Integrating seed microbiome knowledge into restoration and ex situ conservation of native Australian plants
Allison A. Mertin A B C * , Merize Philpott B , Linda L. Blackall C , Kris French B , Edward C. Y. Liew A and Marlien M. van der Merwe
A B C * , Merize Philpott B , Linda L. Blackall C , Kris French B , Edward C. Y. Liew A and Marlien M. van der Merwe  A
A
A Research Centre for Ecosystem Resilience, Australian Institute of Botanical Science, Royal Botanic Gardens and Domain Trust, Mrs Macquaries Road, Sydney, NSW 2000, Australia.
B Centre for Sustainable Ecosystem Solutions, School of Earth, Atmospheric and Life Sciences, University of Wollongong, Wollongong, NSW 2522, Australia.
C School of BioSciences, University of Melbourne, Parkville, Vic. 3010, Australia.
Australian Journal of Botany - https://doi.org/10.1071/BT22109
Submitted: 21 September 2022 Accepted: 14 June 2023 Published online: 12 July 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: Seeds harbour a diversity of microbes, which in some plants aid with germination and establishment. Seeds form a critical part in the lifecycle of plants and a role in many conservation and restoration activities.
Aims: Because this is an emerging field in seed biology, we aim to highlight the key research gaps of interest to seed on the basis of restoration and ex situ conservation.
Methods: We identify knowledge gaps associated with the seed endophytic microbiome of native Australian plants through undertaking a literature review. Additionally, culturing methods were used to identify the fungal seed endophytes of five native Australian species.
Key results: We identified a diversity of taxa within the native seed and show three taxa that are common to all study hosts. Sampling seed from additional hosts at a site and additional sites of a host species showed new fungal diversity. Our literature review showed that little information is available on native seed microbiomes and we identified four key areas where research gaps exist, linking with seed-based restoration practices.
Conclusions: We provide evidence that there is a complex and diverse seed microbiome within some Australian native plants and suggest ways that it could be integrated into restoration and conservation practices.
Implications: We propose that by taking into consideration the presence of a seed microbiome and its potential impacts on plant health, seed microbiomes could be used as one method to restore microbial diversity into an ecosystem and to contribute to the seedling microbiome and plant health at restored sites.
Keywords: endophyte, horizontal transmission, microbial diversity, mycobiome, restoration, seed bank, seed fungal endophytes, temporal changes, vertical transmission.
Introduction
There is a growing awareness of the importance of the microbial community in ecosystem function, productivity, and health (Finlay et al. 1997; Graham et al. 2016). We know that the loss of microbial diversity can alter ecosystem processes such as litter decomposition, organic-matter mineralisation, nitrogen uptake and primary production (Delgado-Baquerizo et al. 2016), and it can reduce the ability of plants to respond to stresses and changes in the environment, reducing the resilience and adaptive potential of the ecosystem (Shade et al. 2012). Considering microbial diversity in ecosystem restoration is therefore of critical importance. However, there is a distinct lack of baseline microbiome information for natural ecosystems, with most of our knowledge originating from surveys of agricultural plants (Clay 1987; Clay and Holah 1999; Arnold and Herre 2003; Afkhami and Rudgers 2008; Compant et al. 2011; Barret et al. 2015; Giauque and Hawkes 2016; Klaedtke et al. 2016; Adam et al. 2018), forestry species (Helander et al. 1994; Arnold et al. 2007; Deckert et al. 2019; Bergmann and Busby 2021) and soil microbiomes (Peay et al. 2007; Heneghan et al. 2008; Muñoz-Rojas et al. 2016; Gellie et al. 2017; Schöler et al. 2017; Leff et al. 2018; Saleem et al. 2019; Ngugi et al. 2020).
Integration of microbial communities in restoration within Australia is still in the early stages of application, with current approaches predominantly using soil microbes (Moreira-Grez et al. 2019; Alfonzetti et al. 2022; D’Agui et al. 2022; Dadzie et al. 2022) despite microbes being present also within all tissues of a plant. All the microbes associated with a plant form the plant holobiont, which is composed of epiphytic (exterior microbes), and endophytic (those within the living tissue of a plant) microbes (Fig. 1; Vandenkoornhuyse et al. 2015), and of the plant holobiont, endophytes are thought to comprise the majority of the community (Hardoim et al. 2015).
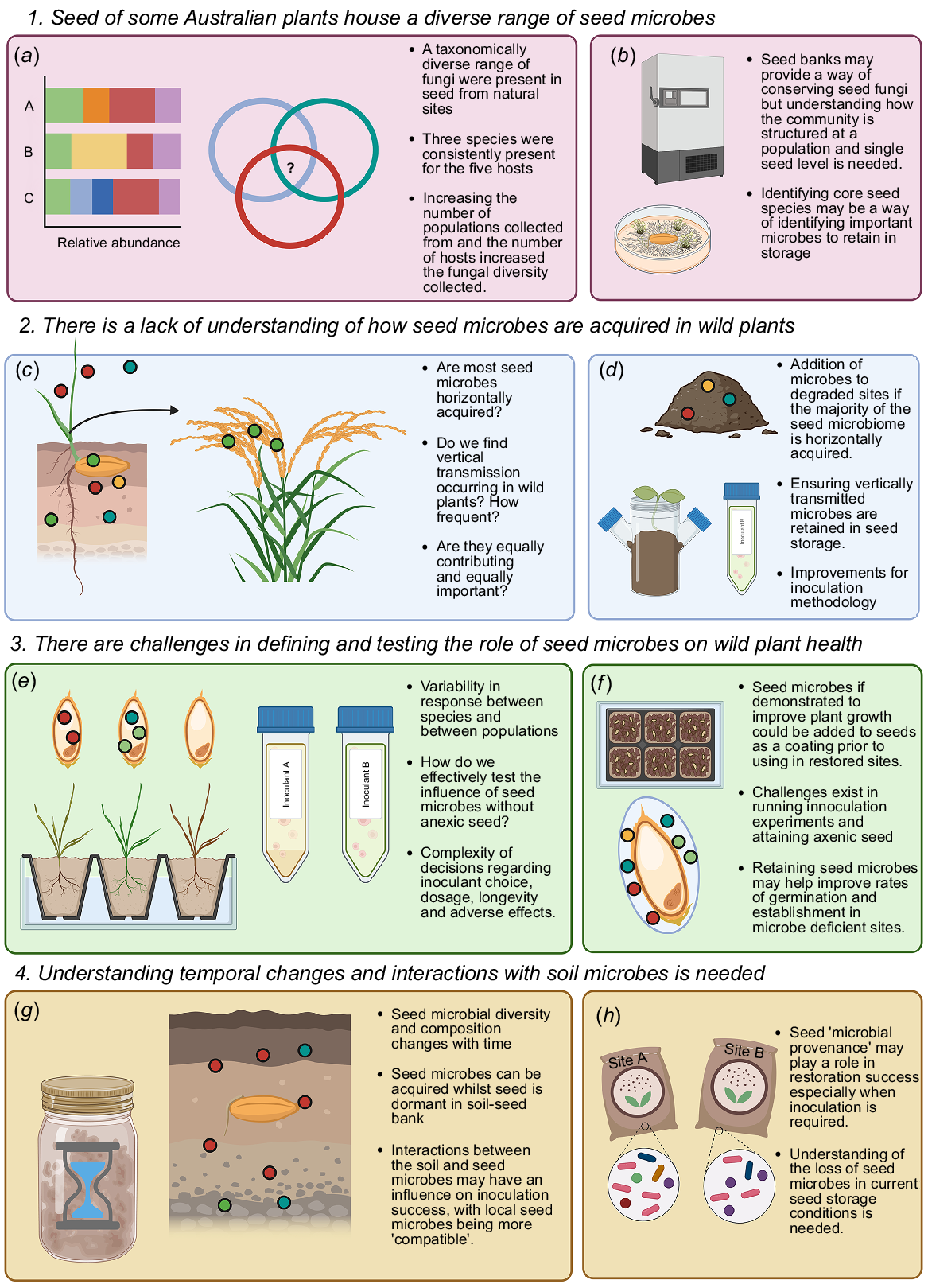
(a) Different spheres of a plant where microbes such as fungi and bacteria can exist as endophytes forming part of the plant holobiont, (b) different components of a seed within which seed endophytes can occur, (c) seed endophytes can also occur between the cells of the seed tissue.
Endophytes can occur within all tissues of a host plant, with the diversity, composition, and function of microbiomes of seed the least known of all the assemblages occupying different plant organs (Shade et al. 2017). Yet, seeds contain the embryo that gives rise to the dominant sporophyte generation and specialised tissue to protect and sustain the early development of the new generation. Seeds are either contained within fruit that facilitates dispersal or have special structures such as wings to aid dispersal, thus being the primary mechanism for the plant to migrate and colonise new areas. Furthermore, seed can remain dormant in the community in a soil seed bank or within a canopy in the case of serotinous seed, waiting for suitable conditions to release and germinate.
Research has shown that a taxonomically diverse range of bacteria and fungi occur within all the tissue types within seed (Truyens et al. 2015; Verma and White 2019; Zheng and Gong 2019). Biodiversity and Ecosystem Functioning hypotheses (Loreau et al. 2001) suggest that by soil and plants having a diversity of microbes, they harbour a wider range of potential functions, and this higher functional diversity allows for them to respond to a wider range of potential abiotic and biotic stressors (Loreau et al. 2001; Prosser et al. 2007; Saleem et al. 2019; Taylor et al. 2020; Morales Moreira et al. 2023). Supporting this hypothesis, research has found that a diverse seed microbiome provides disease resistance against soil pathogens and other pathogenic organisms in Oryza sativa (Verma et al. 2018; Matsumoto et al. 2021), Solanum lycopersicum (Morella et al. 2019), Brassica napus (Rybakova et al. 2017) and the invasive species Phragmites australis (White et al. 2018). Field experiments showed that Triticum aestivum seeds under drought conditions had a more diverse and higher number of culturable endophytes than did seeds from plants under rainfed conditions (Hone et al. 2021). Likewise, a high fungal diversity was significantly correlated with high seed viability and germination in Musa spp. seeds (Hill et al. 2021). Furthermore, greenhouse inoculation studies have shown that the addition of a more species rich microbial consortia improves plant growth more than less diverse or even single-species inoculants in Triticum aestivum (Sahib et al. 2020) and Solanum lycopersicum (Singh et al. 2015).
As seed also provides a vector for seed-borne pathogens (Sheppard 1998; Goko et al. 2021; Martín et al. 2022), phytosanitary procedures are aimed towards the use of physical and chemical treatments of seeds to remove seed microbes (Mancini and Romanazzi 2014), resulting in changes in the composition of the seed microbiome (Solanki et al. 2019). Recent studies have raised concern regarding this practice because of the loss of beneficial seed endophytes (Ayesha et al. 2021), resulting in reduced seedling vigour (Vasanthakumari et al. 2019), reduced emergence (You et al. 2020) and reduced germination and survival (Leroy et al. 2019). Some seed endophytes can show benefits to their host plant, especially during early lifecycle stages (Shahzad et al. 2018). Many plant species used in restoration have low rates of germination and establishment (Merritt et al. 2007; Turner et al. 2013; Erickson 2015; Erickson et al. 2016); the diversity of microbes isolated from seed begs the question whether this challenge may be overcome by considering seed endophytes and their roles during these early stages of restoration, similar to how seed microbes are utilised in agriculture for improving seedling growth (Cardarelli et al. 2022; Paravar et al. 2023), as a herbivory deterrent (Czarnoleski et al. 2010; Laihonen et al. 2022) and to provide resilience against drought (Hubbard et al. 2012; Abideen et al. 2022; Siddique et al. 2022) and salt stress (Walitang et al. 2018; Desai et al. 2023).
The seed microbiome may comprise a flexible and a core microbiome, with the latter being defined as taxa that are consistently abundant and prevalent across individuals and populations, providing key components of the composition of the microbial community (Shade and Handelsman 2012). Flexible taxa, the microbial taxa adapted to specific local constraints, contribute to the seed microbiome and are reflective of the surrounding site, from where they can be acquired horizontally from the surrounding soil and as aerosols from the surrounding air (Abdelfattah et al. 2023).
Identification of ‘core’ seed microbiota can help identify microbial taxa and functions that may be particularly important for host fitness (Simonin et al. 2022). Identification of core seed microbes may assist restoration and conservation work in highlighting what species are particularly important to retain in seed-bank collections of these hosts, or they may provide good candidates when inoculation or transplantations is required to restore microbes to a degraded site. By identifying the core microbiome and identifying which of the core is possibly lost during seed processing and storage, we may be able to augment the seed with those lost. We understand that seed storage is crucial to maintain seed viability, but we do not have any knowledge whether some of the microbes lost during the storage period may actually be able to help germination and early establishment.
Seed plays an important role in conservation and restoration of degraded lands, with seed forming the basis of many large-scale restoration and conservation projects. Seed used in restoration can either be harvested from natural sites, regenerated sites or from seed-production areas (SPAs), where seed is grown in bulk for large-scale restoration projects (Nevill et al. 2016). Following harvesting, the seed can undergo processing and storage, depending on various factors, seed longevity and demand (Frischie et al. 2020). Long-term storage of seeds in seed vaults or seed banks is the basis of ex situ collections for many orthodox native Australian species (Richardson et al. 1997). Conservation of microbial diversity through seed banks as ‘incidental’ fungal banks has been proposed (Berg and Raaijmakers 2018; Hill et al. 2021). However, utilising seed banks as incidental fungal banks requires seed-sampling strategies that also represent seed microbial diversity. With this in mind, we wonder what ‘luggage’ the seed carries with it. Can it help to establish the next generation of plants? Given the importance of seed in conservation and restoration efforts, should we consider the seed microbiome in the steps preceding the utilisation and storage of these propagules?’
In this paper, we searched the literature for what is known about seed microbiomes of native species, particularly for Australian species, that could facilitate restoration and conservation. We sought to help direct new research to enable Australia to develop a restoration programme that maximises the conservation of plant species, incorporating appropriate seed microbiomes to facilitate that conservation. We also present some preliminary results from our on-going research into Australian native species to determine whether a core seed microbiome might be present for all study species, or for particular taxa, because this could focus microbiome conservation. We investigated variability in microbiomes among closely related and distantly related taxa and also investigated whether sampling at different sites resulted in similar seed microbiomes for a particular taxon. Finally, using the literature and our preliminary results, we identified key knowledge gaps that we believe are applicable to restoration and conservation.
Materials and methods
We undertook a literature search using the database Scopus (Elsevier, Atlanta, USA) and the search term ‘seed endophytes’ OR ‘seed microbiome’ by using the abstract, title and keyword limiter, as at September 2022. We summarised the published information by the host plant species and the origin of the seeds used (wild, cultivated, germplasm collection). We focused on wild seed, particularly seed of Australian native species. We were particularly interested in information that would contribute to restoration activities.
We also isolated and identified seed endophytic fungi from the following five Australian native plants: Banksia ericifolia (Proteaceae), B. serrata (Proteaceae), Petrophile pulchella (Proteaceae), Microlaena stipoides (Poaceae) and Themeda triandra (Poaceae), following the methods of Mertin et al. (2022). Seed were collected from B. ericifolia, B. serrata and P. pulchella during June–August 2021 and for T. triandra and M. stipoides in December to late January 2022. The two grass species are commonly used in restoration and while seed are readily available from commercial suppliers, the work here was conducted on wild-collected seed. Wild seed was collected from serotinous cones from the three Proteaceae species. Apart from P. pulchella, where only one site was sampled, mature seed were collected from multiple sites and, at each site, 10–12 maternal plants were sampled. To isolate fungal endophytes, seeds were serially immersed in 4% bleach for 10 min, distilled water for 2 min, 70% ethanol for 5 min, then a final rinse of distilled water for 2 min, thrice. Each surface-sterilised seed was plated onto 1/4-strength potato dextrose agar (PDA) medium in 60 mm Petri dishes and dark incubated for 28 days at 25°C. Efficacy of the surface sterilisation method was tested by the plate imprint method of Reissinger et al. (2001) by using an arbitrary subset of 15 seeds from each host at each site. This involved pressing the surface of each tested sterilised seed onto 1/4-strength PDA containing 2% lactic acid and incubating it in the dark for 7 days. Only seeds where no growth was observed on the agar plate were surface sterilised effectively. Unique morphotypes were pure-cultured via hyphal tipping and DNA was extracted from the mycelium of every fungal isolate using the DNA Fast Prep kit (Q-biogene Inc., CA, USA) as per the manufacturer’s instructions. Approximately 700 base pairs of the internal transcribed spacer (ITS) region of rRNA were amplified using the primers ITS1F/ITS4 (Gardes and Bruns 1993) and Sanger-sequenced following the methods of Mertin et al. (2022).
The resultant dataset contains the ITS sequences for 2069 pure-cultured isolates. These isolates were clustered into operational taxonomic units (OTUs) by using the software CD-HIT est. (Li and Godzik 2006) on the basis of 97% sequence similarity. Taxonomy was assigned to each OTU on the basis of percentage identity scores by using the NCBI BLASTn plugin in Geneious v.11.1.5 against the UNITE database (Abarenkov et al. 2010). We used this dataset to ascertain the taxonomic breadth of seed endophytes in native seed from the five plant species and identified core and shared taxa among the hosts by undertaking a heatmap analysis by using Bray Curtis dissimilarity measures, implemented in the software R v.4.1.2, (R Core Team 2021) using the package Phyloseq v.1.38.0 (McMurdie and Holmes 2013).
As seed banks have been proposed as ‘incidental fungal banks’ (Berg and Raaijmakers 2018; Hill et al. 2021), we used our data to investigate what strategies may affect the fungal diversity incidentally collected through seed banking. We asked whether sampling of additional sites of a host species and sampling additional hosts at a site captures additional seed microbial species. We analysed a subset of our dataset, comprising fungal species from the seed of Banksia serrata, B. ericifolia and Petrophile pulchella collected from multiple individuals from four, six and one site respectively. All three hosts co-occurred at the Royal National Park site (RNP), with the two banksias co-occurring at most sites. The grasses were not included in this analysis, because they did not co-occur with the other host species.
Results
The literature provided only a small window into the diversity of seed endophytes of native Australian species (Supplementary material Table S1). Knowledge from natural ecosystem hosts is less common than for agricultural species, with few studies looking at Australian native host species in particular. Of the 141 publications identified through our Scopus search, 81 were about agricultural species, with Zea mays, Triticum aestivum, Oryza sativa and Hordeum vulgare comprising 32 of the 81 publications (Table S1). Of the 31 papers on wild species, six were focussed on wild-crop relatives, three were metal- or salt-accumulating plants, and seven were from invasive species. Two papers identified seed endophytes from a range of species stored in seed banks. Of the peer reviewed papers surveying wild species, four investigated endophytes from Australian native plant species, namely, Glycine clandestine, a wild soybean relative, (Chandel et al. 2022) and the aquatic seagrass Halophila ovalis (Tarquinio et al. 2021), our own work on the Australian endemic genus Banksia (Mertin et al. 2022) and the work of Aldous et al. (1999) on a range of native Australian grasses. Our work on B. serrata and B. ericifolia, where seeds and maternal lines were kept separate, found that culturable fungi were present across all sites, individuals and species (van der Merwe 2021; Mertin et al. 2022).
We identified the following knowledge gaps in our literature search that have implications to seed conservation and restoration:
An absence of papers from wild species addressing the transmission pathways of fungi to seed.
Despite the beneficial effect of seed microbes being shown for many crop plants, we found only a few papers testing plant-growth benefits for wild species, with none of these on native Australian plants.
Few studies have looked at the interactions between the seed and soil microbes during the period of dormancy, where the seed is within the soil seed bank and the effect of seed provenance on these interactions.
Most studies gathered information only at one time point, limiting our knowledge of seed fungal dynamics and temporal patterns. This is particularly important, because seeds can lay dormant as soil seed banks or within seed vaults for long periods of time.
For the five Australian native species we studied, we found a diverse range of seed fungi, belonging to multiple phyla (Table S2) The majority of the 161 fungal species we identified from the five hosts belonged to Ascomycota, with Sordariomycetes, Dothideomycetes and Eurotiomycetes the dominant fungal classes. The Basidiomycota, Mucoromycota and Mortierellamycota were present but less diverse. In particular, Basidiomycota was represented only by two classes, Ustilaginomycetes and Agaricomycetes. Species of Pencillium, Banksiamyces and Alternaria were the most abundant fungal genera present (Table S2). Nineteen species of Penicillium were found among the five hosts, with P. glabrum, P. citreonigrum and P. citrinum being the most abundant.
More species of fungal seed endophytes were collected as we sampled more hosts at a site. At the Royal National Park site (RNP), we identified 19 fungal species from 10 individuals of P. pulchella (Fig. 2). With the addition of seeds from B. serrata (from the same family but different genus), we identified an additional 13 fungal species. With the addition of a third host B. ericifolia (same family), we again identified 11 fungal species not identified in the previous two hosts. However, when considering the abundance of specific fungal taxa, we found that the highly abundant fungal species of the two banksias are very similar but different from that of Petrophile pulchella. (Fig. 2), suggesting that perhaps host relatedness plays a role in structuring site communities and that abundant fungal taxa may be captured through sampling seed from close relatives. These results indicated that by sampling seed of multiple hosts at the same site, we are more likely to capture the fungal seed microbiome of a site.
Bar plots indicating the relative abundance of fungal species among sites (RNP, MTB, PTG, BLK, BBY and LNC) of each host (Petrophile pulchella, Banksia serrata and B. ericifolia) and among hosts at a site (RNP). Each rectangle within a column shows the relative abundance of that fungal species as a percentage of total fungal species for that sampling group (column). Orange rectangles indicate new seed fungal species identified when (a) additional host species were sampled at Royal National Park (RNP). Green represents when (b) additional sites of B. serrata were sampled and blue represents when (c) additional sites of B. ericifolia were sampled. Arrow shows the order of site and host sampling.
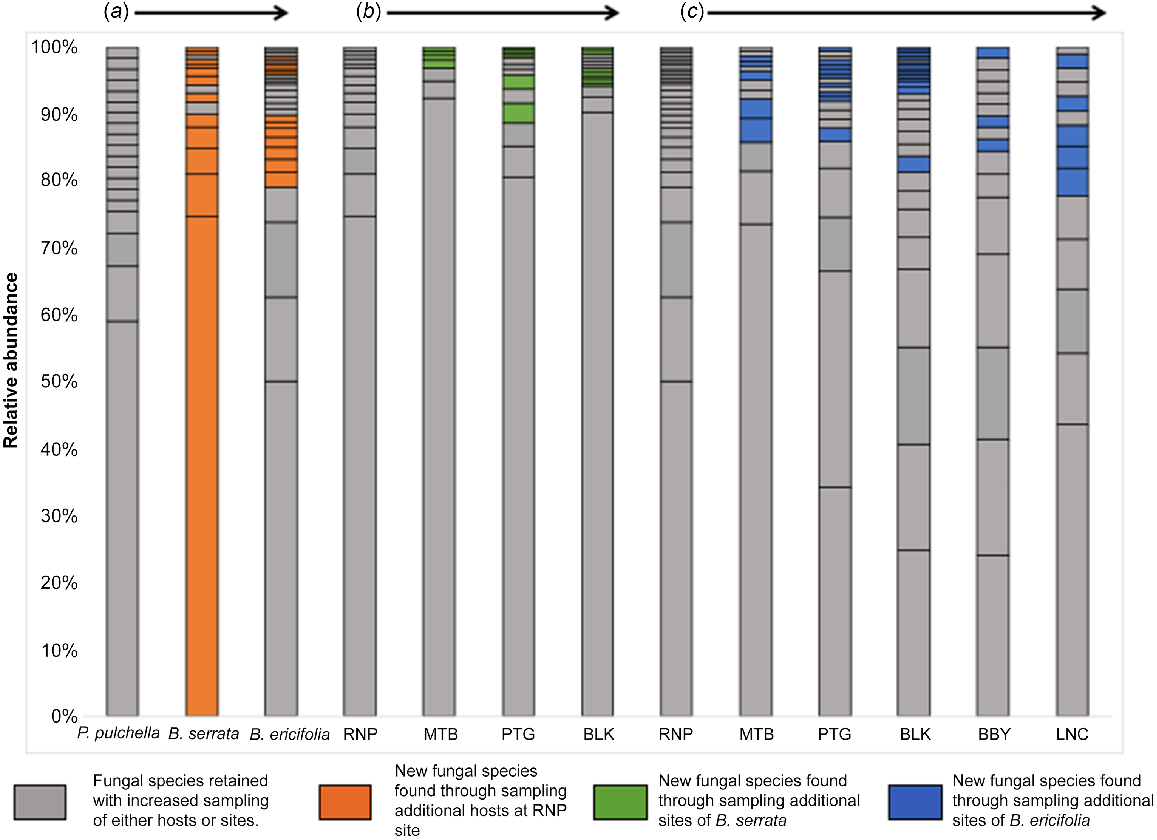
We also investigated whether we capture more seed fungal species by sampling multiple sites of a host. We found that for B. serrata, new fungal species were discovered by sampling additional sites (Fig. 2). However, additional taxa from new sites were largely rare taxa, with the more abundant fungal species present at all sites. For B. ericifolia, our analyses indicated that even after four sites, new fungal species were uncovered, although, as within B. serrata, most of the highly abundant taxa are captured with sampling of only two sites. Thus, if we want to capture the complete fungal seed microbiome of a species, we need to sample seed from multiple sites.
Among the 161 fungal species identified across the five host species, three fungal species were shared, namely, Pestalotiopsis knightiae, Talaromyces chloroloma and Penicillium glabrum. As T. chloroloma was found in very low abundance (<0.25% of all sequences for each host) in the two banksias, we suggest that this is not a core species; however, the more abundant P. knightiae and P. glabrum are suggested to comprise core species for all five study plant hosts (Fig. 3). Furthermore, both grasses shared 14 taxa, with Alternaria alternata, Epicoccum sorghinum, Sarocladium strictum and Diaporthe eucalyptorum comprising the core microbiome because these taxa were consistently found in high abundance in these hosts.
Heatmap showing the relative abundance (as a percentage of total isolates for each host species) of each of the fungal species identified (rows) among each of the five host species of Banksia serrata, B. ericifolia, Themeda triandra, Microlaena stipoides and Petrophile pulchella (columns). Fungal species found across all hosts have red taxon labels, those shared between the two grasses are green, and those shared between the two banksias are orange.
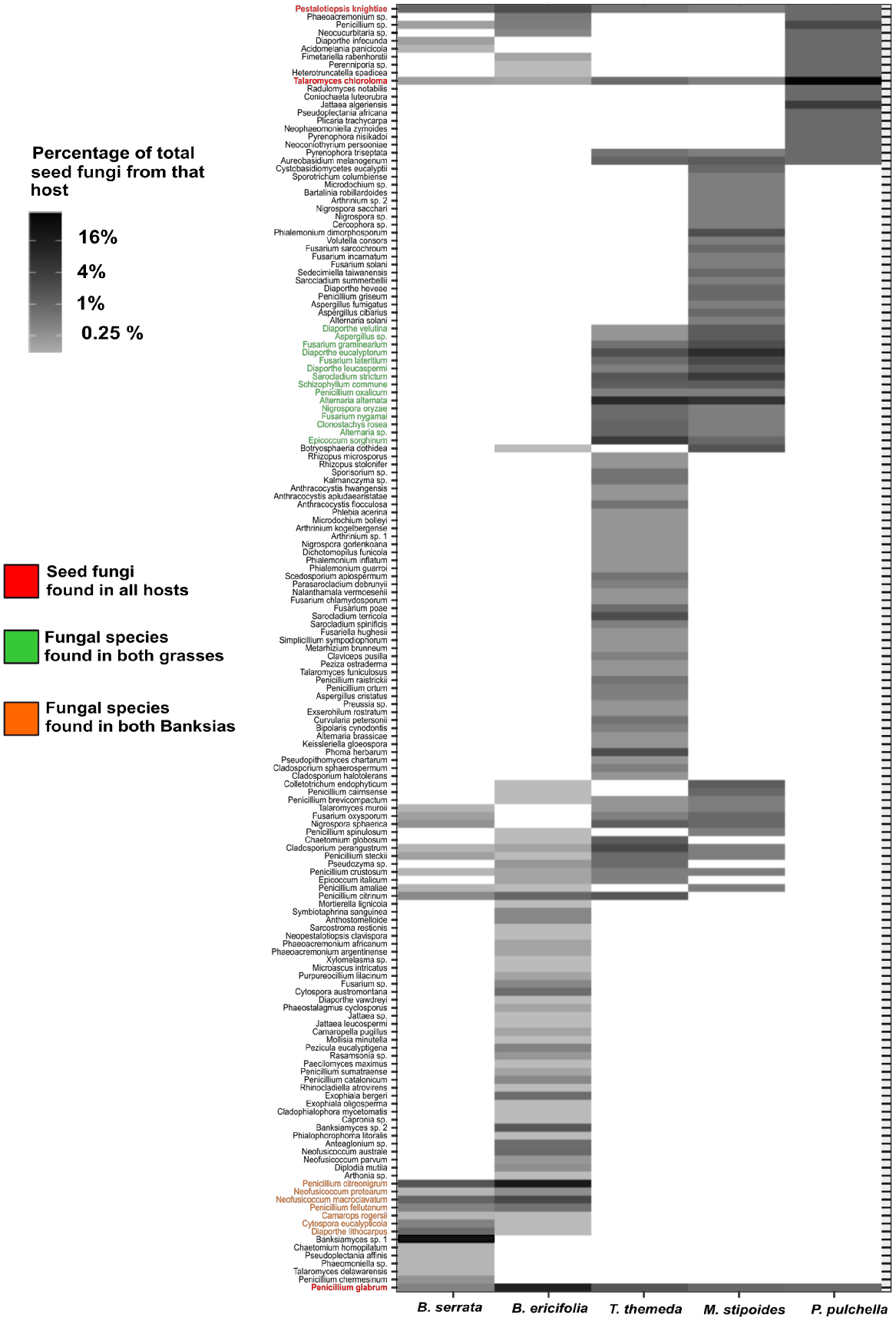
We did not identify a core microbiome shared among the three woody hosts of B. serrata, B. ericifolia and P. pulchella. Although Petrophile pulchella had nine taxa in common with B. ericifolia, they were in low abundance in B. ericifolia and, as such, are not core species of both these hosts. Seven fungal species were shared between the two banksias, which were also absent from the grasses. In particular, P. citreonigrum and Neofusicoccum macroclavatum were found in both banksias in high abundance and prevalence, and, as such, would be considered core taxa (Fig. 3).
Discussion: implications for restoration and conservation
Preliminary findings for some native Australian plants
In our work, we identified a taxonomically diverse range (161 species) of fungal taxa cultured from the seed of five native Australian plants collected from natural bushland areas. The fungi represented a range of different genera, although were dominated by species of Penicillium, Banksiamyces and Alternaria. Penicillium species have been associated with producing mycotoxins such as citromycetin (Grove and Brian 1951), citroviridin (Nishie et al. 1988) and citrinin (Flajs and Peraica 2009) and, hence, have been thought to play a key role in defence against pathogens. They also have been associated with causing seed disease and decay (Duduk et al. 2017).
Likewise, species of Banksiamyces and Alternaria have been associated with causing decay in lignified tissue of the host plant (Beaton and Weste 1984; Sharma et al. 2016), and some species of Alternaria have been associated with causing disease on cotton (Le and Gregson 2019), pistachio (Ash and Lanoiselet 2001) and canola (Al-Lami et al. 2019). In other instances, Alternaria has been associated with promoting plant growth through increasing root growth (Zhou et al. 2018) and increasing seed size (Lindblom et al. 2018). Banksiamyces has only recently been isolated from seed (Mertin et al. 2022), being previously known only as a saprophyte on senescent Banksia infructescences (Beaton and Weste 1982). Having this diversity of fungi and a diverse range of roles (pathogens, saprotrophs and beneficial) within the population may provide a complementarity of traits, which on the basis of biodiversity and ecosystem functioning hypotheses, may assist with responding to a range of potential abiotic and biotic stressors (Loreau et al. 2001; Prosser et al. 2007; Saleem et al. 2019; Taylor et al. 2020; Morales Moreira et al. 2023).
The fungal species that we have isolated from native Australian plant seeds shared some taxa with those that are frequently isolated from crop species. A recent meta-analysis of seed microbes from 50 different plant species (Simonin et al. 2022) identified that species of Alternaria were dominant in multiple crop families, with Alternaria also being common within our dataset. Banksiamyces was dominant in the banksias, but was not identified within the meta analysis and may represent a genera of fungi that is a host specialist on banksias, as suggested by Sommerville and May (2006).
Conserving seed microbiome diversity, whether in seed banks or retaining it through seed production area procedures also requires some understanding of how the communities are structured. As many of our analyses were focussed on comparing the seed fungi present between the different host species and among different populations of a host species, further work is required to understand how seed fungal communities are structured at the single-seed level. Studies of seed from both crop and natural ecosystem hosts have identified that only a few culturable fungal species are present within each seed. Mertin et al. (2022), in Banksia seed, found that 97% of seeds contained only one culturable fungus, whereas studies of individual bean seeds using culture-independent approaches identified that multiple fungal species belonging to at least three different fungal families were present in individual seeds (Bintarti et al. 2022). The differences in results by using these techniques suggests that perhaps a proportion of seed endophytes are not culturable or are slow-growing, meaning that faster-growing species may outcompete them in culture. It may also suggest that different plants house different communities and different sizes of community.
The first step in integrating native Australian seed microbiome knowledge into restoration efforts requires an understanding of the prevalence and diversity of seed endophytes within native Australian species that are frequently used in restoration. Having ‘baseline data’ of how prevalent seed fungi are, what fungal taxa are present and similarities and differences among multiple populations of a host in these communities, will allow for us to be more informed about the inherent variability of these communities, resulting in better decisions in developing appropriate restoration programs.
Currently, seed collection guidelines for seed-bank storage and restoration purposes advocate collecting a genetically diverse seed collection of a plant species to help improve the success of restoration and improve the future adaptive potential of plantings (Broadhurst et al. 2008; Bragg et al. 2022). The number of populations and individuals that are sampled are based on the dispersal distances of that species, or the genetic distances between populations (Harrison et al. 2021). Results from this study suggest that we need multiple sites of a host to document the complete diversity of seed microbial communities, although sampling of only a few sites captures the highly abundant endophytes.
There were the following three fungal species that were shared among the five host species: Pestalotiopsis knighteae, Talaromyces choloroloma and Penicillium glabrum. Pestalotiopsis knighteae was first described from isolates from Knightia. Pestalotiopsis is a species-rich genus that is widely distributed throughout tropical and temperate regions (Maharachchikumbura et al. 2014) and is commonly isolated as a pathogen and an endophyte. Talaromyces chloroloma was first isolated from soils of the South African fynbos (Visagie and Jacobs 2012), and Penicillium glabrum is known also as a ubiquitous fungus, commonly associated with soil, decaying organic matter and as storage rots of seeds or pathogens of fruit and vegetables (Barreto et al. 2011). In our previous work solely looking at B. serrata and B. ericifolia, we identified core fungal species that were present and abundant within the four populations of each host sampled (Fig. 4), although the two hosts shared few fungal species overall, despite them co-occurring at a site. Each host had unique core microbes, and a diversity of other microbes that were sometimes unique to that site.
(a) Column graph showing the percentage of total fungal isolates that were assigned to each fungal species from the seed of Banksia ericifolia and B. serrata from the following four sites in the greater Sydney region: Blackheath (BLK), Mount Banks (MTB), Patonga (PTG) and Wattamolla (RNP). Each fungal species is represented by a different colour. (b) An isolate of the core community of B. ericifolia, assigned as Penicillium glabrum, and (c) an isolate of the core community of B. serrata, assigned as Banksiamyces sp. 1. Photographs by A. Mertin.
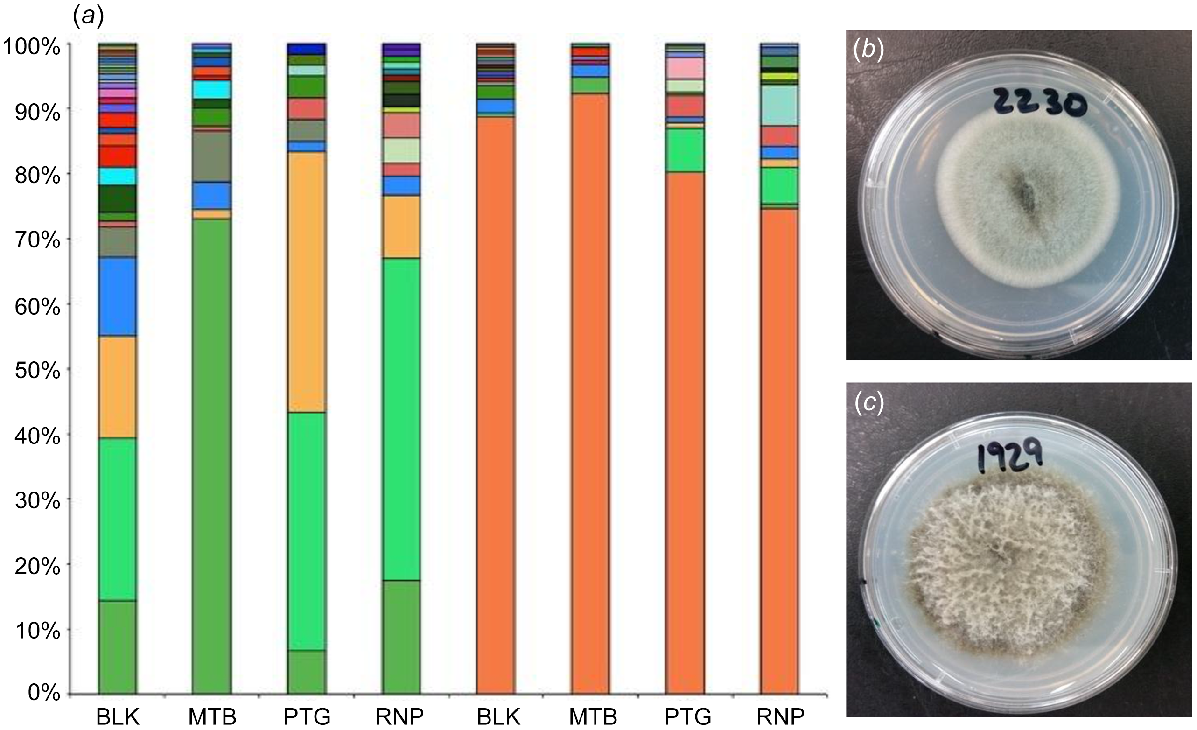
Because it may not be possible to conserve all of the seed diversity via seed banking, with studies demonstrating that seed storage methods result in the loss of microbes (Chandel et al. 2021; Dutta et al. 2022; Martín et al. 2022), identifying these core microbes may direct restoration efforts into what fungal species to focus on conserving. These microbes may be of particular value when using the direct seeding approach in degraded soils where the microbiome is poor, because the seed would provide the only source of microbes for the seedling until the aboveground parts of the plant have grown. There are likely to be trade-offs in just focussing on retaining the core fungal species in seed collections, because rare and transient taxa may provide functions that supplement or fulfil particular niches in the community (Chen et al. 2020). (Fig. 5a and b)
Gaps in our understanding of seed microbiomes
Our data indicated that seed from healthy ecosystems may carry with them a diverse fungal community, but our data do not provide any information on how this community established within the seed. Our literature review also identified a lack of studies on how fungi are transmitted to wild seed. Understanding how plants acquire microorganisms, in particular how the microbes get into the seed and where they localise, can help design better ways to inoculate seeds with microbes that may provide an alternative approach to adding soil microbes to degraded sites in restoration. Understanding whether endogenous or exogenous microbes are the main contributors to the seed microbiome may also inform how important retaining each of these contributing factors is in the maintenance of a diverse microbiome for restoration projects (Fig. 5c, d).
The following three main pathways have been reported for how seed microbes can be acquired in crop plants: (i) vertically via transfer of the microbiota from vegetative tissues to the ovule and then to the developing embryo (e.g. Epichloë in grasses; Liu et al. 2017), (ii) vertically through floral pathways via the stigma of maternal plants (Hodgson et al. 2014; Nelson 2018), which is utilised as a way to introduce beneficial microbes to progeny seed (Mitter et al. 2017), and (iii) horizontally whereby seeds are colonised from the external environment via fungal spores and propagules (e.g. Brassica napus seed; Morales Moreira et al. 2021). This can include via wind as aerosols or, more commonly, direct colonisation from soil microbes when the seed is dormant in the soil seed bank (Gallery et al. 2007; Klaedtke et al. 2016; Dai et al. 2020). The relative proportion of species transmitted vertically to horizontally is largely unknown, although studies have indicated that most of the microbiome of crops is assembled horizontally from the surrounding environment and only a subset is vertically transmitted (U’ren et al. 2009; Sarmiento et al. 2017; Abdelfattah et al. 2023). In our comparisons of the fungi cultured from the five host species, we found that very few taxa were shared. In particular, among the three hosts that co-occurred (B. serrata, B. ericifolia and P. puchella), we found no ‘core’ taxa, although two core taxa were shared between the banksias. Having few taxa shared among hosts at a site suggests that perhaps a large proportion of the seed fungi are maternally inherited or that particular aspects of a host’s life history or physiology may dictate what fungi colonise the seed. The similarity in taxa between the two banksias suggests that host life history and perhaps relatedness may play a role in this and provides an avenue for future research.
Strict maternal vertical transmission during floral and early seed development is seen as rare as fresh seeds typically contain few fungal species (Ridout et al. 2019). Despite this, vertical transmission, where a subset of the initial seed microbiome is still present after multiple generations of seed, has been identified in multiple monocot and dicot crops, such as radish and bean (Chesneau et al. 2022), wheat (Vujanovic et al. 2019), rice (Kim et al. 2022) and the model plants Arabidopsis thaliana and Brachypodium distachyon (Johnston-Monje et al. 2021). Endophytes that are vertically transmitted from parent to offspring during seed development may have greater influence on offspring because these associations have the potential to persist throughout the seedling and the adult lifestyle stages (Liu et al. 2013). Therefore, identifying vertically transmitted members of the seed microbiome, and ensuring that they are retained through seed processing and storage methods used in restoration activities may help improve plant growth and have these beneficial microbes be passed on to future generations of plants and their progeny.
Our literature review showed that within crop species, seed microbes play an important role in the spermosphere (the soil and microbes around a dormant or germinating seed; Nelson 2004), breaking seed dormancy (Delgado-Sánchez et al. 2011), reducing seed predation (Zhang et al. 2012), promoting germination (Philipson and Christey 1986; Clay 1987; Delgado-Sánchez et al. 2013) and promoting seedling establishment (Wäli et al. 2009). Where some species are seen as beneficial to their host, other endophytes have also been implicated as latent pathogens and latent saprotrophs causing disease and decay during times of host stress or senescence (Carroll 1988). As such, seed endophytes play a broader role in the health of the ecosystem. Saprotrophs play a role in degrading organic matter, which then can be recycled within the ecosystem.
The role of seed microbes in seedling survival and performance in wild plants has also been demonstrated, with seeds containing microbes that contribute to drought tolerance (Jeong et al. 2021; Lactuca serriola), seedling germination and development (Sánchez-López et al. 2018; Shearin et al. 2018; Crotalaria pumila and Phragmites australis), increasing shoot growth (Choi et al. 2022; Capsella bursa-pastoris), and improved cadmium tolerance, increased seed yield, total chlorophyll content, antioxidant production and lipid peroxidation (Parmar et al. 2022; Dysphania ambrosoidoides). Many of these studies have data only for the juvenile stages of the plant and, as such, the long-term effects on plant growth into the adult stages are still unknown. Results from our study indicated that the fungi found in high abundance within our dataset (Penicillium glabrum, Alternaria alternata and Banksiamyces) have been reported in the literature as showing a wide range of roles within their host. For example, Penicillium glabrum has been associated with increased plant growth in low-nutrient environments (Khan et al. 2008), but also with seed decay and disease (Barret et al. 2016) and, so, the potential role of these seed endophytes may vary depending on the host plant and the environmental context. Future research into experimentally testing the strain variation in plant growth promoting potential and disease potential of fungi from native seed is needed.
There are experimental challenges in understanding the role of seed microbes. When a comparative design is being undertaken, comparing the growth of seedlings with or without seed microbes, getting axenic seed to use as a negative control treatment without affecting germination or growth is challenging. It is often achieved through the use of immersion in bleach (Verma and White 2019) or through the use of fungicides and antibiotics (Leroy et al. 2019; Vasanthakumari et al. 2019), but in these instances it is acknowledged that there is no guarantee of microbe-free seed. Even seed produced in hermetic microcosms still had vertically transmitted microbes present (Johnston-Monje et al. 2021). Inoculation experiments are particularly challenging as variables such as the inoculant species and diversity, cell concentration, inoculation method and maintaining inoculum viability when coating or pelleting a seed can all affect the experimental outcome (Paravar et al. 2023). Undertaking experiments in field trials introduces an additional range of abiotic (Lopes et al. 2021) and biotic factors, such as the resident soil microbes that need to be considered (O’Callaghan et al. 2022) (Fig. 5e and f).
Microbial interactions between soil and seed microbes that occur on and around a germinating seed play an important role in how a plant acquires its initial microbiome (Schiltz et al. 2015). In restored sites (especially those where emphasis on reinstating organic matter and soil microbes are not emphasised), there is not always a soil microbiome to ‘grab’ from, and hence the seed microbiome forms the only source of microbes for the seedling until the aboveground parts of the plant are present (Araújo et al. 2014; Banerjee et al. 2019). Similarly, the surrounding vegetation can be so depauperate that horizontal transmission is absent (Henning et al. 2021). As with seeds, soils also contain both beneficial and detrimental fungi (Morgan et al. 2005). Some fungi within seed can directly protect the seed from soil pathogens by producing secondary metabolites and chemicals, or protect indirectly by competing for space and nutrients within the seed (Matsumoto et al. 2021), whereas others are associated with causing disease (Baker and Smith 1966). Hence, having a diverse range of microbes present in the mature seed used for plantings may be advantageous.
It is not known what effect using local versus non-local seeds may have on the plant microbiome. Introducing ‘non-local’ seed microbes to a resident soil microbial community may have an effect on the seed–soil microbe interactions, with the potential for flow-on effects to the seedling microbiome. This concept is supported by studies that have shown that inoculating soil with local soil microbes can be more beneficial to seedling growth than is inoculating with a non-local beneficial mycorrhizal inoculant (Paluch et al. 2013; Dadzie et al. 2022). Soil microbes can have a strong influence on seed microbes, and it has been suggested that using local soil from the location of the source plant can enhance the conservation of the seed microbiota (Chandel et al. 2022). This suggests that microbial provenance plays a role in seed–soil microbial interactions and plant-host benefits.
Understanding the implications of seed microbe provenance and seed and soil microbial interactions may contribute to the field of seed enhancement technologies. Microbes isolated from seed may perform better than soil microbes because they can withstand the physio-chemical changes that happen in the seed during germination, with some having the ability to translocate to other parts of the plant (Abdelfattah et al. 2023) or be vertically transmitted (Barret et al. 2015). Bio-priming or coating seeds with seed-borne microbes could be incorporated into the suite of seed-enhancement technologies (Pedrini et al. 2020) already being tested in large-scale direct seeding applications. Understanding whether seed-borne microbes are ‘compatible’ with local soil microbes would also contribute to increased success using this approach, because soil inoculations can often fail due to the resident soil microbes effect (O’Callaghan et al. 2022; Fig. 5g, h).
Long-term seed storage may occur as natural canopy-held seed banks or soil seed banks. In these natural settings, over 90% of available seeds are faced with predation from vertebrates and insects (Kolb et al. 2007; Dalling et al. 2011), while also dealing with soil-borne pathogenic fungi (Kluger et al. 2008). It is from these natural settings that much of our understanding of how endophyte communities change over time comes. It has been suggested that an increase in endophyte diversity with age may be due to a large part of the endophyte community being horizontally transmitted, and this diversity accumulating over time (Arnold and Lutzoni 2007). This is demonstrated by endophytic communities differing in both abundance and diversity between canopy-held young and old Banksia ericifolia seeds (van der Merwe 2021).
Our literature review revealed that little is known about the extent of seed microbial change over extensive periods of time within the soil seed bank. Temporal changes in microbiomes that occur while seed is stored in the natural soil seed bank have restoration implications. This is a consideration during mining operations where topsoil containing the soil seed bank is stored unplanted for extended periods of time (Moreira-Grez et al. 2019), resulting in the decline of carbon content and a shift in the composition of the soil microbiome (Golos and Dixon 2014; Muñoz-Rojas et al. 2016). The implications of these changes could significantly influence the composition of the seed microbiome, although this change remains unknown. Identifying conserved species in seeds as they age may advance our understanding of age-related viability of seeds and survival potential. This may aid in restoration and conservation efforts, shifting the focus to important microbes, which need to be retained during storage.
Long-term exsitu storage conditions of most seeds focus on increasing the longevity of seeds by reducing storage temperatures and seed moisture content (De Vitis et al. 2020). Seed endophyte Epichloë viability diminishes significantly with high temperature and humidity levels, as well as with length of storage (Rolston et al. 1986; Gundel et al. 2009; Hume et al. 2013). Shifts in the microbial composition during storage favours the retention of particular microbes able to withstand those temperature and humidity conditions (Chandel et al. 2021). It is unknown what effect these changes have on the seedling microbiome and seedling survival and provides future avenues for research in this emerging research area (Fig. 5g and h).
Conclusions
Incorporating microbial diversity into restoration projects requires some understanding of how this diversity is structured, providing management units to better survey, monitor, conserve and utilise these hidden but ubiquitous communities to improve restoration outcomes. Our understanding of how seed microbial communities are structured, the role within their host plants, how the communities change over time and the best methods to retain their diversity under different storage methods are all key knowledge gaps currently within the field.
This paper provides a baseline reference system of the fungal endophyte diversity present within five native Australian species in a range of natural ecosystems, and provides a small glimpse into the hidden diversity in the seed of Australian natural ecosystems. The broad taxonomic diversity of the fungal species recovered within our work suggests that there is a large amount of hidden microbial diversity within Australian natural ecosystems, which we believe needs to be considered in exsitu seed storage, seed production areas, seed collection and processing methods, and direct seeding restoration projects. Building our knowledge on how these complex communities are structured, what their functions are and how they can change over time requires integration of knowledge from multiple disciplines such as microbiology, ecology, plant genetics and restoration ecology. Integrating this knowledge into conservation and restoration practices is an iterative process that will provide an insight into the hidden diversity within the seed of native Australian plants.
References
Abarenkov K, Nilsson RH, Larsson K-H, et al. (2010) The UNITE database for molecular identification of fungi – recent updates and future perspectives. The New Phytologist 186, 281-285.
| Crossref | Google Scholar |
Abdelfattah A, Tack AJM, Lobato C, Wassermann B, Berg G (2023) From seed to seed: the role of microbial inheritance in the assembly of the plant microbiome. Trends in Microbiology 31, 346-355.
| Crossref | Google Scholar |
Abideen Z, Cardinale M, Zulfiqar F, Koyro H-W, Rasool SG, Hessini K, Darbali W, Zhao F, Siddique KHM (2022) Seed endophyte bacteria enhance drought stress tolerance in Hordeum vulgare by regulating, physiological characteristics, antioxidants and minerals uptake. Frontiers in Plant Science 13, 980046.
| Crossref | Google Scholar |
Adam E, Bernhart M, Müller H, Winkler J, Berg G (2018) The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant and Soil 422, 35-49.
| Crossref | Google Scholar |
Afkhami ME, Rudgers JA (2008) Symbiosis lost: imperfect vertical transmission of fungal endophytes in grasses. The American Naturalist 172, 405-416.
| Crossref | Google Scholar |
Al-Lami HFD, You MP, Barbetti MJ (2019) Incidence, pathogenicity and diversity of Alternaria spp. associated with alternaria leaf spot of canola (Brassica napus) in Australia. Plant Pathology 68, 492-503.
| Crossref | Google Scholar |
Aldous DE, Isaacs S, Mebalds MI (1999) Endophytes in the genus Neotyphodium are not found in Australian native grasses. Australasian Plant Pathology 28, 183-186.
| Crossref | Google Scholar |
Alfonzetti M, Tetu SG, Mills CH, Gallagher RV (2022) Assessing the efficacy of extruded seed pellets and microbial amendments for native revegetation. Restoration Ecology 31, e13857 10.1111/rec.13857.
| Google Scholar |
Araújo ASF, Borges CD, Tsai SM, Cesarz S, Eisenhauer N (2014) Soil bacterial diversity in degraded and restored lands of Northeast Brazil. Antonie van Leeuwenhoek 106, 891-899.
| Crossref | Google Scholar |
Arnold AE, Herre EA (2003) Canopy cover and leaf age affect colonization by tropical fungal endophytes: ecological pattern and process in Theobroma cacao (Malvaceae). Mycologia 95, 388-398.
| Crossref | Google Scholar |
Arnold AE, Lutzoni F (2007) Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 88, 541-549.
| Crossref | Google Scholar |
Arnold E, Henk D, Eells R, Lutzoni F, Vilgalys R (2007) Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 99, 185-206.
| Crossref | Google Scholar |
Ash GJ, Lanoiselet VM (2001) First report of Alternaria alternata causing late blight of pistachio (Pistacia vera) in Australia. Plant Pathology 50, 803.
| Crossref | Google Scholar |
Ayesha MS, Suryanarayanan TS, Nataraja KN, Prasad SR, Shaanker RU (2021) Seed treatment with systemic fungicides: time for review. Frontiers in Plant Science 12, 654512.
| Crossref | Google Scholar |
Baker KF, Smith SH (1966) Dynamics of seed transmission of plant pathogens. Annual Review of Phytopathology 4, 311-332.
| Crossref | Google Scholar |
Banerjee S, Walder F, Büchi L, Meyer M, Held AY, Gattinger A, Keller T, Charles R, van der Heijden MGA (2019) Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. The ISME Journal 13, 1722-1736.
| Crossref | Google Scholar |
Barret M, Briand M, Bonneau S, Préveaux A, Valière S, Bouchez O, Hunault G, Simoneau P, Jacques M-A (2015) Emergence shapes the structure of the seed microbiota. Applied Environmental Microbiology 81, 1257-1266.
| Crossref | Google Scholar |
Barret M, Guimbaud J-F, Darrasse A, Jacques M-A (2016) Plant microbiota affects seed transmission of phytopathogenic microorganisms. Molecular Plant Pathology 17, 791-795.
| Crossref | Google Scholar |
Barreto MC, Houbraken J, Samson RA, Frisvad JC, San-Romão MV (2011) Taxonomic studies of the Penicillium glabrum complex and the description of a new species P. subericola. Fungal Diversity 49, 23-33.
| Crossref | Google Scholar |
Beaton G, Weste G (1982) Banksiamyces gen. nov., a discomycete on dead Banksia cones. Transactions of the British Mycological Society 79, 271-277.
| Crossref | Google Scholar |
Beaton G, Weste G (1984) A new species of Banksiamyces on Banksia saxicola (Proteaceae). Transactions of the British Mycological Society 83, 533-535.
| Crossref | Google Scholar |
Berg G, Raaijmakers JM (2018) Saving seed microbiomes. The ISME Journal 12, 1167-1170.
| Crossref | Google Scholar |
Bergmann GE, Busby PE (2021) The core seed mycobiome of Pseudotsuga menziesii var. menziesii across provenances of the Pacific Northwest, USA. Mycologia 113, 1169-1180.
| Crossref | Google Scholar |
Bintarti AF, Sulesky-Grieb A, Stopnisek N, Shade A (2022) Endophytic microbiome variation among single plant seeds. Phytobiomes Journal 6, 45-55.
| Crossref | Google Scholar |
Bragg JG, van der Merwe M, Yap JYS, Borevitz J, Rossetto M (2022) Plant collections for conservation and restoration: can they be adapted and adaptable? Molecular Ecology Resources 22, 2171-2182.
| Crossref | Google Scholar |
Broadhurst LM, Lowe A, Coates DJ, Cunningham SA, McDonald M, Vesk PA, Yates C (2008) Seed supply for broadscale restoration: maximizing evolutionary potential. Evolutionary Applications 1, 587-597.
| Crossref | Google Scholar |
Cardarelli M, Woo SL, Rouphael Y, Colla G (2022) Seed treatments with microorganisms can have a biostimulant effect by influencing germination and seedling growth of crops. Plants 11, 259.
| Crossref | Google Scholar |
Carroll G (1988) Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecology 69, 2-9.
| Crossref | Google Scholar |
Chandel A, Mann R, Kaur J, Norton S, Edwards J, Spangenberg G, Sawbridge T (2021) Implications of seed vault storage strategies for conservation of seed bacterial microbiomes. Frontiers in Microbiology 12, 784796.
| Crossref | Google Scholar |
Chandel A, Mann R, Kaur J, Norton S, Auer D, Edwards J, Spangenberg G, Sawbridge T (2022) The role of soil microbial diversity in the conservation of native seed bacterial microbiomes. Microorganisms 10, 750.
| Crossref | Google Scholar |
Chen Q-L, Ding J, Zhu D, Hu H-W, Delgado-Baquerizo M, Ma Y-B, He J-Z, Zhu Y-G (2020) Rare microbial taxa as the major drivers of ecosystem multifunctionality in long-term fertilized soils. Soil Biology and Biochemistry 141, 107686.
| Crossref | Google Scholar |
Chesneau G, Laroche B, Préveaux A, Marais C, Briand M, Marolleau B, Simonin M, Barret M (2022) Single seed microbiota: assembly and transmission from parent plant to seedling. mBio 13, e01648-e01622.
| Crossref | Google Scholar |
Choi B, Jeong S, Kim E (2022) Variation of the seed endophytic bacteria among plant populations and their plant growth-promoting activities in a wild mustard plant species, Capsella bursa-pastoris. Ecology and Evolution 12, e8683.
| Crossref | Google Scholar |
Clay K (1987) Effects of fungal endophytes on the seed and seedling biology of Lolium perenne and Festuca arundinacea. Oecologia 73, 358-362.
| Crossref | Google Scholar |
Clay K, Holah J (1999) Fungal endophyte symbiosis and plant diversity in successional fields. Science 285, 1742-1744.
| Crossref | Google Scholar |
Compant S, Mitter B, Colli-Mull JG, Gangl H, Sessitsch A (2011) Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microbial Ecology 62, 188-197.
| Crossref | Google Scholar |
Czarnoleski M, Olejniczak P, Mikołajczak P, Lembicz M, Kozłowski J (2010) Fungal endophytes protect grass seedlings against herbivory and allow economical seed production. Evolutionary Ecology Research 12, 769-777.
| Google Scholar |
Dadzie FA, Moles AT, Erickson TE, Slavich E, Muñoz-Rojas M (2022) Native bacteria and cyanobacteria can influence seedling emergence and growth of native plants used in dryland restoration. Journal of Applied Ecology 59, 2983-2992.
| Crossref | Google Scholar |
Dai Y, Li XY, Wang Y, Li CX, He Y, Lin HH, Wang T, Ma XR (2020) The differences and overlaps in the seed-resident microbiome of four Leguminous and three Gramineous forages. Microbial Biotechnology 13, 1461-1476.
| Crossref | Google Scholar |
Dalling JW, Davis AS, Schutte BJ, Elizabeth Arnold A (2011) Seed survival in soil: interacting effects of predation, dormancy and the soil microbial community. Journal of Ecology 99, 89-95.
| Crossref | Google Scholar |
De Vitis M, Hay FR, Dickie JB, Trivedi C, Choi J, Fiegener R (2020) Seed storage: maintaining seed viability and vigor for restoration use. Restoration Ecology 28, S249-S255.
| Crossref | Google Scholar |
Delgado-Baquerizo M, Maestre FT, Reich PB, Jeffries TC, Gaitan JJ, Encinar D, Berdugo M, Campbell CD, Singh BK (2016) Microbial diversity drives multifunctionality in terrestrial ecosystems. Nature Communications 7, 10541.
| Crossref | Google Scholar |
Delgado-Sánchez P, Ortega-Amaro MA, Jiménez-Bremont JF, Flores J (2011) Are fungi important for breaking seed dormancy in desert species? Experimental evidence in Opuntia streptacantha (Cactaceae). Plant Biology 13, 154-159.
| Crossref | Google Scholar |
Delgado-Sánchez P, Jiménez-Bremont JF, de la Luz Guerrero-González M, Flores J (2013) Effect of fungi and light on seed germination of three Opuntia species from semiarid lands of central Mexico. Journal of Plant Research 126, 643-649.
| Crossref | Google Scholar |
Desai S, Mistry J, Shah F, Chandwani S, Amaresan N, Supriya NR (2023) Salt-tolerant bacteria enhance the growth of mung bean (Vigna radiata L.) and uptake of nutrients, and mobilize sodium ions under salt stress condition. International Journal of Phytoremediation 25, 66-73.
| Crossref | Google Scholar |
Duduk N, Lazarević M, Žebeljan A, Vasić M, Vico I (2017) Blue mould decay of stored onion bulbs caused by Penicillium polonicum, P. glabrum and P. expansum. Journal of Phytopathology 165, 662-669.
| Crossref | Google Scholar |
Dutta S, Choi SY, Lee YH (2022) Temporal dynamics of endogenous bacterial composition in rice seeds during maturation and storage, and spatial dynamics of the bacteria during seedling growth. Frontiers in Microbiology 13, 877781.
| Crossref | Google Scholar |
D’Agui HM, van der Heyde ME, Nevill PG, Mousavi-Derazmahalleh M, Dixon KW, Moreira-Grez B, Valliere JM (2022) Evaluating biological properties of topsoil for post-mining ecological restoration: different assessment methods give different results. Restoration Ecology 30, e13738.
| Crossref | Google Scholar |
Erickson TE, Merritt DJ, Turner SR (2016) Overcoming physical seed dormancy in priority native species for use in arid-zone restoration programs. Australian Journal of Botany 64, 401-416.
| Crossref | Google Scholar |
Finlay BJ, Maberly SC, Cooper JI (1997) Microbial diversity and ecosystem function. Oikos 80, 209-213.
| Crossref | Google Scholar |
Flajs D, Peraica M (2009) Toxicological properties of citrinin. Archives of Industrial Hygiene and Toxicology 60, 457-464.
| Crossref | Google Scholar |
Frischie S, Miller AL, Pedrini S, Kildisheva OA (2020) Ensuring seed quality in ecological restoration: native seed cleaning and testing. Restoration Ecology 28, S239-S248.
| Crossref | Google Scholar |
Gallery RE, Dalling JW, Arnold AE (2007) Diversity, host affinity, and distribution of seed-infecting fungi: a case study with Cecropia. Ecology 88, 582-588.
| Crossref | Google Scholar |
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2, 113-118.
| Crossref | Google Scholar |
Gellie NJC, Mills JG, Breed MF, Lowe AJ (2017) Revegetation rewilds the soil bacterial microbiome of an old field. Molecular Ecology 26, 2895-2904.
| Crossref | Google Scholar |
Giauque H, Hawkes CV (2016) Historical and current climate drive spatial and temporal patterns in fungal endophyte diversity. Fungal Ecology 20, 108-114.
| Crossref | Google Scholar |
Goko ML, Murimwa JC, Gasura E, Rugare JT, Ngadze E (2021) Identification and characterisation of seed-borne fungal pathogens associated with maize (Zea mays L.). International Journal of Microbiology 2021, 6702856.
| Crossref | Google Scholar |
Golos PJ, Dixon KW (2014) Waterproofing topsoil stockpiles minimizes viability decline in the soil seed bank in an arid environment. Restoration Ecology 22, 495-501.
| Crossref | Google Scholar |
Graham EB, Knelman JE, Schindlbacher A, et al. (2016) Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Frontiers in Microbiology 7, 214.
| Crossref | Google Scholar |
Grove JF, Brian PW (1951) Identity of frequentic acid and citromycetin. Nature 167, 995.
| Crossref | Google Scholar |
Gundel PE, Martínez-Ghersa MA, Garibaldi LA, Ghersa CM (2009) Viability of Neotyphodium endophytic fungus and endophyte-infected and noninfected Lolium multiflorum seeds. Botany 87, 88-96.
| Crossref | Google Scholar |
Hardoim PR, Van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiology and Molecular Biology Reviews 79, 293-320.
| Crossref | Google Scholar |
Helander ML, Sieber TN, Petrini O, Neuvonen S (1994) Endophytic fungi in Scots pine needles: spatial variation and consequences of simulated acid rain. Canadian Journal of Botany 72, 1108-1113.
| Crossref | Google Scholar |
Heneghan L, Miller SP, Baer S, Callaham MA, Jr, Montgomery J, Pavao-Zuckerman M, Rhoades CC, Richardson S (2008) Integrating soil ecological knowledge into restoration management. Restoration Ecology 16, 608-617.
| Crossref | Google Scholar |
Henning JA, Kinkel L, May G, Lumibao CY, Seabloom EW, Borer ET (2021) Plant diversity and litter accumulation mediate the loss of foliar endophyte fungal richness following nutrient addition. Ecology 102, e03210.
| Crossref | Google Scholar |
Hill R, Llewellyn T, Downes E, Oddy J, MacIntosh C, Kallow S, Panis B, Dickie JB, Gaya E (2021) Seed banks as incidental fungi banks: fungal endophyte diversity in stored seeds of banana wild relatives. Frontiers in Microbiology 12, 643731.
| Crossref | Google Scholar |
Hodgson S, de Cates C, Hodgson J, Morley NJ, Sutton BC, Gange AC (2014) Vertical transmission of fungal endophytes is widespread in forbs. Ecology and Evolution 4, 1199-1208.
| Crossref | Google Scholar |
Hone H, Mann R, Yang G, Kaur J, Tannenbaum I, Li T, Spangenberg G, Sawbridge T (2021) Profiling, isolation and characterisation of beneficial microbes from the seed microbiomes of drought tolerant wheat. Scientific Reports 11, 11916.
| Crossref | Google Scholar |
Hubbard M, Germida J, Vujanovic V (2012) Fungal endophytes improve wheat seed germination under heat and drought stress. Botany 90, 137-149.
| Crossref | Google Scholar |
Jeong S, Kim T-M, Choi B, Kim Y, Kim E (2021) Invasive Lactuca serriola seeds contain endophytic bacteria that contribute to drought tolerance. Scientific Reports 11, 13307.
| Crossref | Google Scholar |
Johnston-Monje D, Gutiérrez JP, Lopez-Lavalle LAB (2021) Seed-transmitted bacteria and fungi dominate juvenile plant microbiomes. Frontiers in Microbiology 12, 737616.
| Crossref | Google Scholar |
Khan SA, Hamayun M, Yoon H, et al. (2008) Plant growth promotion and Penicillium citrinum. BMC Microbiology 8, 231.
| Crossref | Google Scholar |
Kim H, Jeon J, Lee KK, Lee Y-H (2022) Longitudinal transmission of bacterial and fungal communities from seed to seed in rice. Communications Biology 5, 772.
| Crossref | Google Scholar |
Klaedtke S, Jacques M-A, Raggi L, Préveaux A, Bonneau S, Negri V, Chable V, Barret M (2016) Terroir is a key driver of seed-associated microbial assemblages. Environmental Microbiology 18, 1792-1804.
| Crossref | Google Scholar |
Kluger CG, Dalling JW, Gallery RE, Sanchez E, Weeks-Galindo C, Arnold AE (2008) Host generalists dominate fungal communities associated with seeds of four neotropical pioneer species. Journal of Tropical Ecology 24, 351-354.
| Crossref | Google Scholar |
Kolb A, Ehrlén J, Eriksson O (2007) Ecological and evolutionary consequences of spatial and temporal variation in pre-dispersal seed predation. Perspectives in Plant Ecology, Evolution and Systematics 9, 79-100.
| Crossref | Google Scholar |
Laihonen M, Saikkonen K, Helander M, Vázquez de Aldana BR, Zabalgogeazcoa I, Fuchs B (2022) Epichloë endophyte-promoted seed pathogen increases host grass resistance against insect herbivory. Frontiers in Microbiology 12, 786619.
| Crossref | Google Scholar |
Le DP, Gregson A (2019) Alternaria leaf spot of cotton seedlings grown in New South Wales, Australia is predominantly associated with Alternaria alternata. Australasian Plant Pathology 48, 209-216.
| Crossref | Google Scholar |
Leff JW, Bardgett RD, Wilkinson A, Jackson BG, Pritchard WJ, De Long J, Oakley S, Mason KE, Ostle N, Johnson D, Baggs E (2018) Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits. The ISME Journal 12, 1794-1805.
| Crossref | Google Scholar |
Leroy C, Maes AQ, Louisanna E, Séjalon-Delmas N (2019) How significant are endophytic fungi in bromeliad seeds and seedlings? Effects on germination, survival and performance of two epiphytic plant species. Fungal Ecology 39, 296-306.
| Crossref | Google Scholar |
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658-1659.
| Crossref | Google Scholar |
Lindblom SD, Wangeline AL, Valdez Barillas JR, Devilbiss B, Fakra SC, Pilon-Smits EAH (2018) Fungal endophyte Alternaria tenuissima can affect growth and selenium accumulation in its hyperaccumulator host Astragalus bisulcatus. Frontiers in Plant Science 9, 1213.
| Crossref | Google Scholar |
Liu Y, Zuo S, Zou Y, Wang J, Song W (2013) Investigation on diversity and population succession dynamics of endophytic bacteria from seeds of maize (Zea mays L., Nongda108) at different growth stages. Annals of Microbiology 63, 71-79.
| Crossref | Google Scholar |
Liu J, Nagabhyru P, Schardl CL (2017) Epichloë festucae endophytic growth in florets, seeds, and seedlings of perennial ryegrass (Lolium perenne). Mycologia 109, 691-700.
| Crossref | Google Scholar |
Lopes MJdS, Dias-Filho MB, Gurgel ESC (2021) Successful plant growth-promoting microbes: Inoculation methods and abiotic factors. Frontiers in Sustainable Food Systems 5, 606454.
| Crossref | Google Scholar |
Loreau M, Naeem S, Inchausti P, et al. (2001) Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294, 804-808.
| Crossref | Google Scholar |
Maharachchikumbura SSN, Hyde KD, Groenewald JZ, Xu J, Crous PW (2014) Pestalotiopsis revisited. Studies in Mycology 79, 121-186.
| Crossref | Google Scholar |
Mancini V, Romanazzi G (2014) Seed treatments to control seedborne fungal pathogens of vegetable crops. Pest Management Science 70, 860-868.
| Crossref | Google Scholar |
Martín I, Gálvez L, Guasch L, Palmero D (2022) Fungal pathogens and seed storage in the dry state. Plants 11, 3167.
| Crossref | Google Scholar |
Matsumoto H, Fan X, Wang Y, et al. (2021) Bacterial seed endophyte shapes disease resistance in rice. Nature Plants 7, 60-72.
| Crossref | Google Scholar |
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217.
| Crossref | Google Scholar |
Merritt DJ, Turner S, Clarke S, Dixon KW (2007) Seed dormancy and germination stimulation syndromes for Australian temperate species. Australian Journal of Botany 55, 336-344.
| Crossref | Google Scholar |
Mertin AA, Laurence MH, van der Merwe M, French K, Liew ECY (2022) The culturable seed mycobiome of two Banksia species is dominated by latent saprotrophic and multi-trophic fungi. Fungal Biology 126, 738-745.
| Crossref | Google Scholar |
Mitter B, Pfaffenbichler N, Flavell R, et al. (2017) A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Frontiers in Microbiology 8, 11.
| Crossref | Google Scholar |
Morales Moreira ZP, Helgason BL, Germida JJ (2021) Environment has a stronger effect than host plant genotype in shaping spring Brassica napus seed microbiomes. Phytobiomes Journal 5, 220-230.
| Crossref | Google Scholar |
Morales Moreira ZP, Chen MY, Ortuno DLY, Haney CH (2023) Engineering plant microbiomes by integrating eco-evolutionary principles into current strategies. Current Opinion in Plant Biology 71, 102316.
| Crossref | Google Scholar |
Moreira-Grez B, Muñoz-Rojas M, Kariman K, Storer P, O’Donnell AG, Kumaresan D, Whiteley AS (2019) Reconditioning degraded mine site soils with exogenous soil microbes: plant fitness and soil microbiome outcomes. Frontiers in Microbiology 10, 1617.
| Crossref | Google Scholar |
Morella NM, Zhang X, Koskella B (2019) Tomato seed-associated bacteria confer protection of seedlings against foliar disease caused by Pseudomonas syringae. Phytobiomes Journal 3, 177-190.
| Crossref | Google Scholar |
Morgan JAW, Bending GD, White PJ (2005) Biological costs and benefits to plant–microbe interactions in the rhizosphere. Journal of Experimental Botany 56, 1729-1739.
| Crossref | Google Scholar |
Muñoz-Rojas M, Erickson TE, Dixon KW, Merritt DJ (2016) Soil quality indicators to assess functionality of restored soils in degraded semiarid ecosystems. Restoration Ecology 24, S43-S52.
| Crossref | Google Scholar |
Nelson EB (2004) Microbial dynamics and interactions in the spermosphere. Annual Reviews of Phytopathology 42, 271-309.
| Crossref | Google Scholar |
Nelson EB (2018) The seed microbiome: origins, interactions, and impacts. Plant and Soil 422, 7-34.
| Crossref | Google Scholar |
Nevill PG, Tomlinson S, Elliott CP, Espeland EK, Dixon KW, Merritt DJ (2016) Seed production areas for the global restoration challenge. Ecology and Evolution 6, 7490-7497.
| Crossref | Google Scholar |
Ngugi MR, Fechner N, Neldner VJ, Dennis PG (2020) Successional dynamics of soil fungal diversity along a restoration chronosequence post-coal mining. Restoration Ecology 28, 543-552.
| Crossref | Google Scholar |
Nishie K, Cole RJ, Dorner JW (1988) Toxicity of citreoviridin. Research Communications in Chemical Pathology and Pharmacology 59, 31-52.
| Google Scholar |
O’Callaghan M, Ballard RA, Wright D (2022) Soil microbial inoculants for sustainable agriculture: limitations and opportunities. Soil Use and Management 38, 1340-1369.
| Crossref | Google Scholar |
Paluch EC, Thomsen MA, Volk TJ (2013) Effects of resident soil fungi and land use history outweigh those of commercial mycorrhizal inocula: testing a restoration strategy in unsterilized soil. Restoration Ecology 21, 380-389.
| Crossref | Google Scholar |
Paravar A, Piri R, Balouchi H, Ma Y (2023) Microbial seed coating: an attractive tool for sustainable agriculture. Biotechnology Reports 37, e00781.
| Crossref | Google Scholar |
Parmar S, Sharma VK, Li T, Tang W, Li H (2022) Fungal seed endophyte FZT214 improves Dysphania ambrosioides cd tolerance throughout different developmental stages. Frontiers in Microbiology 12, 783475.
| Crossref | Google Scholar |
Peay KG, Bruns TD, Kennedy PG, Bergemann SE, Garbelotto M (2007) A strong species–area relationship for eukaryotic soil microbes: island size matters for ectomycorrhizal fungi. Ecology Letters 10, 470-480.
| Crossref | Google Scholar |
Pedrini S, Balestrazzi A, Madsen MD, Bhalsing K, Hardegree SP, Dixon KW, Kildisheva OA (2020) Seed enhancement: getting seeds restoration-ready. Restoration Ecology 28, S266-S275.
| Crossref | Google Scholar |
Philipson MN, Christey MC (1986) The relationship of host and endophyte during flowering, seed formation, and germination of Lolium perenne. New Zealand Journal of Botany 24, 125-134.
| Crossref | Google Scholar |
Prosser JI, Bohannan BJM, Curtis TP, et al. (2007) The role of ecological theory in microbial ecology. Nature Reviews Microbiology 5, 384-392.
| Crossref | Google Scholar |
R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at https://www.R-project.org/
Reissinger A, Vilich V, Sikora RA (2001) Detection of fungi in planta: effectiveness of surface sterilization methods. Mycology Research 105, 563-566.
| Crossref | Google Scholar |
Ridout ME, Schroeder KL, Hunter SS, Styer J, Newcombe G (2019) Priority effects of wheat seed endophytes on a rhizosphere symbiosis. Symbiosis 78, 19-31.
| Crossref | Google Scholar |
Rolston MP, Hare MD, Moore KK, Christensen MJ (1986) Viability of Lolium endophyte fungus in seed stored at different moisture contents and temperatures. New Zealand Journal of Experimental Agriculture 14, 297-300.
| Crossref | Google Scholar |
Rybakova D, Mancinelli R, Wikström M, Birch-Jensen A-S, Postma J, Ehlers R-U, Goertz S, Berg G (2017) The structure of the Brassica napus seed microbiome is cultivar-dependent and affects the interactions of symbionts and pathogens. Microbiome 5, 104.
| Crossref | Google Scholar |
Sahib MR, Pervaiz ZH, Williams MA, Saleem M, DeBolt S (2020) Rhizobacterial species richness improves sorghum growth and soil nutrient synergism in a nutrient-poor greenhouse soil. Scientific Reports 10, 15454.
| Crossref | Google Scholar |
Saleem M, Hu J, Jousset A (2019) More than the sum of its parts: microbiome biodiversity as a driver of plant growth and soil health. Annual Review of Ecology, Evolution, and Systematics 50, 145-168.
| Crossref | Google Scholar |
Sarmiento C, Zalamea P-C, Dalling JW, Davis AS, Stump SM, U’Ren JM, Arnold AE (2017) Soilborne fungi have host affinity and host-specific effects on seed germination and survival in a lowland tropical forest. Proceedings of the National Academy of Sciences 114, 11458-11463.
| Crossref | Google Scholar |
Schiltz S, Gaillard I, Pawlicki-Jullian N, Thiombiano B, Mesnard F, Gontier E (2015) A review: what is the spermosphere and how can it be studied? Journal of Applied Microbiology 119, 1467-1481.
| Crossref | Google Scholar |
Schöler A, Jacquiod S, Vestergaard G, Schulz S, Schloter M (2017) Analysis of soil microbial communities based on amplicon sequencing of marker genes. Biology and Fertility of Soils 53, 485-489.
| Crossref | Google Scholar |
Shade A, Handelsman J (2012) Beyond the Venn diagram: the hunt for a core microbiome. Environmental Microbiology 14, 4-12.
| Crossref | Google Scholar |
Shade A, Peter H, Allison SD, et al. (2012) Fundamentals of microbial community resistance and resilience. Frontiers in Microbiology 3, 417.
| Crossref | Google Scholar |
Shade A, Jacques M-A, Barret M (2017) Ecological patterns of seed microbiome diversity, transmission, and assembly. Current Opinions in Microbiology 37, 15-22.
| Crossref | Google Scholar |
Shahzad R, Khan AL, Bilal S, Asaf S, Lee I-J (2018) What is there in seeds? Vertically transmitted endophytic resources for sustainable improvement in plant growth. Frontiers in Plant Science 9, 24.
| Crossref | Google Scholar |
Sharma A, Aggarwal NK, Yadav A (2016) First report of lignin peroxidase production from Alternaria alternata ANF238 isolated from rotten wood sample. Bioengineering Bioscience 4, 76-87.
| Crossref | Google Scholar |
Shearin ZRC, Filipek M, Desai R, Bickford WA, Kowalski KP, Clay K (2018) Fungal endophytes from seeds of invasive, non-native Phragmites australis and their potential role in germination and seedling growth. Plant and Soil 422, 183-194.
| Crossref | Google Scholar |
Sheppard J (1998) Seed-borne pathogens of vegetable and flower seeds: their devastation, identification and control. Seed Technology 187-197.
| Google Scholar |
Siddique S, Naveed M, Yaseen M, Shahbaz M (2022) Exploring potential of seed endophytic bacteria for enhancing drought stress resilience in maize (Zea mays L.). Sustainability 14, 673.
| Crossref | Google Scholar |
Simonin M, Briand M, Chesneau G, Rochefort A, Marais C, Sarniguet A, Barret M (2022) Seed microbiota revealed by a large-scale meta-analysis including 50 plant species. New Phytologist 234, 1448-1463.
| Crossref | Google Scholar |
Singh M, Awasthi A, Soni SK, Singh R, Verma RK, Kalra A (2015) Complementarity among plant growth promoting traits in rhizospheric bacterial communities promotes plant growth. Scientific Reports 5, 15500.
| Crossref | Google Scholar |
Solanki MK, Abdelfattah A, Britzi M, Zakin V, Wisniewski M, Droby S, Sionov E (2019) Shifts in the composition of the microbiota of stored wheat grains in response to fumigation. Frontiers in Microbiology 10, 1098.
| Crossref | Google Scholar |
Sommerville K, May T (2006) Some taxonomic and ecological observations on the genus Banksiamyces. The Victorian Naturalist 123, 366-375.
| Google Scholar |
Sánchez-López AS, Pintelon I, Stevens V, et al. (2018) Seed endophyte microbiome of Crotalaria pumila unpeeled: identification of plant-beneficial methylobacteria. International Journal of Molecular Sciences 19, 291.
| Crossref | Google Scholar |
Tarquinio F, Attlan O, Vanderklift MA, Berry O, Bissett A (2021) Distinct endophytic bacterial communities inhabiting seagrass seeds. Frontiers in Microbiology 12, 703014.
| Crossref | Google Scholar |
Taylor BN, Simms EL, Komatsu KJ (2020) More than a functional group: diversity within the legume–rhizobia mutualism and its relationship with ecosystem function. Diversity 12, 50.
| Crossref | Google Scholar |
Truyens S, Weyens N, Cuypers A, Vangronsveld J (2015) Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environmental Microbiology Reports 7, 40-50.
| Crossref | Google Scholar |
Turner SR, Steadman KJ, Vlahos S, Koch JM, Dixon KW (2013) Seed treatment optimizes benefits of seed bank storage for restoration-ready seeds: the feasibility of prestorage dormancy alleviation for mine-site revegetation. Restoration Ecology 21, 186-192.
| Crossref | Google Scholar |
U’ren JM, Dalling JW, Gallery RE, Maddison DR, Davis EC, Gibson CM, Arnold AE (2009) Diversity and evolutionary origins of fungi associated with seeds of a neotropical pioneer tree: a case study for analysing fungal environmental samples. Mycological Research 113, 432-449.
| Crossref | Google Scholar |
Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A (2015) The importance of the microbiome of the plant holobiont. New Phytologist 206, 1196-1206.
| Crossref | Google Scholar |
Vasanthakumari MM, Shridhar J, Madhura RJ, Nandhitha M, Kasthuri C, Janardhana B, Nataraja KN, Ravikanth G, Uma Shaanker R (2019) Role of endophytes in early seedling growth of plants: a test using systemic fungicide seed treatment. Plant Physiology Reports 24, 86-95.
| Crossref | Google Scholar |
Verma SK, Kingsley KL, Bergen MS, Kowalski KP, White JF (2018) Fungal disease prevention in seedlings of rice (Oryza sativa) and other grasses by growth-promoting seed-associated endophytic bacteria from invasive Phragmites australis. Microorganisms 6, 21.
| Crossref | Google Scholar |
Visagie C, Jacobs K (2012) Three new additions to the genus Talaromyces isolated from Atlantis sandveld fynbos soils. Persoonia - Molecular Phylogeny and Evolution of Fungi 28, 14-24.
| Crossref | Google Scholar |
Vujanovic V, Islam MN, Daida P (2019) Transgenerational role of seed mycobiome – an endosymbiotic fungal composition as a prerequisite to stress resilience and adaptive phenotypes in Triticum. Scientific Reports 9, 18483.
| Crossref | Google Scholar |
Walitang DI, Kim C-G, Kim K, Kang Y, Kim YK, Sa T (2018) The influence of host genotype and salt stress on the seed endophytic community of salt-sensitive and salt-tolerant rice cultivars. BMC Plant Biology 18, 51.
| Crossref | Google Scholar |
White JF, Kingsley KI, Kowalski KP, Irizarry I, Micci A, Soares MA, Bergen MS (2018) Disease protection and allelopathic interactions of seed-transmitted endophytic pseudomonads of invasive reed grass (Phragmites australis). Plant and Soil 422, 195-208.
| Crossref | Google Scholar |
Wäli PR, Helander M, Saloniemi I, Ahlholm J, Saikkonen K (2009) Variable effects of endophytic fungus on seedling establishment of fine fescues. Oecologia 159, 49-57.
| Crossref | Google Scholar |
You MP, Lamichhane JR, Aubertot J-N, Barbetti MJ (2020) Understanding why effective fungicides against individual soilborne pathogens are ineffective with soilborne pathogen complexes. Plant Disease 104, 904-920.
| Crossref | Google Scholar |
Zhang XX, Li CJ, Nan ZB, Matthew C (2012) Neotyphodium endophyte increases Achnatherum inebrians (drunken horse grass) resistance to herbivores and seed predators. Weed Research 52, 70-78.
| Crossref | Google Scholar |
Zheng Y, Gong X (2019) Niche differentiation rather than biogeography shapes the diversity and composition of microbiome of Cycas panzhihuaensis. Microbiome 7, 152.
| Crossref | Google Scholar |
Zhou LS, Tang K, Guo SX (2018) The plant growth-promoting fungus (PGPF) Alternaria sp. A13 markedly enhances Salvia miltiorrhiza root growth and active ingredient accumulation under greenhouse and field conditions. International Journal of Molecular Sciences 19, 270.
| Crossref | Google Scholar |