonomic change. Otherwise, they may be rendered meaningless when, for example, progress reveals more than one species exist at a site from where seed was pooled.
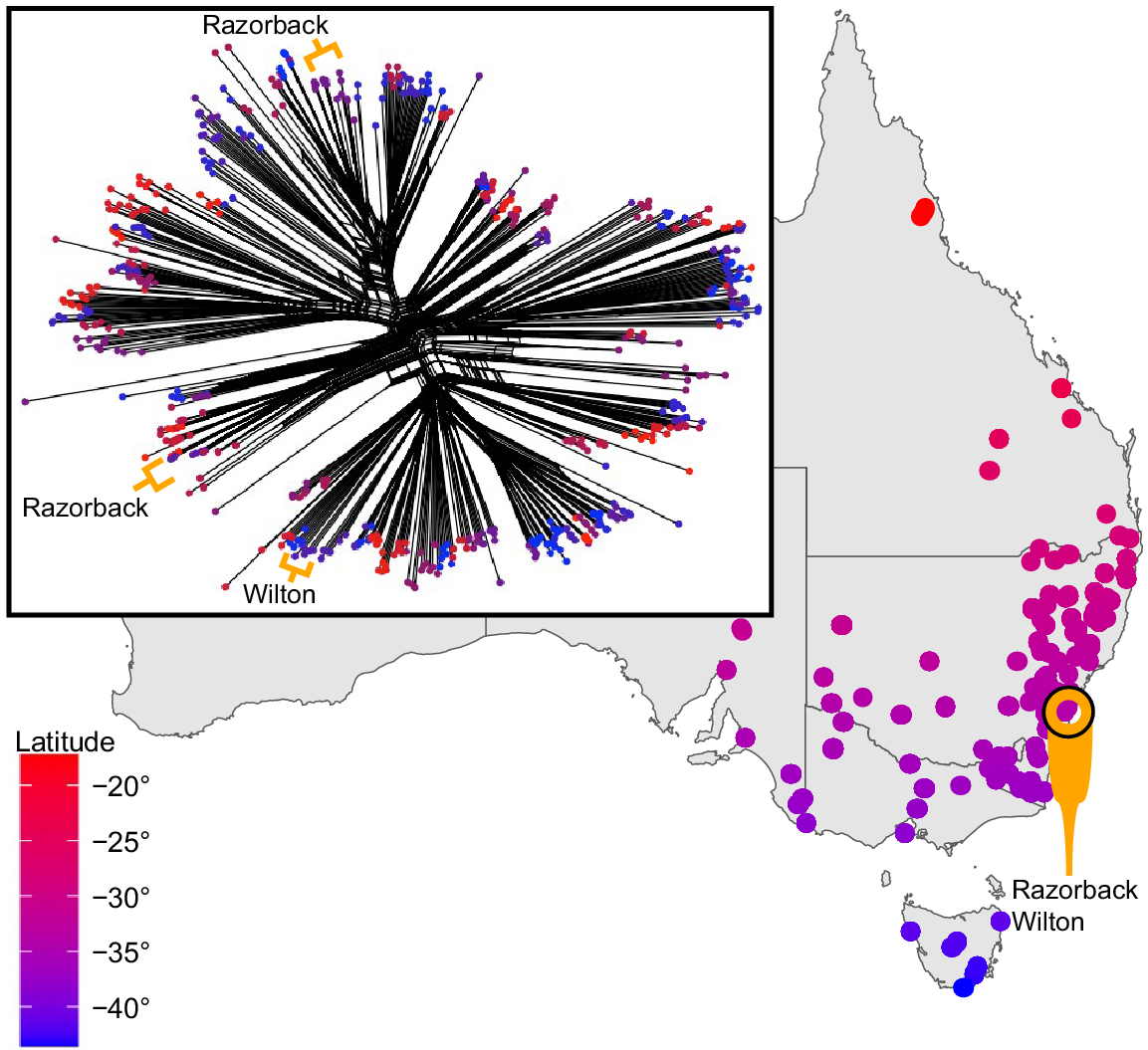
Species complexes pose a particular problem to pooled seed collections and can easily be avoided with separate maternal line collections. Here we use Ajuga australis R.Br. (Lamiaceae), the only species of its genus recognised in Australia, as a means to validate our argument. It is a perennial rosette-forming forb that generally inhabits riparian areas and drainages across a wide number of habitats on the eastern side of the Australian continent, from northern Queensland to as far as southern Tasmania (Harley et al. 2004). It is indisputably clear that A. australis is composed of several distinct morphological forms that, usually based on anecdotal examination, appear associated with a particular habitat. However, closer observation of this morphological diversity demonstrates a bewildering continuum between these morphotypes, none of which have been examined in context with genetic or environmental factors. This challenge was evident even when four Australian species were recognised (Bentham 1834, 1870; Stapf 1933) and then later synonymised until a comprehensive investigation of the taxon is made (Eichler 1965; Conn 1999). An investigation using genomic data of comprehensive sampled Australian Ajuga representing the breadth of known morphological and geographic diversity of the species (Wilson 2022) and multiple individuals from a sampling site (over 500 samples from 90 sites) indicated that there is geographical overlap in genetically distinct groups. Preliminary results from a phylogenetic network and principal component analysis shows that two recognisably distinct morphotypes collected at one site (samples within a radius of 30 m) are clearly reconciled as two distinct genetic entities (Fig. 1). Furthermore, at a site not more than 5 km away, a third morphotype was demonstrated as genetically distinct from the other two entities (Fig. 1). Such a result from a localised area suggests that care in the examination and description of this species is a priority. Consequently, seed collections lacking maternal detail for taxa associated with unresolved taxonomic groups are particularly susceptible to capturing an inaccurate representation of taxonomic lineages and genetic diversity. Ajuga australis is just one example of many species complexes awaiting detailed systematic work.
Genetic relationship and the corresponding locations of Ajuga australis sens. lat. sample sites representative of the known morphological diversity and geographic distribution in Australia. A single nucleotide polymorphism (SNP) dataset was analysed to visualise genetic relationships using the NeighborNet method employed in Splitstree v. 4.14.6 (Huson et al. 2008) to generate a network. This is useful for visualising evolutionary histories in groups with substantial reticulation (Huson and Bryant 2006). The total dataset comprised 12 321 SNPs (99.6% had a reproducibility scores ≥ 96%). The proportion of missing data for the samples was between 8.02 and 79.9%, with a mean of 17.8%. Colour is indicative of latitudinal distribution, and results show how the Razorback and Wilton sites, which are located approximately 10 km apart, consist of three genetic profiles situated in distinct areas of the genetic distribution of Ajuga australis.
Reason 2: The impact of erroneous plant identifications can be minimised
Plant identification can be difficult and many plant species can only be identified by specialists who understand the intricate variation in the morphological characters involved. Often, floral characters are the most important morphological characters differentiating between species and these are often absent at the time of seed collecting. Although best practice would be to visit a population prior to seed collecting, bagging individuals and collecting these later, resources (time and money) to follow this procedure are not available to all seed collectors.
Examples of challenging field identification in the absence of floral characters are prominent within the Orchidaceae family. Many Australian orchid genera comprise taxa that are impossible to identify without floral morphology due to similar vegetative characters and sympatric distribution, even when plants are in bud or have been recently pollinated (e.g. ‘Key to members of the Thelymitra nuda complex in Australia’ as outlined by Jeanes (2013)). This greatly increases the chance of species misidentification when in situ seed is collected without prior identification of each flowering individual.
Orchids represent over 16% of nationally listed threatened flora in Australia (Australian Government 2023), with multiple research institutes establishing dedicated orchid conservation initiatives involving seed bank storage, ex situ propagation, and reintroduction/translocation programs (see Reiter et al. (2021) as an example). Species misidentification from in situ seed collections has been observed across multiple orchid genera, including Genoplesium/Corunastylis, Diuris, Prasophyllum, Caladenia, and Thelymitra (R. Dimon, pers. obs.). Although relatively uncommon, this has resulted in downstream challenges for both historic and ongoing species-specific conservation outcomes, especially in establishing representative insurance populations for ex situ and translocation strategies. The issue most commonly arises from ex situ seed collections of threatened species growing sympatrically (and sometimes hybridising) with a population of a common species, which becomes indistinguishable post-pollination.
Orchidaceae have some of the smallest seeds of all angiosperms, producing thousands to millions of seeds per pod (Arditti and Ghani 2000), which means mature or near-dehiscing seed pods cannot be combined in the same collection bag in the field and later separated again post-collection. For times when pooled seed collection has occurred and inaccurate species identification has been recognised, every individual seedling that has been propagated under the seed accession needs to be flowered in a nursery environment before species identification and further conservation initiatives can occur. This process can take over 5 years for some orchid species and involve considerable time and nursery space, which could have been avoided by keeping separate maternal line collections. Fortunately, current practices for orchid seed collections are resolved by identifying and tagging all individuals across the population during regular surveys and accessioning individual seed pods when collected, which allows any potential mis-identified collections to be traced back to a single origin, thus allowing all other ‘bona-fide’ seed collections from a population to be utilised for their targeted conservation initiatives.
Reason 3: Facilitate the separation of polyploid races
Genome size and number of chromosomes vary among and within families and species, and this is impactful at many scales of plant evolution and ecology (Pellicer and Leitch 2019). The duplication of the genome (polyploidisation) can result in individuals with different genome sizes being unable to produce fertile offspring with each other. Polyploid races refer to intra-specific variations in genome size found within taxonomic species. For some well-studied species, experts can tell individuals with differing genome sizes apart using morphological characters. In some cases this has led to the description of new species, such as the tetraploid Stylidium dilatatum W.D.Jackson and R.J.E.Wiltshire, distinct from the broader species complexes Stylidium graminifolium Sw. ex Willd (Jackson and Wiltshire 2001). Essentially, polyploid races can be treated as different evolutionary races. However, for many species we do not know if multiple ploidy races exist and if they overlap in distribution. Genomic data is providing valuable information, but this remains restricted to species with population level data through projects such as Restore and Renew (Rossetto et al. 2019; Hogbin 2022) and preliminary work on reference genome sizes. For example, sites with mixed ploidy were identified within Allocasuarina littoralis based on reference samples provided by Phillip Rose (Rose et al. 2015).
Detailed work on Themeda triandra has characterised the distribution of various ploidy races (Hayman 1960), with the ratio of diploids to tetraploids at a site varying between zero and one (Ahrens et al. 2020). In general, the diploids and the tetraploids only rarely interbreed, and research has shown that each ploidy race has different adaptive regimes with the tetraploid outcompeting the diploid in certain environmental conditions (Godfree et al. 2017). Separate maternal line seed collections of this species will enable researchers to identify and select diploid or tetraploid individuals for specific conditions without additional detailed work.
Reason 4: Facilitate isolation of hybrid maternal lines
The term hybridisation refers to the natural crossing of two different genotypes as either intra-specific admixture (when the pollen donor and the recipient are from the same species) or inter-specific hybridisation (when the pollen donor plant and the receptor plant do not belong to the same species). Two issues present themselves during seed collections when dealing with inter-specific hybridisation, and both are easier to isolate and deal with downstream when separate maternal lines are maintained. The first is that seed from a non-hybrid or ‘pure’ mother plant may be the product of an unidentified inter-species hybridisation event. The second is that adult hybrid plants (offspring from inter-specific hybridisation) are not always easy to distinguish in the field but can often produce seed (viable or not) that can impact negatively on the quality of a pooled seed collection. The fitness of F1 (first generation) and F2 (second generation) hybrid seed can range dramatically from non-viable seed to seedlings that do not reach maturity (see the example of Senecio madagascariensis and Senecio pinnatifolius; Prentis et al. 2007), or individuals that are very vigorous (hybrid vigour) and either fertile or sterile. All of these can have negative consequences for conservation and restoration outcomes. For example, if hybrid seedlings are introduced into a population of a rare species, they may outcompete the rare species (in the case of hybrid vigour), or they may overwhelm the genetic constitution of the rare species, leading to the extinction of pure parental genomes through later generation hybrids and back crosses (Hegde et al. 2006; Muhlfeld et al. 2014; Todesco et al. 2016).
Hybridisation occurs extensively in plants; relevant examples include both naturally occurring hybrids such as Allocasuarina defungens × A. littoralis (Rose et al. 2015) and Astrotricha crassifolia × Astrotricha longifolia/Astrotricha floccosa (Warman 2018 confirmed by Yap and Rossetto 2020) and hybrids enabled by anthropogenic activity such as introductions (Senecio; Prentis et al. 2007) and habitat fragmentation (Eucalyptus aggregata, Field et al. 2008). A study on the threatened Eucalyptus tetrapleura ascertained that hybridisation with co-occurring congeners Eucalyptus siderophloia and Eucalyptus fibrosa is common in some sub-populations (Rutherford et al. 2019). Morphology did not provide clues to the origin of the hybrid individuals and only with genetic data could individuals be classified as hybrids or ‘pure’. This work highlights the dangers of genetic swamping of the pure individuals and the importance of using the correct material in management plans, particularly in translocations.
A very similar scenario was found for the threatened species Eucalyptus sp. Cattai, but here hybrids were particularly common amongst the seedlings and only a small number of adult hybrids were identified in the field (Rutherford et al. 2022). The use of separate maternal line seed collections (sampled specifically for the study) in germination trials suggested that some parents are more prone to hybridisation than others (Yap et al. 2019). This information could only be extracted from the seed because separate maternal line collections were available for the work. This work demonstrates how separate maternal line collections can help reduce the risk of introducing hybrid seedlings into both translocations and living collections maintained for conservation purposes; and also minimise kinship in translocations (see Reason number 5).
Hybrids as such are neither ‘good’ nor ‘bad’ (Hirashiki et al. 2021). Hybrids can be valuable sources of novel genotypes, have been used in agricultural breeding programs (Warschefsky et al. 2014; Labroo et al. 2021) and horticulture (Leonhardt and Criley 1999) and are potentially a powerful tool in reintroductions (Parmesan et al. 2023). Having the option to include or exclude them in research, conservation or other applications is a positive outcome from separate maternal line seed collections.
Reason 5: Isolating non-local and local provenance maternal lines
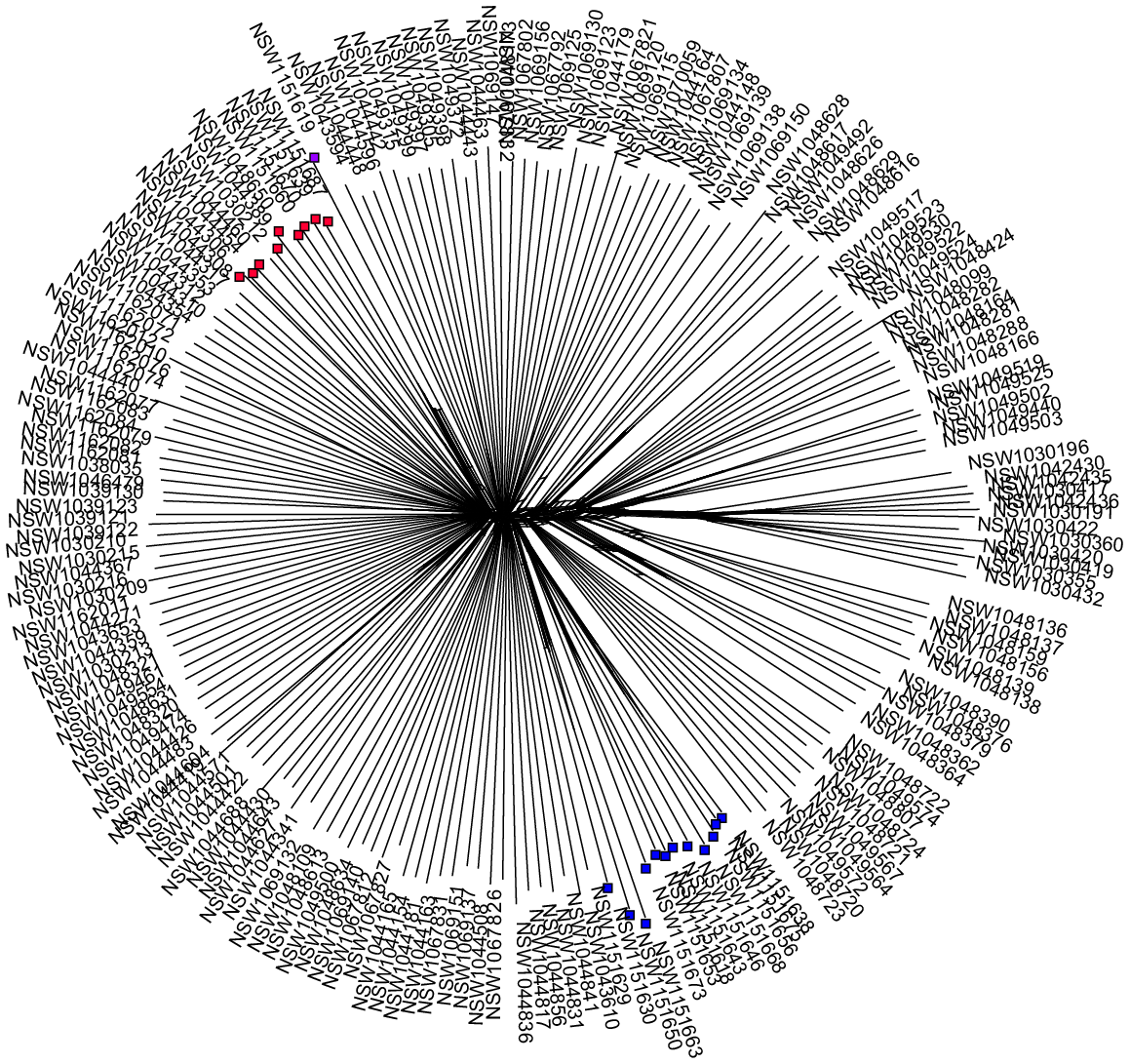
Recent anthropogenic activity, including restoration conservation and horticulture, have introduced genotypes into new environments; sometimes where other members of the same species occur. In some cases, such as studies investigating local adaptation or conservation work where local provenance may want to be conserved, there is a need to facilitate easy separation of the introduced maternal lines and the local maternal lines. As an example, the planting of non-local provenance material is recognised as a potential threat to the genetic integrity of an endangered population of Eucalyptus camaludensis in the Hunter catchment, New South Wales, Australia (listed under the NSW Biodiversity Conservation Act 2016). A restoration project attempted to collect seed across the local provenance for a restoration program that aimed to optimise local genetic provenance in the revegetated populations. Separate maternal lines were maintained throughout the propagation process and each maternal tree was genotyped. Genomic data across the distribution of the species allowed us to identify that one of the maternal trees was genetically distinct from the other collections and clustered with material from over 400 km away in another catchment (the Lachlan River Catchment) (Fig. 2). It was subsequently deemed to be planted. Given that separate maternal lines were maintained, the non-local provenance stored seed and propagated plants were easily identified and could be excluded from the restoration plantings. Given the potential threat of planting the non-local provenance material (swamping of local genotypes), had maternal lines not been maintained, the entire collection of seed from multiple mothers may have been abandoned and deemed useless, instead only a single seed lot and a few dozen seedlings were abandoned. Although keeping the maternal lines separate took more time and effort during the early phase of the project, in the long term it was time- and cost-saving.
A splitstree (Huson 1998; Huson et al. 2008) of Eucalyptus camaldulensis samples genotyped using DArTseq, showing the clustering of Hunter Valley maternal trees (blue nodes), Lachlan River catchment trees (red nodes), and the planted Hunter Valley maternal tree (purple node). This illustrates that although collected from a local tree, the maternal line does not represent a local provenance and should not be treated as such in conservation, restoration and research activities.
Reason 6: Ability to select rare lineages or alleles in a collection
The relative evolutionary contribution of lineages can be impacted by the dominant reproductive mode (vegetative or sexual), the mating system (self-fertilisation or outcrossed), and the level of kinship of maternal parents at a site, all of which can vary across species and sites (Goodwillie et al. 2005; Barrett 2015; Whitehead et al. 2018; Bragg et al. 2021; Lu-Irving et al. 2023). For plants that can reproduce or expand through clonality, selfing and biparental inbreeding, the distribution of genotypes at a site can be highly uneven. Some genotypes can be presented in only low numbers, and others dominate. For outcrossing species, distribution of families at a site can depend on pollinators and seed dispersal mechanisms (Feigs et al. 2022). Sampling strategies to capture allelic variation focus on collecting seed from a large number of maternal lines (e.g. Brown and Marshall 1995; Hoban 2019). However, if we consider the downstream utilisation of the seed (conservation gardens in living collections, translocation, seed production areas or revegetation projects), there are three issues with pooled material regardless of how well the collection captures the genetic diversity at a site: 1. taking a random selection of seed can easily miss some less frequent alleles and this can result in under representation of allelic variation in all of the above applications, 2. taking a random selection can easily select individuals that are closely related and this can lead to inbreeding or problems with self-incompatibility in all of the above applications, and 3. the genetic component of the above applications will have to be far more intensive and require sequencing of a much larger number of individuals to identify rare alleles or select individuals to minimise relatedness (kinship).
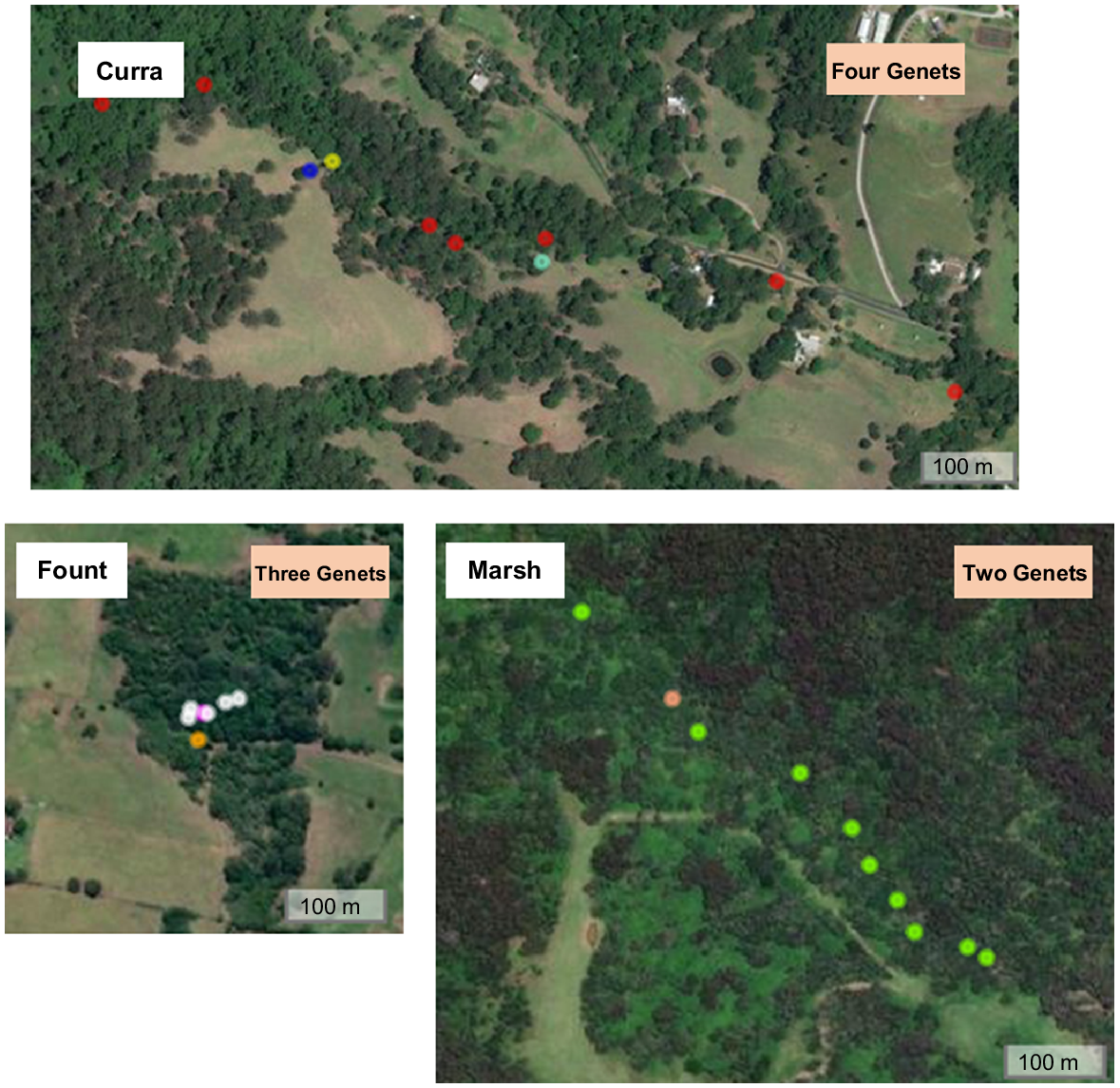
Clonal reproduction and expansion, where individuals give rise to new individual plants in the absence of sexual recombination, can result in multiple genetically identical individuals at a site. Although there are numerous known examples of plants that reproduce through clonal reproduction, there are likely many that we are unaware of (Klimes et al. 1997; Barrett 2015). A conservation genomic study conducted for management of the rare and threatened species, Daphnandra johnsonii Schodde, a rainforest tree in New South Wales (Australia), indicated that clonality is common in this species, preventing successful production of seeds, but at some sites more than one genet are present (Rossetto et al. 2021). The number of genets at a site differ, the genets do not occur in equal numbers and their spatial distribution on the landscape does not provide clues as to which genet an individual belongs to (Fig. 3). At one site (‘Marsh’) 10 trees were genotyped and amongst these, two genets were identified, with nine and one individual trees (ramets) representing each genet (green and orange dots respectively in lower right of Fig. 3). The distance between ramets belonging to the same genet were sometimes in the hundreds of metres (such as at ‘Curra’), whereas in other instances genomic data revealed distinct genets among spatially proximate trees (such as at ‘Fount’). If seed were collected at any of these sites and pooled, each of the rare genotypes would be very hard to separate from the rest; on the other hand, separate maternal line collections would allow for a much faster and more cost-effective way of identifying the genetically distinct accessions.
Each image represents an aerial view of a site of Daphnandra johnsonii, with the coloured dots indicating the location of genotyped individuals at each site. At each site, genets identified with SNP data are coloured differently. No genet was shared between sites. The genets were determined from single nucleotide polymorphisms data (SNPs) following opreviously described workflows (Bragg et al. 2020). Kinship (relatedness) estimates were used for assessing the degree of clonality across the plants of D. johnsonii and identifying the genets. The estimates were calculated using the PLINK Method of Moments estimator implemented in SNPrelate (Zheng et al. 2012).
A similar scenario would involve species where biparental inbreeding and self-fertilisation are the prevalent mating systems at a site (Lu-Irving et al. 2023). At sites where there are high levels of selfing, but multiple families present, the level of relatedness (kinship) can change dramatically across the local site and some genotypes or alleles can occur in very low frequency (such as for Acacia purpureopetala, van der Merwe et al. 2021a). Pooling seed from such sites will result in rare alleles getting lost amongst the more common alleles. Pooled accession will be a problem where the seed needs to be applied to a conservation translocation where plants are selected to minimise kinship amongst individuals used in planting (Bragg et al. 2020) to reduce the potential effects of inbreeding and inbreeding depression and maximise mixing of novel genotypes. It will be necessary to genotype far more individuals prior to planting to minimise relatedness, but if separate maternal lines were maintained then genotyping only a few of each accession will provide details about relatedness.
Reason 7: Informs the effect of trait variation across maternal lines
Functional traits involved in seed germination (such as optimum germination temperature, germination success, germination rate and seed viability) vary across species, populations, and, importantly, maternal lines (Roach and Wulff 1987; Donohue 2009).
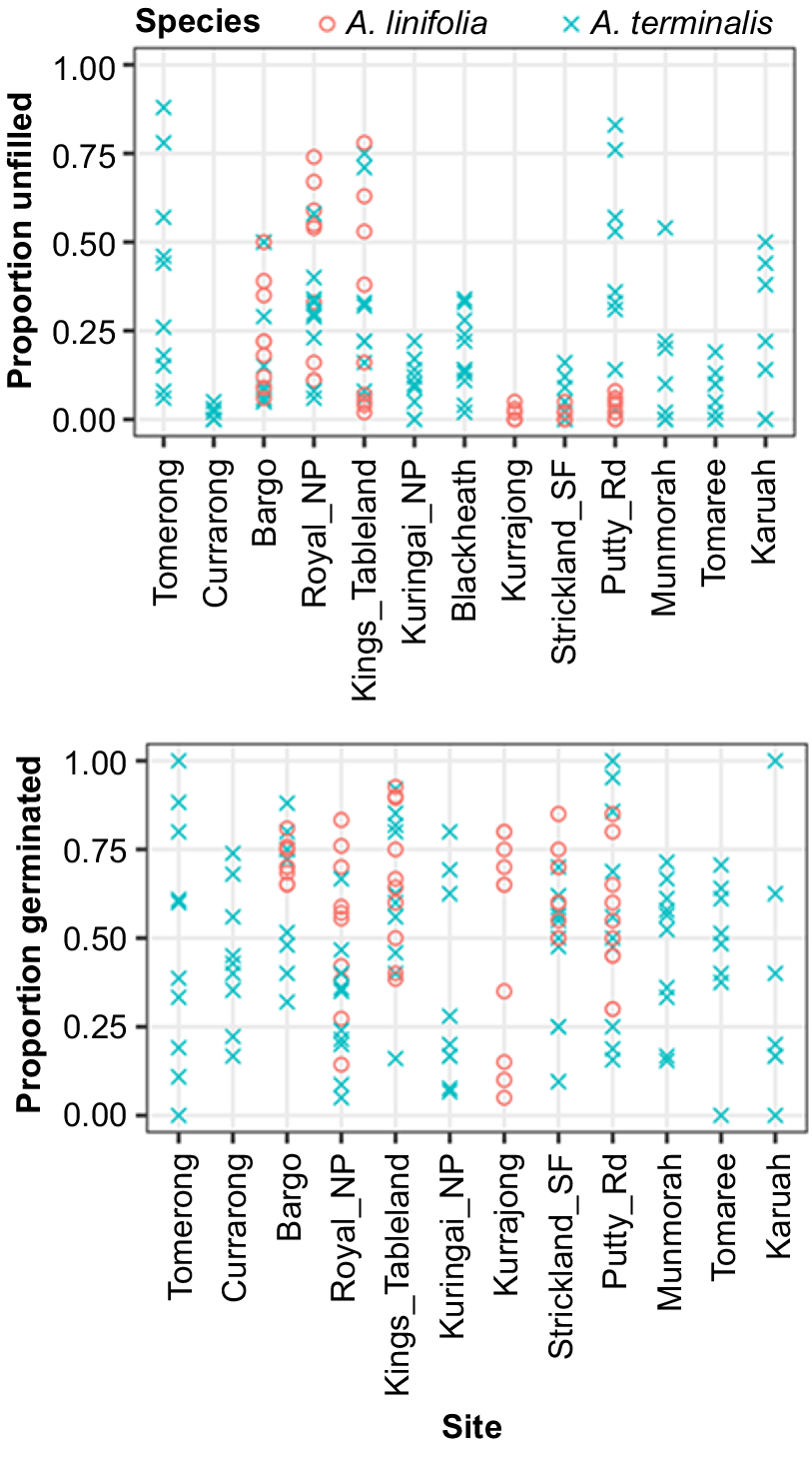
We collected seed for Acacia linifolia (Vent.) Willd and Acacia terminalis (Salisb.) J.F.Macbr. across multiple sites and maternal lines. After removing visibly undeveloped and deformed seed we used a medical diagnostic X-ray machine to detect unfilled seed (predated and seed without intact embryos). We found that the proportion of unfilled seeds varied substantially by site and maternal line (Fig. 4). Plants at some sites suffered little seed predation and produced consistently high proportions of filled seeds with intact embryos (e.g. A. terminalis at Currarong and A. linifolia at Kurrajong and Putty Rd.). However, at other sites, rates of seed predation were highly variable among maternal lines, with some plants producing higher proportions of filled seed than others (e.g. proportions of filled seed produced by A. terminalis plants from Tomerong varied from 6 to 88%). As a result, numbers of seed from maternal lines after removal of unfilled seed were not equal. We followed this up by tracking the maternal lines during germination and again found large variation across maternal lines with some maternal lines at near 100% germination and others 0% (Fig. 4 bottom). Thus, not all the maternal lines contribute equally to a seed collection and pooled seed collections can create the false impression that each maternal line is represented equally in a collection; whereas in fact some are completely absent, and others only appear in low frequency. This can also exacerbate conditions for Reason number 8, discussed below.
These graphs represent the proportion of seeds per mother that were filled (top) and from the filled seeds germinated (bottom). Seeds were from two species of Acacia collected from multiple sites. The blue crosses each represent seed from a maternal line from a site for A. terminalis and the red circles represent A. linifolia. NP, national park; SF, state forest.
Reason 8: Ability to document and understand the effects of imposed selection
Each step of seed processing and storage can affect genotypes differently and impose artificial selective filters on seed collections (Nagel et al. 2015), potentially leading to loss of genetic diversity (genetic erosion). If seed from different maternal lines is pooled, this loss will be undetected and a decrease in seed viability could be perceived as an even loss across all the maternal lines presented in the collection. Non-random mortality during storage (Bennington and McGraw 1995; sometimes referred to as the ‘invisible fraction’ sensuGrafen 1988) can also impact research interpretations derived from such seed collections (for more details see Weis 2018). Although little work has been done to understand genetic erosion in germplasm collections of non-crop species, detailed studies of crops have highlighted the uneven loss of genotypes or cultivars during storage and processing including samples from divergent angiosperm lineages such as flax (Balouchi et al. 2017), rice (Kameswara Rao and Jackson 1997; Lee et al. 2019), barley (Nagel et al. 2015) and cucumber (Hu et al. 2010). Maintaining separate maternal lines during storage can increase our understanding of loss of lineages during storage when retesting of viability and germination are completed. This information can inform recollecting to capture lost alleles and, when possible and needed, change processing or storage conditions to maintain maximum genetic diversity.
Reason 9: Expedite breeding programs using heritable traits
Trait-based breeding programs have been vital in the development of new crop and forestry lines, incorporating traits available in elite germplasm and other genetic resources (Cooper et al. 2021). In some ex situ collections, it is helpful to predict or manage plant traits. For example, if a plant species is threatened with extinction due to a disease, it might be useful to identify resistant individuals (if they exist) and promote their contribution to future generations. If a collection is being used to produce seeds for restoration across landscapes, it might be useful to predict environmental tolerances of progeny and to distribute them among planting sites accordingly. To use trait information in this way, the trait must be heritable, and predicting trait values usually requires trait measurements for a set of individuals with some known genetic relationships to each other. This underscores the potential importance of tracking seeds in maternal lines, which promotes opportunities to generate datasets with trait measurements for panels of half-siblings.
For example, the introduction of Myrtle rust (Austropuccinia psidii) into Australia has caused numerous previously common plant species to become threatened with extinction (Fensham and Radford-Smith 2021) and is impacting populations of keystone species in important habitats, such as Melaleuca quinquenervia in swamp vegetation (Pegg et al. 2018). An extensive separate maternal line seed collection of M. quinquenervia enabled the estimation of rust resistance traits, including in many half-siblings. This information enabled the establishment of a seed production area at the Australian Botanic Garden Mount Annan that is deliberately enriched with individuals from families that exhibited high levels of resistance, while also maintaining substantial genetic diversity. Progeny from this planting might eventually be used to restore swamp habitat with relatively rust resistant seeds. These activities were enabled by the availability of seeds with known familial relationships for use in experiments and planting (i.e. separate maternal line seed collections) and relevant genetic and resistance data.
Reason 10: Contain contamination (pest and diseases)
Microbes can exist as pathogens within and on seeds causing seed decay and plant disease (Agarwal and Sinclair 1996). Seed pathogenic fungi and bacteria greatly affect seed quality, and can cause diseases that impact rates of germination and establishment (Aftab et al. 2008). They can also remain dormant but viable within stored seed for long periods of time (Brodal and Asdal 2021). Some seed pathogens can act as vectors, with some transmitted from generation to generation, providing a means by which a disease can become established at a site (Elmer 2001).
Seeds have been shown to be associated with both beneficial, saprophytic and pathogenic species (Tarquinio et al. 2021; Chandel et al. 2022; Mertin et al. 2022). These micro-organisms enter seed via different pathways and as such can be restricted to a single maternal plant or site (Gebeyaw 2020). Contact of non-infected with infected seed can provide a means by which pathogens transmit between individuals (Nallathambi et al. 2020). Recent work indicates that microbes can stay viable in seed collections for extended periods of time.
‘Maternal effects’ can shape seed fungal communities. In particular, the maternal source of a seed explained a larger part of the variation in seed community composition than environment in Quercus petraea (Fort et al. 2021). Additionally, the importance of microbial inheritance in forming the initial plant microbiome, and the role of the maternal plant within this, suggest that the maternal plant plays a key role in the formation and transmission of seed and seedling microbial communities (Abdelfattah et al. 2023). Keeping maternal lines separate within seed collections may retain communities more reflective of the natural processes that can shape seed communities. Not only will separate maternal line seed collections contain potential pathogens but also facilitate more research in this emerging field.
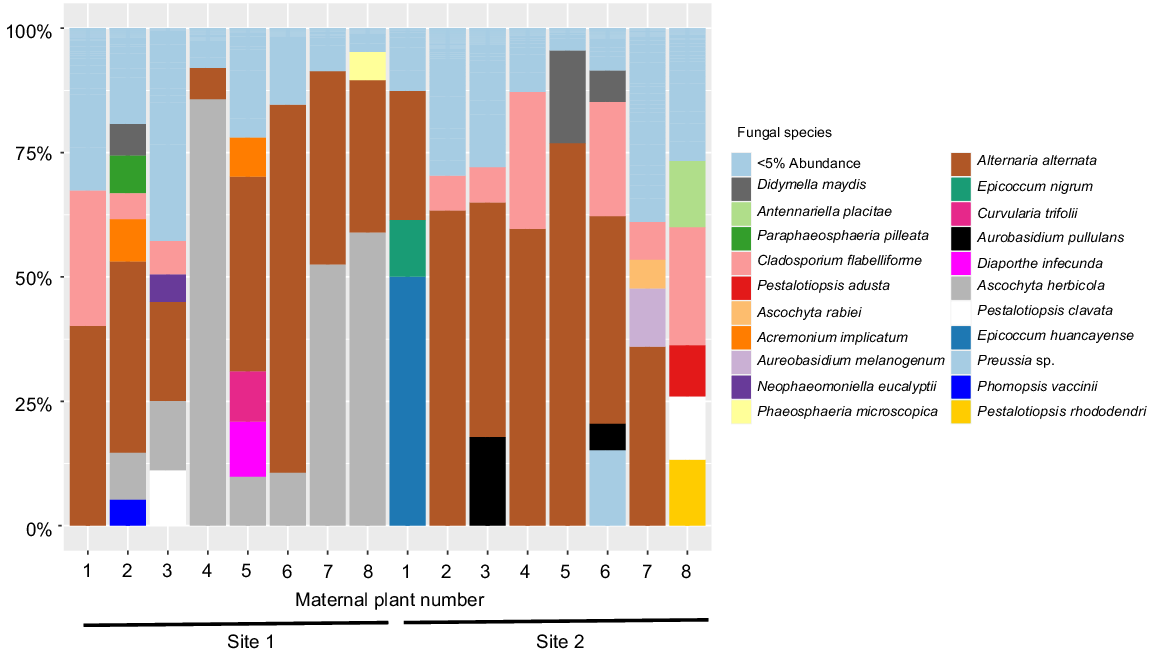
We investigated microbe communities in Themeda triandra seed and present the results for two sites (Fig. 5). We found that the communities vary in composition, with some maternal plants housing pathogenic species and others not. For example, in Site 1 most plants house Alternaria alternata and Ascochyta herbicola, but only seed from maternal plant five houses the plant pathogens Diaporthe infecunda (Moreira et al. 2020) and Curvularia trifolii (Falloon 1976) (Fig. 5). Similarly, in Site 2 Maternal Plant 1 was the only one to house the plant pathogens Epicoccum nigrum and Epicoccum huancayense, (Taguiam et al. 2021) with them forming the dominant part of the community for that maternal line. These results indicate that seed communities differ between maternal plants, highlighting the importance of the maternal plant in shaping seed communities.
Relative abundance (%) of seed fungal species within 50 mg of Themeda triandra seed pooled to each maternal plant (n = 8) at two sites in New South Wales, Australia. Data is based on ~300 bp of the ITS2 (internal transcribed spacer 2) region, with species identity determined by comparison with the UNITE database (Abarenkov et al. 2010). The potentially pathogenic fungus, Curvularia trifolii (fucshia colour) was only identified in Maternal Plant 5 at Site 1.
Synthesis
We outlined 10 reasons why the maintenance of separate maternal lines increases the value and applications of seed collections. Most of our reasoning comes down to the fact that when dealing with a pooled seed collection it is difficult (if not impossible) to sort a pooled collection into evolutionary lineages (genotypes, families, species). We can retrospectively use genetic data to separate out the lineages but this is destructive, and wasteful of time, space and cost.
Some of the listed reasons are directly linked or can escalate each other, for example, we would like to avoid using incorrect taxa (Reasons 1, 2 and 4) in conservation translocations where we also want to maximise genetic diversity but avoid high rates of kinship (Reason 6). Another example would be where we would like to set up a seed production area for a species where mixed genome sizes have been recorded (such as A. littoralis); we need to use only diploid individuals (Reason 4) but to avoid inbreeding we want to avoid individuals that are closely related (Reason 6). Reasons 6, 7 and 8 all play an important role in determining how much genetic diversity is held within a seed collection and how we can monitor and assess changes in diversity across the life of a collection.
Unlike pooled collections, we can use separate maternal line collections effectively to evaluate and monitor genetic diversity in a seed collection. Seedlings produced during standard viability tests that are routinely conducted in many seedbanks can be used for DNA extractions and sequencing. Fortunately, the genomic tools and workflows needed to assess genetic diversity and representativeness have become increasingly accessible (Rossetto et al. 2019, 2021). Conservation and research collection managers can screen small numbers of seed from each separately stored maternal line, and use the data to establish the identity and origin (species, hybrid or non-hybrid, genome size and provenance, Reasons 1, 2, 3, 4 and 5) of the accession, estimate the genetic variation (based on allelic variation and relatedness) for each maternal line and quantify the overall collection (all maternal lines for a site, species or area; Reason 6). This information can then form the backbone for monitoring the collection (Reasons 7 and 8) and planning augmentation or recollection if needed. This will also facilitate screening for heritable traits (Reason 9) and tracing them back to a specific lineage or mother.
A wide array of scientific studies utilises seeds in experiments (seed biology, ecophysiology, resurrection ecology, microbiology, to name a few) and for optimal interpretative power the experimental design should be representative of the standing diversity within the species or population of the species, rather than represent a mixed (Reasons 1, 2, 3, 4 and 5 above) or small non-representative gene pool (Reasons 6, 7 and 8 above). This can only be achieved if a genetically representative collection of individuals is available for the experimental work. Although genetic data should ideally inform the representativeness of a collection, the availability of separate maternal lines is, at a minimum, a good starting point for many studies.
Prior to establishing either a translocation population or a seed orchard, genetic data should be used to select the individuals from the collection to be planted. The availability of separate maternal line collections can greatly expedite this process and decrease the costs associated with obtaining genetic data because only a small representation of each maternal line will need to be used and genotyped. When using seed in restoration and conservation applications we want to be able to avoid inbreeding and maximise evolutionary potential or even select for a specific trait (Bragg et al. 2022). With separate maternal line seed collections, we can avoid using seed from the same mother as well as spatially separate maternal lines during planting. After careful monitoring it will also be possible to augment restored populations with individuals from selected maternal lines.
There is much to learn from the elite germplasm collections available for crop development and this body of work has already established that separate maternal line collections can best facilitate multiple downstream applications (Nagel et al. 2015; Jia et al. 2017; Solberg et al. 2020). While the goals and constraints of conservation germplasm collections differ from those of crop improvement collections, the value of separating maternal lines has already begun to be recognised and incorporated into seed collecting guidelines for plant conservation and restoration (e.g. Guerrant et al. 2014; Center for Plant Conservation 2019; van der Merwe et al. 2021b). Resources (time, money and seed availability) for conservation collections of wild species are often limited, but the investment of additional resources to label, track and store separate maternal lines yields high returns by unlocking myriad benefits and applications that enhance the ultimate value of the collections. To optimise use of limited resources to achieve the best conservation outcomes, we need to maximise our efforts by constantly increasing the quality of our germplasm collections. One step forward would be to maintain separate maternal lines.
Declaration of funding
Results from multiple projects were used to provide examples. TCW received funding from Australian Biological Resources Study (RG19-17) for the work on Ajuga australis; AM received funding from the Botany Foundation for the work on seed microbes; MvdM received funding from the NSW Environmental Trust (2016 RD 0084), which contributed towards the seed work on Acacia; SY and MR received funding from NSW Government the Saving our Species program for work on Daphnandra johnsii; JGB received funding from Australian Research Council that included the Melaleuca quinquenervia myrtle rust work (ARC Linkage grant LP180100721).
References
Abarenkov K, Henrik Nilsson R, Larsson K-H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U (2010) The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytologist 186, 281-285.
| Crossref | Google Scholar |
Abdelfattah A, Tack AJM, Lobato C, Wassermann B, Berg G (2023) From seed to seed: the role of microbial inheritance in the assembly of the plant microbiome. Trends in Microbiology 31, 346-355.
| Crossref | Google Scholar |
Aftab M, Freeman A, Bretag T (2008) Seed health testing in pulse crops. Department of Environment and Primary Industries, Melbourne, Victoria. Available at https://apo.org.au/node/56574 [Verified 19 April 2023]
Ahrens CW, James EA, Miller AD, Scott F, Aitken NC, Jones AW, Lu-Irving P, Borevitz JO, Cantrill DJ, Rymer PD (2020) Spatial, climate and ploidy factors drive genomic diversity and resilience in the widespread grass Themeda triandra. Molecular Ecology 29(20), 3872-3888.
| Crossref | Google Scholar |
Arditti J, Ghani AKA (2000) Tansley Review No. 110. Numerical and physical properties of orchid seeds and their biological implications. New Phytologist 145, 367-421.
| Crossref | Google Scholar |
Australian Government (2023) Species profile and threats database, EPBC Act List of threatened flora. Available at http://www.environment.gov.au/cgi-bin/sprat/public/publicthreatenedlist.pl?wanted=flora [Verified 2 May 2023]
Balouchi H, Baladi S, Moradi A, Dehnavi MM (2017) The influence of temperature and moisture content on seed longevity of two genotypes of Linum usitatissimum. Seed Science and Technology 45, 130-138.
| Crossref | Google Scholar |
Barrett SCH (2015) Influences of clonality on plant sexual reproduction. Proceedings of the National Academy of Sciences 112, 8859-8866.
| Crossref | Google Scholar |
Bennington CC, McGraw JB (1995) Phenotypic selection in an artificial population of Impatiens pallida: the importance of the invisible fraction. Evolution 49, 317-324.
| Crossref | Google Scholar |
Bragg JG, Cuneo P, Sherieff A, Rossetto M (2020) Optimizing the genetic composition of a translocation population: Incorporating constraints and conflicting objectives. Molecular Ecology Resources 20, 54-65.
| Crossref | Google Scholar |
Bragg JG, Yap J-YS, Wilson T, Lee E, Rossetto M (2021) Conserving the genetic diversity of condemned populations: optimizing collections and translocation. Evolutionary Applications 14, 1225-1238.
| Crossref | Google Scholar |
Bragg JG, van der Merwe M, Yap J-YS, Borevitz J, Rossetto M (2022) Plant collections for conservation and restoration: can they be adapted and adaptable? Molecular Ecology Resources 22, 2171-2182.
| Crossref | Google Scholar |
Brodal G, Asdal Å (2021) Longevity of plant pathogens in dry agricultural seeds during 30 years of storage. Microorganisms 9, 2175.
| Crossref | Google Scholar |
Bucharova A, Bossdorf O, Scheepens JF, Salguero-Gómez R (2023) Sustainable seed harvesting in wild plant populations. bioRxiv 2023.01.12.523821.
| Crossref | Google Scholar |
Byrne PF, Volk GM, Gardner C, Gore MA, Simon PW, Smith S (2018) Sustaining the future of plant breeding: the critical role of the USDA-ARS national plant germplasm system. Crop Science 58, 451-468.
| Crossref | Google Scholar |
Chandel A, Mann R, Kaur J, Tannenbaum I, Norton S, Edwards J, Spangenberg G, Sawbridge T (2022) Australian native Glycine clandestina seed microbiota hosts a more diverse bacterial community than the domesticated soybean Glycine max. Environmental Microbiome 17, 56.
| Crossref | Google Scholar |
Chapman T, Miles S, Trivedi C (2019) Capturing, protecting and restoring plant diversity in the UK: RBG Kew and the Millennium Seed Bank. Plant Diversity 41, 124-131.
| Crossref | Google Scholar |
Cooper M, Powell O, Voss-Fels KP, Messina CD, Gho C, Podlich DW, Technow F, Chapman SC, Beveridge CA, Ortiz-Barrientos D, Hammer GL (2021) Modelling selection response in plant-breeding programs using crop models as mechanistic gene-to-phenotype (CGM-G2P) multi-trait link functions. In silico Plants 3, diaa016.
| Crossref | Google Scholar |
Crawford AD, Monks L, Shade A, Wood JA, Errington G, Stevens A, Norton SL, Coates D (2021) Chapter 15: Maintenance, utilization and information storage. In ‘Plant germplasm conservation in Australia: strategies and guidelines for developing, managing and utilising ex situ collections’. 3rd edn. (Eds AJ Martyn Yenson, CA Offord, PF Meagher, T Auld, D Bush, DJ Coates, LE Commander, LK Guja, SL Norton, RO Makinson, R Stanley, N Walsh, D Wrigley, L Broadhurst) pp. 449–476. (Australian Network for Plant Conservation: Canberra, ACT)
Donohue K (2009) Completing the cycle: maternal effects as the missing link in plant life histories. Philosophical Transactions of the Royal Society B: Biological Sciences 364, 1059-1074.
| Crossref | Google Scholar |
Elmer WH (2001) Seeds as vehicles for pathogen importation. Biological Invasions 3, 263-271.
| Crossref | Google Scholar |
Engels J, Engelmann F (1998) Introductory statement. Seed Science Research 8, 1-2.
| Google Scholar |
Everingham SE, Offord CA, Sabot MEB, Moles AT (2021) Time-traveling seeds reveal that plant regeneration and growth traits are responding to climate change. Ecology 102(3), e03272.
| Crossref | Google Scholar |
Falloon RE (1976) Curvularia trifolii as a high-temperature turfgrass pathogen. New Zealand Journal of Agricultural Research 19, 243-248.
| Crossref | Google Scholar |
Feigs JT, Holzhauer SIJ, Huang S, Brunet J, Diekmann M, Hedwall P-O, Kramp K, Naaf T (2022) Pollinator movement activity influences genetic diversity and differentiation of spatially isolated populations of clonal forest herbs. Frontiers in Ecology and Evolution 10, 908258.
| Crossref | Google Scholar |
Fensham RJ, Radford-Smith J (2021) Unprecedented extinction of tree species by fungal disease. Biological Conservation 261, 109276.
| Crossref | Google Scholar |
Field DL, Ayre DJ, Whelan RJ, Young AG (2008) Relative frequency of sympatric species influences rates of interspecific hybridization, seed production and seedling performance in the uncommon Eucalyptus aggregata. Journal of Ecology 96(6), 1198-1210.
| Crossref | Google Scholar |
Fort T, Pauvert C, Zanne AE, Ovaskainen O, Caignard T, Barret M, Compant S, Hampe A, Delzon S, Vacher C (2021) Maternal effects shape the seed mycobiome in Quercus petraea. New Phytologist 230, 1594-1608.
| Crossref | Google Scholar |
Gebeyaw M (2020) Review on: impact of seed-borne pathogens on seed quality. American Journal of Plant Biology 5, 77-81.
| Crossref | Google Scholar |
Godfray HCJ (2002) Challenges for taxonomy. Nature 417, 17-19.
| Crossref | Google Scholar |
Godfree RC, Marshall DJ, Young AG, Miller CH, Mathews S (2017) Empirical evidence of fixed and homeostatic patterns of polyploid advantage in a keystone grass exposed to drought and heat stress. Royal Society Open Science 4(11), 170934.
| Crossref | Google Scholar |
Goodwillie C, Kalisz S, Eckert CG (2005) The evolutionary enigma of mixed mating systems in plants: occurrence, theoretical explanations, and empirical evidence. Annual Review of Ecology, Evolution, and Systematics 36, 47-79.
| Crossref | Google Scholar |
Griffith MP, Calonje M, Meerow AW, Tut F, Kramer AT, Hird A, Magellan TM, Husby CE (2015) Can a botanic garden cycad collection capture the genetic diversity in a wild population? International Journal of Plant Sciences 176, 1-10.
| Crossref | Google Scholar |
Guerrant EO, Jr., Havens K, Vitt P (2014) Sampling for effective ex situ plant conservation. International Journal of Plant Sciences 175, 11-20.
| Crossref | Google Scholar |
Hao L, Xu W, Qi G, Xin T, Xu Z, Lei H, Song J (2022) GAGE is a method for identification of plant species based on whole genome analysis and genome editing. Communications Biology 5, 947.
| Crossref | Google Scholar |
Harley RM, Atkins S, Budantsev A, Cantino PD, Conn B, Grayer RJ, Harley MM, De Kok R, Krestovskaja T, Morales A, Paton AJ, Ryding O, Upson T (2004) Labiatae. In ‘Flowering Plants - Dicotyledons’. The families and genera of vascular plants. Vol. 7. (Ed. JW Kadereit) pp. 167–275. (Springer: Berlin) doi:10.1007/978-3-642-18617-2_11
Hayman DL (1960) The distribution and cytology of the chromosome races of Themeda australis in southern Australia. Australian Journal of Botany 8, 58-68.
| Crossref | Google Scholar |
Hegde SG, Nason JD, Clegg JM, Ellstrand NC (2006) The evolution of California’s wild radish has resulted in the extinction of its progenitors. Evolution 60(6), 1187-1197.
| Google Scholar |
Hirashiki C, Kareiva P, Marvier M (2021) Concern over hybridization risks should not preclude conservation interventions. Conservation Science and Practice 3, e424.
| Crossref | Google Scholar |
Hoban S (2019) New guidance for ex situ gene conservation: sampling realistic population systems and accounting for collection attrition. Biological Conservation 235, 199-208.
| Crossref | Google Scholar |
Hoffmann AA, Sgrò CM (2011) Climate change and evolutionary adaptation. Nature 470, 479-485.
| Crossref | Google Scholar |
Hogbin T (2022) Restore and Renew – making genomic and climatic information freely available to restoration practitioners. Available at https://recer.org.au/2022/11/restore_and_renew_restoration_genomics/ [Accessed 2 May 2023]
Hu J, Li J, Liang F, Liu L, Si S (2010) Genetic relationship of a cucumber germplasm collection revealed by newly developed EST-SSR markers. Journal of Genetics 89, e28-e32.
| Google Scholar |
Huson DH (1998) SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14(1), 68-73.
| Crossref | Google Scholar |
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23, 254-267.
| Crossref | Google Scholar |
Huson DH, Kloepper T, Bryant D (2008) SplitsTree 4.0-Computation of phylogenetic trees and networks. Bioinformatics 14, 68-73.
| Google Scholar |
Jackson WD, Wiltshire RJE (2001) Historical taxonomy and a resolution of the Stylidium graminifolium complex (Stylidiaceae) in Tasmania. Australian Systematic Botany 14(6), 937-969.
| Crossref | Google Scholar |
Jeanes JA (2013) An overview of the Thelymitra nuda (Orchidaceae) complex in Australia including the description of six new species. Muelleria: An Australian Journal of Botany 31, 3-30.
| Crossref | Google Scholar |
Jia J, Li H, Zhang X, Li Z, Qiu L (2017) Genomics-based plant germplasm research (GPGR). The Crop Journal 5, 166-174.
| Crossref | Google Scholar |
Kameswara Rao N, Jackson MT (1997) Variation in seed longevity of rice cultivars belonging to different isozyme groups. Genetic Resources and Crop Evolution 44, 159-164.
| Crossref | Google Scholar |
Labroo MR, Studer AJ, Rutkoski JE (2021) Heterosis and hybrid crop breeding: a multidisciplinary review. Frontiers in Genetics 12, 643761.
| Crossref | Google Scholar |
Lee J-S, Velasco-Punzalan M, Pacleb M, Valdez R, Kretzschmar T, McNally KL, Ismail AM, Cruz PCS, Sackville Hamilton NR, Hay FR (2019) Variation in seed longevity among diverse Indica rice varieties. Annals of Botany 124, 447-460.
| Crossref | Google Scholar |
León-Lobos P, Way M, Aranda PD, Lima-Junior M (2012) The role of ex situ seed banks in the conservation of plant diversity and in ecological restoration in Latin America. Plant Ecology & Diversity 5(2), 245-258.
| Crossref | Google Scholar |
Li D-Z, Pritchard HW (2009) The science and economics of ex situ plant conservation. Trends in Plant Science 14, 614-621.
| Crossref | Google Scholar |
Li XX, Sun RF, Fang ZY, Liu TJ, Wang JL, Shen D, Wang HP, Song JP, Sun YY, Wang LR, Jiang SL (2016) Germplasm resources of horticultural crops and sustainable development of horticultural industry in China. Acta Horticulturae 1129, 1-8.
| Crossref | Google Scholar |
Lu-Irving P, Bragg JG, Rossetto M, King K, O’Brien M, van der Merwe MM (2023) Capturing genetic diversity in seed collections: an empirical study of two congeners with contrasting mating systems. Plants 12(3), 522.
| Crossref | Google Scholar |
Martyn Yenson AJ, Offord CA, Meagher PF, Auld T, Bush D, Coates DJ, Commander LE, Guja LK, Norton SL, Makinson RO, Stanley R, Walsh N, Wrigley D, Broadhurst L (2021) ‘Plant germplasm conservation in Australia: strategies and guidelines for developing, managing and utilising ex situ collections.’ 3rd edn. (Australian Network for Plant Conservation: Canberra, ACT)
Mayr E, Bock WJ (2002) Classifications and other ordering systems. Journal of Zoological Systematics and Evolutionary Research 40, 169-194.
| Crossref | Google Scholar |
Mertin AA, Laurence MH, van der Merwe M, French K, Liew ECY (2022) The culturable seed mycobiome of two Banksia species is dominated by latent saprotrophic and multi-trophic fungi. Fungal Biology 126(11-12), 738-745.
| Crossref | Google Scholar |
Moreira RR, Caus G, Gomes Figueiredo JA, May De Mio LL (2020) Phomopsis rot caused by Diaporthe infecunda on fruit and flowers of Passiflora edulis in Brazil. Australasian Plant Pathology 49, 141-145.
| Crossref | Google Scholar |
Muhlfeld CC, Kovach RP, Jones LA, Al-Chokhachy R, Boyer MC, Leary RF, Lowe WH, Luikart G, Allendorf FW (2014) Invasive hybridization in a threatened species is accelerated by climate change. Nature Climate Change 4(7), 620-624.
| Crossref | Google Scholar |
Nagel M, Kranner I, Neumann K, Rolletschek H, Seal CE, Colville L, Fernández-Marín BE, Börner A (2015) Genome-wide association mapping and biochemical markers reveal that seed ageing and longevity are intricately affected by genetic background and developmental and environmental conditions in barley. Plant, Cell & Environment 38, 1011-1022.
| Crossref | Google Scholar |
Nelson GJ (1971) “Cladism” as a philosophy of classification. Systematic Zoology 20, 373-376.
| Crossref | Google Scholar |
O’Donnell K, Sharrock S (2017) The contribution of botanic gardens to ex situ conservation through seed banking. Plant Diversity 39(6), 373-378.
| Crossref | Google Scholar |
Parmesan C, Singer MC, Wee B, Mikheyev S (2023) The case for prioritizing ecology/behavior and hybridization over genomics/taxonomy and species’ integrity in conservation under climate change. Biological Conservation 281, 109967.
| Crossref | Google Scholar |
Pegg GS, Lee DJ, Carnegie AJ (2018) Predicting impact of Austropuccinia psidii on populations of broad leaved Melaleuca species in Australia. Australasian Plant Pathology 47, 421-430.
| Crossref | Google Scholar |
Pellicer J, Leitch IJ (2020) The Plant DNA C-values database (release 7.1): an updated online repository of plant genome size data for comparative studies. New Phytologist 226, 301-305.
| Crossref | Google Scholar |
Prentis PJ, White EM, Radford IJ, Lowe AJ, Clarke AR (2007) Can hybridization cause local extinction: a case for demographic swamping of the Australian native Senecio pinnatifolius by the invasive Senecio madagascariensis? New Phytologist 176, 902-912.
| Crossref | Google Scholar |
Prober SM, Doerr VAJ, Broadhurst LM, Williams KJ, Dickson F (2019) Shifting the conservation paradigm: a synthesis of options for renovating nature under climate change. Ecological Monographs 89, e01333.
| Crossref | Google Scholar |
Rauschkolb R, Li Z, Godefroid S, Dixon L, Durka W, Májeková M, Bossdorf O, Ensslin A, Scheepens JF (2022) Evolution of plant drought strategies and herbivore tolerance after two decades of climate change. New Phytologist 235, 773-785.
| Crossref | Google Scholar |
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conservation Biology 17, 230-237.
| Crossref | Google Scholar |
Reiter N, Dimon R, Freestone M (2021) Case Study 11.5: Saving orchids from extinction: the RBGV orchid conservation program ex situ collection. In ‘Plant germplasm conservation in Australia: strategies and guidelines for developing, managing and utilising ex situ collections’. 3rd edn. (Eds AJ Martyn Yenson, CA Offord, PF Meagher, T Auld, D Bush, DJ Coates, LE Commander, LK Guja, SL Norton, RO Makinson, R Stanley, N Walsh, D Wrigley, L Broadhurst) pp. 355–357. (Australian Network for Plant Conservation: Canberra, ACT)
Roach DA, Wulff RD (1987) Maternal effects in plants. Annual Review of Ecology and Systematics 18(1), 209-235.
| Crossref | Google Scholar |
Rose PF, Wilson KL, Telford IRH, Lamont RW, Bruhl JJ (2015) Multiple lines of evidence clarify limits of Allocasuarina thalassoscopica, A. defungens and A. littoralis (Allocasuarina sect. Cylindropitys, Casuarinaceae. Australian Systematic Botany 27, 257-281.
| Crossref | Google Scholar |
Rossetto M, Bragg J, Kilian A, McPherson H, van der Merwe M, Wilson PD (2019) Restore and Renew: a genomics-era framework for species provenance delimitation. Restoration Ecology 27, 538-548.
| Crossref | Google Scholar |
Rossetto M, Yap J-YS, Lemmon J, Bain D, Bragg J, Hogbin P, Gallagher R, Rutherford S, Summerell B, Wilson TC (2021) A conservation genomics workflow to guide practical management actions. Global Ecology and Conservation 26, e01492.
| Crossref | Google Scholar |
Rutherford S, van der Merwe M, Wilson PG, Kooyman RM, Rossetto M (2019) Managing the risk of genetic swamping of a rare and restricted tree. Conservation Genetics 20(5), 1113-1131.
| Crossref | Google Scholar |
Rutherford S, Wilson TC, Yap J-YS, Lee E, Errington G, Rossetto M (2022) Evolutionary processes in an undescribed eucalypt: implications for the translocation of a critically endangered species. Annals of Botany 130(4), 491-508.
| Crossref | Google Scholar |
Solberg SØ, Yndgaard F, Andreasen C, Von Bothmer R, Loskutov IG, Asdal Å (2020) Long-term storage and longevity of orthodox seeds: a systematic review. Frontiers in Plant Science 11, 1007.
| Crossref | Google Scholar |
Sommerville KD, Martyn AJ, Offord CA (2013) Can seed characteristics or species distribution be used to predict the stratification requirements of herbs in the Australian Alps? Botanical Journal of the Linnean Society 172, 187-204.
| Crossref | Google Scholar |
Taguiam JD, Evallo E, Balendres MA (2021) Epicoccum species: ubiquitous plant pathogens and effective biological control agents. European Journal of Plant Pathology 159, 713-725.
| Crossref | Google Scholar |
Tarquinio F, Attlan O, Vanderklift MA, Berry O, Bissett A (2021) Distinct endophytic bacterial communities inhabiting seagrass seeds. Frontiers in Microbiology 12, 703014.
| Crossref | Google Scholar |
Todesco M, Pascual MA, Owens GL, Ostevik KL, Moyers BT, Hübner S, Heredia SM, Hahn MA, Caseys C, Bock DG, Rieseberg LH (2016) Hybridization and extinction. Evolutionary Applications 9(7), 892-908.
| Crossref | Google Scholar |
van der Merwe MM, Yap J-YS, Wilson PD, Murphy HT, Ford A (2021a) All populations matter: conservation genomics of Australia’s iconic purple wattle, Acacia purpureopetala. Diversity 13, 139.
| Crossref | Google Scholar |
van der Merwe M, Broadhurst L, Coates D, Rossetto M (2021b) Chapter 3 Genetic guidelines for acquiring and maintaining collections for ex situ conservation. In ‘Plant germplasm conservation in Australia: strategies and guidelines for developing, managing and utilising ex situ collections’. 3rd edn. (Eds AJ Martyn Yenson, CA Offord, PF Meagher, T Auld, D Bush, DJ Coates, LE Commander, LK Guja, SL Norton, RO Makinson, R Stanley, N Walsh, D Wrigley, L Broadhurst) pp. 55–83. (Australian Network for Plant Conservation: Canberra, ACT)
Warschefsky E, Penmetsa RV, Cook DR, Von Wettberg EJB (2014) Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. American Journal of Botany 101, 1791-1800.
| Crossref | Google Scholar |
Weis AE (2018) Detecting the “invisible fraction” bias in resurrection experiments. Evolutionary Applications 11, 88-95.
| Crossref | Google Scholar |
White FJ, Ensslin A, Godefroid S, Faruk A, Abeli T, Rossi G, Mondoni A (2023) Using stored seeds for plant translocation: the seed bank perspective. Biological Conservation 281, 109991.
| Crossref | Google Scholar |
Whitehead MR, Lanfear R, Mitchell RJ, Karron JD (2018) Plant mating systems often vary widely among populations. Frontiers in Ecology and Evolution 6, 38.
| Crossref | Google Scholar |
Wilson T (2022) Progress report on the systematics of Australian Ajugoideae. Australasian Systematic Botany Society Newsletter 191, 14-18.
| Google Scholar |
Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS (2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28(24), 3326-3328.
| Crossref | Google Scholar |


