Unravelling the evolutionary history of Eucalyptus cordata (Myrtaceae) using molecular markers
Peter A. Harrison A B , Rebecca C. Jones A , René E. Vaillancourt A , Robert J. E. Wiltshire A and Brad M. Potts AA School of Biological Sciences and National Centre for Future Forest Industries, University of Tasmania, Private Bag 55, Hobart, Tas. 7001, Australia.
B Corresponding author. Email: P.A.Harrison@utas.edu.au
Australian Journal of Botany 62(2) 114-131 https://doi.org/10.1071/BT14019
Submitted: 6 February 2014 Accepted: 16 March 2014 Published: 5 May 2014
Abstract
We studied the evolutionary processes shaping the genetic diversity in the naturally fragmented Eucalyptus cordata, a rare homoblastic tree endemic to the island of Tasmania. A genome-wide scan showed that E. cordata and the endangered heteroblastic E. morrisbyi were closely related, suggesting a neotenous origin of E. cordata from an endemic heteroblastic ancestor. Bayesian cluster analysis based on nuclear microsatellites assayed in 567 E. cordata and E. morrisbyi individuals revealed five genetic clusters. Two clusters comprised populations that correspond to putative ancestral gene pools linking E. cordata and E. morrisbyi. Another cluster included populations that transgressed the drowned Derwent River valley, suggestive of a wider glacial distribution. However, the majority of individuals occurred in the two genetic clusters distributed in the south-west and north-east of the range of E. cordata. The elevated genetic diversity in populations comprising these clusters suggests that they represent two recently fragmented cores of the distribution. Genetic evidence suggests that the newly described, localised E. cordata subspecies quadrangulosa has been recently selected from within the morphologically diverse, south-western cluster. We argue that multiple phases of isolation and drift have led to the contemporary pattern of molecular variation and the scattering of relictual and more recently derived populations across the species distribution.
Additional keywords: Diversity Array Technology (DArT), Eucalyptus, gene flow, genetic drift, heteroblastic, homoblastic, microsatellite, spatial genetic structuring.
Introduction
Eucalyptus cordata Labill. (heart-leaved silver gum) is a rare tree species of the subgenus Symphyomyrtus, endemic to the island of Tasmania. The species is notable compared with other Tasmanian endemics of the series Orbiculares (E. archeri, E. gunnii, E. morrisbyi, and E. urnigera), being homoblastic and retaining the striking, glaucous, opposite and sessile juvenile leaves into reproductive maturity in both wild and ornamental populations, even as a large tree (Brooker 2000; Nicolle 2006; Slee et al. 2006; Fig. 1). Several populations are known where adult foliage does develop on the tops of trees, but flowering still occurs in the distinctive juvenile leaf stage (Potts 1989). Such neotenic homoblasty is relatively rare in eucalypts, but occurs in eucalypts from diverse taxonomic groups (Potts and Wiltshire 1997) as well as in two other species of the series Orbiculares (E. perriniana and E. pulverulenta; Brooker 2000; Nicolle 2006; Slee et al. 2006).

|
Eucalyptus cordata occurs in small, isolated populations across a remarkably diverse range of habitats in south-eastern Tasmania. It grows on dry cliff-tops or hill slopes at lower altitudes and on poorly drained marsh edges at higher altitudes, and has a corresponding diversity of habits from stunted, multi-stemmed shrub to tall, single-stemmed forest trees more than 20 m high (Potts 1989; Williams and Potts 1996). A morphological cline has been described across this range of habitats (Potts 1989), and has been summarised by the recognition of two subspecies (Nicolle et al. 2008). The main discriminating characters between these two subspecies are stem shape and leaf size, which are square (as opposed to round) and larger, respectively, in subspecies quadrangulosa, and there is a genetic basis to this as shown in a glasshouse seedling trial (Potts 1989). Subspecies quadrangulosa occurs at higher altitude and wetter sites in the south-west of the species range (Nicolle et al. 2008).
Eucalyptus cordata has been a key species in studies of chloroplast sharing and reticulate evolution in eucalypts (McKinnon et al. 2004a). Extensive sharing of chloroplast haplotypes of the ‘Southern’ type has been reported among all Symphyomyrtus species occurring in south-eastern Tasmania, and is believed to be the result of hybridisation during species range contractions or expansions in glacial refugia (McKinnon et al. 2001a; McKinnon et al. 2004a). Eucalyptus cordata is one of the only species that are fixed for this chloroplast clade (McKinnon et al. 2004b), and detailed population studies have provided evidence that the chloroplast and rare components of the nuclear genome have been locally captured by the more widespread, co-occurring E. globulus (McKinnon et al. 2004b, 2010).
The only endemic Symphyomyrtus species with contemporary natural distributions confined to this south-eastern forest refugium are E. cordata and the endangered E. morrisbyi. Traditionally, studies have shown that populations within glacial refugia tend to be more genetically diverse than populations that have expanded into more recently available habitat (Comps et al. 2001). However, this pattern may well be case-specific, and studies on eucalypts have suggested that the restricted and fragmented distribution in putative glacial refugia can result in genetically depauperate populations because of genetic erosion (Byrne and Hopper 2008). This is indeed the case with E. morrisbyi (Jones et al. 2005), and it suggests that the disjunct populations of E. cordata may also demonstrate low genetic diversity and high levels of differentiation due to their isolation.
The present study used Diversity Array Technology (DArT), nuclear microsatellites, and chloroplast markers to investigate the population genetic processes that have shaped the genetic architecture of E. cordata. The microsatellite and chloroplast markers were chosen so that the results of this study could be compared with previous works (McKinnon et al. 2004b; Jones et al. 2005, 2013; Rathbone et al. 2007). We used DArT markers, which have wide genome coverage (Sansaloni et al. 2011; Steane et al. 2011), so as to achieve high resolution for the phylogenetic component of the study. The goals of this study were to determine (1) the genetic affinities of E. cordata within series Orbiculares; (2) the genetic affinities of populations within E. cordata; and (3) the evolutionary origin of subspecies quadrangulosa. We present evidence that multiple expansion and contraction events through evolutionary time have shaped the genetic architecture of this species, and that the newly described subspecies quadrangulosa is a neo-endemic.
Materials and methods
Plant material and DNA extraction
To investigate the genetic affinities between populations of E. cordata, total genomic DNA was extracted from fresh leaf tissue collected from 533 individuals from 21 natural populations representing the two subspecies (subspecies quadrangulosa and subspecies cordata) and their intermediates (Table 1, Fig. 2). Additionally, to investigate the genetic affinities between populations of E. cordata and E. morrisbyi, DNA was extracted from fresh leaves collected from 24 individuals from the Risdon Hill population of E. morrisbyi (Fig. 2), and DNA samples that were available from a previous study on the Calverts Hill E. morrisbyi population (Jones et al. 2005) were re-genotyped. Trees were sampled at least two canopy heights apart to avoid sampling clonal and closely related individuals (Jones et al. 2005). All trees were photographed and tagged, with their geographic position recorded. Contemporary population sizes were estimated based on the number of individuals (either clumps of stems or individual stems) observed during sampling. Phenotypic affinities of E. cordata populations were classified according to Nicolle et al. (2008).

|
Total genomic DNA was extracted from 10 mg of ground tissue using a CTAB protocol (Doyle and Doyle 1990) with the modifications of McKinnon et al. (2004b), grinding fresh tissue in liquid nitrogen with either a mortar and pestle or a tungsten carbide bead in a 1.2-mL sample tube placed in a Mixer Mill MM400 (RETSCH, Haan, Germany) for two 1-min periods at 30 Hz. DNA quality and quantity were visually estimated by comparing the Lambda HindIII molecular weight standard (New England BioLabs, Ipswich, MA, USA) against the electrophoretically separated DNA.
Molecular markers and analysis
DArT
The genetic affinities of E. cordata to other species from the series Orbiculares was investigated using genome-wide DArT markers (Sansaloni et al. 2011). This included a subset of E. cordata samples from the current study (including both subspecies and their intermediates), along with representative samples from Tasmanian endemic species (E. gunnii, E. urnigera, E. morrisbyi, E. archeri) and species from mainland Australia (E. saxatilis, E. glaucescens, E. perriniana, E. pulverulenta) of the series Orbiculares (Appendix 1). DNA was prepared following the protocol of Steane et al. (2011). Genome complexity reduction was conducted following Sansaloni et al. (2011), with polymorphisms detected by DArT Pty Ltd (http://www.diversityarrays.com/, verified 3 April 2014) based on the presence or absence of the DArT marker in the sample. The dissimilarity between samples was calculated using an additive Dollo distance (ADD), a distance measure appropriate for the DArT data (Woodhams et al. 2013). The matrix of ADD distances was summarised using a principal coordinate analysis undertaken with GenAlEx version 6.501 (Peakall and Smouse 2012).
Nuclear microsatellite
All E. cordata and E. morrisbyi individuals were genotyped using 13 microsatellite loci. Primer pairs were designed in E. grandis (prefix EMBRA; Brondani et al. 1998; Brondani et al. 2006) and E. globulus (prefix EMCRC; Steane et al. 2001) and were separated into three multiplex mixes (Appendix 2). Microsatellite loci were amplified by polymerase chain reaction (PCR) using a QIAGEN Multiplex PCR kit (QIAGEN, Melbourne, Vic., Australia). Each reaction contained 1 × QIAGEN Multiplex PCR Master Mix, 1 × Q-solution, 5–15 ng DNA template, primers (Appendix 2), and RNase-free water to a final volume of 5 μL. Amplification of microsatellite fragments was performed on a Bio-Rad C1000 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) using two PCR programs. Multiplex mixes A and C used the following program: 95°C for 15 min; 30 cycles of 94°C for 30 s, annealing temperature of 57°C for 90 s, and 72°C for 60 s; a final extension at 60°C for 10 min; finishing with a final hold temperature of 15°C. Amplification of Multiplex mix B used an annealing temperature of 55°C for 90 s. Final PCR product concentration and quality was estimated by comparing a 100-bp molecular weight standard (New England BioLabs) with the electrophoresis separation of the PCR product on a 1% agarose gel pre-stained with Goldview (Guangzhou Geneshun Biotech, Guangzhou, China). PCR products were diluted at a ratio of 1 μL of PCR product to 10 μL of Milli-Q water. One μL of diluted PCR product was dried and sent to the Australian Genome Research Facility (AGRF), South Australia. There, it was resuspended and sized using an AB3730 DNA Analyser (Applied Biosystems, Foster City, CA, USA). Allele sizes were assessed in GeneMapper version 3.7 (Applied Biosystems) using GS500 k(–250)LIZ as a size standard.
To estimate the frequency of null alleles, we used the program INEst (Chybicki and Burczyk 2009). This detected null alleles in EMCRC2 and EMCRC7. However, the removal of these loci did not affect the genetic diversity parameters, so all loci were included. The microsatellite loci were powerful enough to distinguish related and clonal genotypes in E. morrisbyi (Jones et al. 2005) and E. cordata (data not presented). Where clonal genotypes were detected, one representative genotype was retained from each ‘clonal patch’ for further analysis. This resulted in the removal of a sample from Chimney Pot Hill that was 3.6 m from its adjacent clonal sample. It also allowed validation of a large clone ~2 m across on Penguin Island where multiple stems had been specifically sampled from a large lignotuber remnant. Only individuals with allele scores for at least eight loci were retained, resulting in a sample size of 567 individuals from E. cordata and E. morrisbyi. Repeatability of microsatellite sizing was assessed by regenotyping 10% of the E. cordata samples, which showed that the genotyping error rate was only 1.8%.
Genetic affinities between individuals from populations of E. cordata and E. morrisbyi were investigated using the Bayesian methods implemented in STRUCTURE version 2.3.3 (Pritchard et al. 2000; Falush et al. 2003). An admixture model with no a priori population grouping was run, assuming that: (1) there are K populations characterised by allele frequencies that are correlated among populations; (2) individuals have inherited a proportion of their ancestry from each population; (3) there is linkage equilibrium; and (4) populations are in Hardy–Weinberg equilibrium. All STRUCTURE analyses used a burn-in period of 250 000 iterations followed by 100 000 Markov chain Monte Carlo iterations. Twenty independent realisations at each value of K (K = 2 to K = 23) were performed. STRUCTURE Harvester (Earl and vonHoldt 2012) was used to detect the optimum value of K using the post hoc methods of Evanno et al. (2005). Solutions for K were then collapsed into a single representative solution using the Greedy algorithm implemented in CLUMPP version 1.1.2 (Jakobsson and Rosenberg 2007), weighted by the number of individuals in each population.
Population genetic diversity parameters were calculated and averaged across populations and loci by using Genetic Data Analysis version 1.1 (Lewis and Zaykin 2001). This included the observed number of alleles (A), observed (HO) and expected (HE) heterozygosity, and Wright’s fixation index (F). Allelic richness (AR), rarefied to a minimum sample size of 17 individuals per population, and Nei’s (1987) estimate of total heterozygosity (HT) were calculated in FSTAT version 2.9.3.2 (Goudet 2002). Correlations among the genetic diversity measures (HE, HO, AR, and F), altitude, and population size were tested using a Pearson’s product–moment test in the R statistical package (R Core Team 2012). To determine the relationships among populations, Nei’s (1972) genetic distance matrix among all populations was calculated in Genetic Data Analysis. The F-statistics FIS (inbreeding within individuals relative to their population), FIT (inbreeding within individuals relative to all other populations), and FST (inbreeding in populations relative to all other populations) were estimated in FSTAT with 99% bootstrap confidence intervals derived after 1000 replications across loci. A pair-wise FST matrix was also produced using FSTAT, with statistically differentiated populations indicated at the α = 0.05 level after 4200 permutations. The level of differentiation among subspecies cordata, subspecies quadrangulosa and intermediate populations of E. cordata was investigated by pooling populations based on their taxonomic classification and calculating the FST between these three groups in FSTAT. The Mantel test in GenAlEx (version 6.501) was used to determine whether the level of genetic differentiation, based on the matrix of FST/(1 – FST) values (Rousset 1997), was related to the matrix of geographic distances (natural logarithm transformed) among populations.
Chloroplast
The chloroplast genome in Eucalyptus has been shown to be maternally inherited, meaning it can only be dispersed by seed (Byrne et al. 1993; McKinnon et al. 2001b), which, in Eucalyptus, is usually gravity dispersed (Cremer 1977). The hypervariable JLA+ (~630 bp) region of the chloroplast genome was sequenced in 4–7 geographically well-spaced trees for each of the E. cordata and E. morrisbyi populations studied (n = 127); the forward primer rpl2 and reverse primer eucpsbA were used (Freeman et al. 2001). Chloroplast sequences from previous studies of E. cordata (McKinnon et al. 2004b) and E. morrisbyi (Jones et al. 2005) were used where possible, with additional samples sequenced to increase the total sample size to a minimum of five per population. For each DNA sample, ~5–15 ng DNA was used in a final volume of 25 μL PCR reactant mix as in Freeman et al. (2001), except that 2.5 mM MgCl2, 0.15 μM of each primer, 1 unit MangoTaq polymerase (Bioline Australia, Alexandria, NSW), and deionised Milli-Q water to 25 μL was used. Amplification was performed on a Bio-Rad C1000 Thermal Cycler (Bio-Rad Laboratories) using the following PCR cycling program: 95°C for 5 min; 30 cycles consisting of 94°C for 60 s, 61°C for 60 s, and 72°C for 60 s; 72°C for 5 min; finishing with a final hold temperature of 15°C. PCR product concentration and quality was estimated by electrophoresis separation on a 2% agarose gel pre-stained with Goldview (Guangzhou Geneshun Biotech), using a 100-bp molecular weight standard (New England BioLabs). Purification and sequencing of PCR products were performed by Macrogen (Seoul, South Korea) using an AB3730 DNA Analyser (Applied Biosystems).
Forward and reverse sequences were analysed in Sequencher version 4.7 (Gene Codes, Ann Arbor, MI, USA) and CLC Workbench version 4 (CLC Bio, Aarhus, Denmark). The chloroplast haplotypes were then compared with other cpDNA sequences of Symphyomyrtus species held in the School of Biological Sciences DNA database at the University of Tasmania. New chloroplast haplotype sequences were classified and named following the nomenclature of Freeman et al. (2001). Chloroplast haplotype diversity (allelic richness rarefied to four individuals per population, CP AR) and FST were estimated in FSTAT. The Pearson’s correlation of CP AR with the nuclear genetic parameters, altitude, and population size was calculated as described above.
To determine the relative historic rates of pollen and seed migration between populations of E. cordata and E. morrisbyi, the ratio of pollen to seed gene flow was estimated using eqn 5a from Ennos (1994). This equation uses the level of differentiation among populations (FST) in the nuclear and maternally inherited chloroplast markers and takes into account the level of inbreeding by using the FIS derived from the nuclear microsatellite data.
Results
Genetic relationship of E. cordata to other species
The genetic relationship of E. cordata to other species from series Orbiculares was determined using a principal coordinate analysis based on the ADD dissimilarity matrix derived from 3172 non-redundant polymorphic DArT markers assayed in 49 representative samples of 10 species (Fig. 3). The first axis separated the mainland samples of the series Orbiculares from the Tasmanian endemics, with one E. gunnii sample (Sample 31) intermediate between these two groups (Fig. 3). Eucalyptus cordata (Samples 5–21, Fig. 3) showed closer genetic affinities with the Tasmanian endemic species from the series Orbiculares than with the homoblastic species from the mainland (E. pulverulenta, Sample 38, and E. perriniana, Sample 39 in Fig. 3). Axis 2 in the principal coordinate analysis showed a cline in genetic affinities within the Tasmanian endemic group (Fig. 3). At one end of this molecular cline were the majority of E. cordata samples, which were well differentiated from the other Tasmanian endemics. The other Tasmanian endemics were at the other end of this cline and were poorly differentiated from each other in this two-dimensional space, and remained poorly differentiated in the third dimension (data not shown). The E. cordata samples from Brown Mountain (Samples 8 and 18 in Fig. 3), a population that transitions into petiolate adult leaves while still flowering in the juvenile phase, grouped with the other Tasmanian endemics, as did the intermediate samples at Knights Creek and Moogara (Samples 7 and 11 in Fig. 3). However, samples from the heteroblastic E. morrisbyi populations at Calverts Hill (Samples 35 and 36 in Fig. 3) and Risdon Hill (Samples 33 and 34 in Fig. 3) had close affinities to E. cordata and were embedded within the principal coordinate space occupied by the E. cordata samples, which suggested that E. morrisbyi should be included in further analyses of E. cordata.

|
Genetic structuring of E. cordata and E. morrisbyi populations
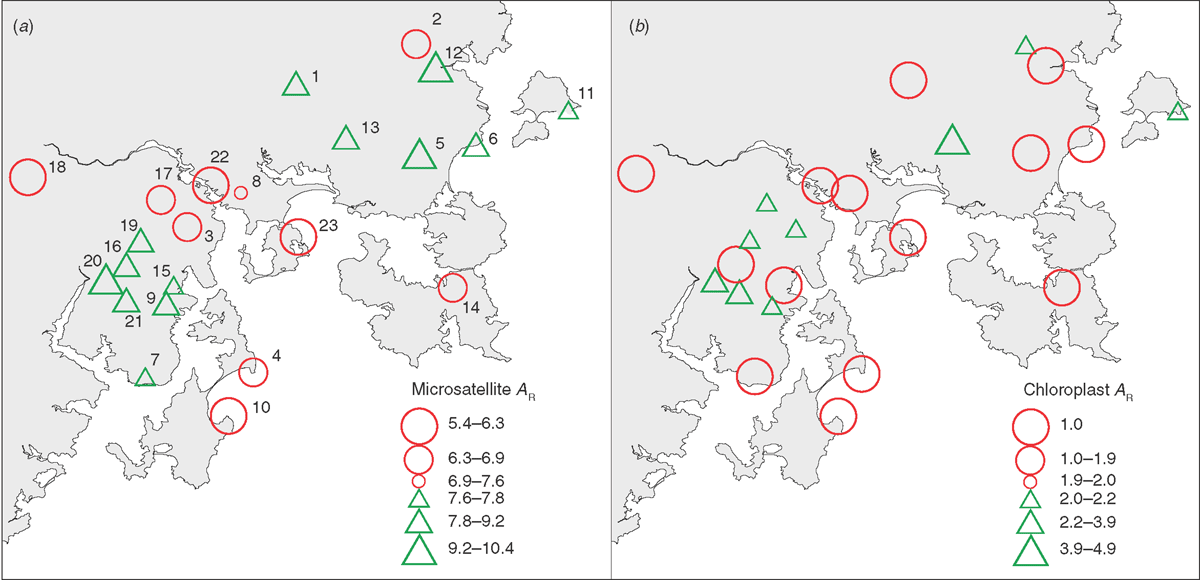
The 13 microsatellite loci used to genotype individuals from E. cordata and E. morrisbyi were highly polymorphic, ranging from 18 to 31 alleles detected per locus (mean A across loci = 24.4, data not shown). Total genetic diversity (HT) averaged across the 13 loci was slightly higher in E. cordata (HT = 0.86) than in E. morrisbyi (HT = 0.80), with many more alleles per loci, on average, detected in E. cordata (A = 24) than in E. morrisbyi (A = 10). The HE and HO within the E. cordata populations averaged 0.79 and 0.74, respectively (Table 1). There was higher genetic diversity in populations of subspecies quadrangulosa than populations of subspecies cordata, as measured by the HE and HO as well as allelic richness (AR) (Table 1). The small E. cordata population on Penguin Island (Population 10 in Fig. 2) and both E. morrisbyi populations (Populations 22, 23) had the lowest genetic diversity (Table 1). Spatial plotting of population genetic diversity revealed two regions of elevated allelic richness, one in the north-east and the other in the south-west of the range of E. cordata (Fig. 4a). Similar patterns were evident in the HE and HO of populations (Table 1). Populations peripheral to these regions of elevated diversity had lower genetic diversity (AR < 7.6), but there were exceptions such as Brown Mountain (Population 1, AR = 8.7, Fig. 4a). Population size could not explain this spatial pattern, with the Pearson’s product–moment correlation test showing no linear relationship between the number of individuals in each population and any of the genetic diversity parameters (Table 2). However, there was a tendency for HO to be significantly lower and the inbreeding levels (F) significantly higher in lower altitude populations of E. cordata (Table 2).
|

|
The populations of E. cordata were well differentiated, with an average FST of 0.09 (range 0.02–0.20; Appendix 3). This compares with an FST of 0.14 between the two E. morrisbyi populations and an average FST between the E. cordata and E. morrisbyi populations of 0.13. Populations of subspecies quadrangulosa were less differentiated (FST = 0.04, n = 3) than the E. cordata intermediates (FST = 0.09, n = 4) and subspecies cordata (FST = 0.10, n = 14). Populations of these subspecies were equally differentiated, on average, from the E. morrisbyi populations, but the smallest pair-wise values of FST involved the subspecies cordata populations from Corbetts Hill (Population 5) and Square Mountain (Population 13). All estimates of pair-wise FST between population pairs were statistically significant, even the lowest pair-wise FST (0.02), which was between the geographically adjacent populations of subspecies quadrangulosa at Snug Plains (21) and Sherwood Hill (20). Most of the higher pair-wise FST estimates involved the small E. cordata population at Penguin Island (10) (average pair-wise FST from other E. cordata populations, 0.16; Appendix 3). Penguin Island is the type locality of E. cordata (Potts 1988) but it is clearly a molecular outlier. An isolation-by-distance model could not explain the pattern of genetic differentiation between populations, with a Mantel test showing no relationship between geographic and genetic distance (Rxy = 0.18, P = 0.08, n = 21).
The genetic affinities of the E. cordata and E. morrisbyi individuals were analysed using STRUCTURE with no a priori population assignment. The post hoc ΔK method of Evanno et al. (2005) showed that the assignment of individuals into five genetic clusters (K = 5) had the highest ΔK value, suggesting this as the most optimal summary of their genetic affinities (Appendix 4). Although other K values had reasonably high ΔK estimates, additional genetic clusters tended only to separate individuals from specific populations and were not as meaningful as K = 5. Most individuals were assigned to two geographically separated clusters and comprised individuals from populations in the north-east (13, 5, 6, 11, 12, and 2) and south-west (17, 19, 16, 20, 21, 15, 9, and 7) of the E. cordata range (Fig. 5). The south-western cluster was more phenotypically diverse than the pure subspecies cordata cluster in the north-east and included individuals from subspecies cordata, subspecies quadrangulosa and their intermediates. The south-western cluster also shared partial genetic affinities with a smaller, third cluster of individuals from subspecies cordata populations at Taranna (14), Chimney Pot Hill (3), and Moogara (18) (Fig. 5). Two smaller clusters reflected affinities between E. cordata and E. morrisbyi, with individuals from the E. cordata populations at Penguin Island (10) and Brown Mountain (1) clustering with the E. morrisbyi population at Calverts Hill (23), whereas individuals from the E. cordata population at Meehan Range (8) clustered with the nearby E. morrisbyi population at Risdon Hill (22) (Fig. 5).

|
Geographic structure and diversity of the chloroplast genome
In total, 27 chloroplast haplotypes were identified following sequencing of the JLA+ chloroplast region in 127 samples from 23 populations of E. cordata and E. morrisbyi (Appendix 5), and these all showed the Southern haplotype (suffix JS; Freeman et al. 2001; McKinnon et al. 2004b). Twenty-four of these chloroplast haplotypes were private to a single population. Only JS81 was shared between E. cordata and E. morrisbyi populations and this sharing involved only O’Briens Hill (9) and the E. morrisbyi population at Risdon Hill (22), which are 30 km apart. Eleven of the 21 E. cordata populations and both E. morrisbyi populations were fixed for a single chloroplast haplotype (Appendix 5), and in four cases, this involved fixation for the most common haplotype (JS43). Nevertheless, higher diversity of chloroplast haplotype was found in E. cordata (HT = 0.89) than in E. morrisbyi (HT = 0.50). The chloroplast haplotype (allelic) richness calculated from this E. cordata dataset was not significantly correlated with the microsatellite allelic richness parameters, altitude or population size (Table 2). The majority of populations had low diversity of the chloroplast genome, except for populations at Square Mountain (13), Sherwood Hill (20) and Snug Plains (21) (Table 1, Fig. 4b). The E. cordata populations were highly differentiated in their chloroplast haplotypes, as indicated by an FST of 0.70. This high level of chloroplast differentiation among populations compared with the differentiation in the nuclear SSRs resulted in an estimation of 22 times more pollen- than seed-mediated gene flow in E. cordata.
Discussion
Our genome-wide genotyping of multiple species from the series Orbiculares showed that E. cordata is more aligned to the Tasmanian endemic species of series Orbiculares than to mainland species, and suggests E. cordata most likely evolved from a common ancestor shared with the Tasmanian endemics. This result confirms findings based on amplified fragment length polymorphism (AFLP) markers that three samples of E. cordata formed a monophyletic group within the Tasmanian Orbiculares species (McKinnon et al. 2008). Similarly, the phylogenetic study of Steane et al. (2011) found that the homoblastic E. cordata was more aligned to the heteroblastic Tasmanian Orbiculares species than to other homoblastic species of the Orbiculares, such as E. perriniana and the mainland E. pulverulenta. Indeed, these molecular studies suggest that these two homoblastic species from the mainland are more aligned to series Viminales than to Orbiculares (McKinnon et al. 2008; Steane et al. 2011). This also implies that neotenic homoblasty in E. cordata has evolved independently from a heteroblastic ancestor, and is consistent with the hypothesis that the retention of the juvenile foliage into reproductive maturity has evolved independently on multiple occasions within Eucalyptus (Potts and Wiltshire 1997). Although most E. cordata populations are homoblastic, trees in the Brown Mountain (1) and Penguin Island (10) populations, still flowering in the juvenile leaf stage, have been observed with adult leaves in the wild (Potts 1988). The heteroblastic characteristic of these populations, coupled with their genetic affinities to individuals from the larger, Calverts Hill (23) population of E. morrisbyi, suggests that these peripheral E. cordata populations may be relicts of the ancestral lineage from which the contemporary E. cordata evolved. This would explain the genetic affinities we detected between the southern and northern extremities of the E. cordata distribution (Link 1, Fig. 6), despite the small population at Penguin Island (10) being a genetic outlier. Penguin Island is the type locality for E. cordata (Potts 1989), and is clearly not typical of the species from a genetic (present study) and phenotypic (Potts 1988) perspective.

|
Although E. cordata is locally distributed as a series of isolated populations, the nuclear microsatellite diversity within populations as measured by HE is similar to that reported for widespread eucalypt species with more continuous distributions (average HE of three species 0.79; Byrne 2008). For example, the HE (0.79) and HO (0.74) in E. cordata populations are similar to those observed in Tasmanian populations of E. obliqua (HE = 0.80, HO = 0.79; Bloomfield et al. 2011), E. pauciflora (HE = 0.80, HO = 0.75; A. Gauli, unpubl. data), and E. globulus (HE = 0.81, HO = 0.70; Jones et al. 2013). By contrast, the two other eucalypt species studied that also occur in Tasmania as small, isolated populations have lower HE and HO than the average observed for the E. cordata populations: E. perriniana (HE = 0.72, HO = 0.62; Rathbone et al. 2007) and E. morrisbyi (HE = 0.69, HO = 0.64; Jones et al. 2005). These observations suggest that, on average, most of the E. cordata populations have not been isolated and/or have persisted as small populations for a long time from an evolutionary perspective. This conclusion is also supported by the moderate level of population differentiation (FST = 0.09), which is below the average reported for regionally (average FST of two species, 0.25) and locally (average FST of two species, 0.15) distributed eucalypt species, and more similar to the mean for widespread species (average FST of two species, 0.06) (Byrne 2008). Loss of heterozygosity for neutral molecular markers requires isolation and persistence of small population sizes for multiple generations (Ellstrand and Elam 1993), and none of these Tasmanian species comprising isolated populations, including E. cordata, have the low population genetic diversity and high population differentiation evident in species such as E. curtisii (FST = 0.30; Smith et al. 2003) and E. caesia (FST = 0.53; Byrne and Hopper 2008), which have persisted as isolated populations in ancient landscapes (Byrne 2008). These species have smaller population sizes than E. cordata, and they have no doubt experienced prolonged bottlenecks through their longer evolutionary history.
A positive relationship between genetic diversity parameters (AR, HE and HO) and population size may be expected due to genetic drift and inbreeding in small populations (Ellstrand and Elam 1993; Leimu et al. 2006). However, the absence of such a relationship is common in eucalypts (reviewed by Butcher et al. 2005), and this was the case for E. cordata. Allelic richness is a highly sensitive genetic diversity parameter and is influenced by the loss of rare alleles following bottlenecks in population size (Maruyama and Fuerst 1985). There is strong evidence that the small E. cordata population at Penguin Island has experienced a persistent bottleneck, based on its low allelic richness (AR = 5.4, compared with the species mean of 8). However, little support was found for major bottlenecks affecting genetic diversity in other small populations, suggesting that these populations have only recently become small and isolated, at least in terms of tree generations.
Eucalypts can have long generational times because of regeneration from underground lignotubers, which can fragment with time and result in disconnected, clonal patches (Tyson et al. 1998; Byrne 2008). In the case of E. morrisbyi, Jones et al. (2005) estimated that clonal patches in the Risdon Hill (22) population were at least 670–1523 years old. Many of the E. cordata populations (particularly of subspecies cordata) similarly comprise individuals that have regenerated vegetatively from large basal lignotubers (Potts 1989). Applying the rate of radial growth of lignotubers reported by Tyson et al. (1998) to the isolated clonal stems detected at Chimney Pot Hill (3), which were separated by 3.6 m, suggests that a single genotype can be at least 529–857 years old. This evidence argues that individuals within many populations of E. cordata could be very long lived, and represent genotypes remaining from previously larger populations that were not affected by small population processes.
Introgressive hybridisation is another possible mechanism for the maintenance of high levels of genetic diversity in the small populations of E. cordata. Hybridisation between co-occurring species has been suggested to increase genetic diversity through the introgression of alleles (Rieseberg et al. 2007) and has been observed in animals (Canestrelli et al. 2010) and in plants (Klier et al. 1991), including eucalypts (Butcher et al. 2002). In several eucalypt studies, the presence of F1 hybrids has been reported to increase with decreasing population size (Field et al. 2009; Larcombe et al. 2014; Potts and Wiltshire 1997), and contemporary and historic hybridisation of E. cordata has been reported (McKinnon et al. 2004b; P. Harrison pers. obs.). However, in the well-studied case of E. cordata–E. globulus hybridisation, McKinnon et al. (2010) provided evidence of cryptic introgression of nuclear markers from E. cordata into the surrounding E. globulus, but there was no evidence of cryptic introgression from E. globulus into pure E. cordata phenotypes.
Although multiple processes may be operating to affect population genetic diversity, we suggest that, on a broad scale, the patterns of molecular genetic diversity and affinities within E. cordata can be largely explained by differences in the time since populations have been isolated. The complex spatial patterns observed reflect the co-existence of relictual and more recently derived populations that have had various degrees of isolation following multiple phases of population extinction or contraction, and expansion. The spatial patterns of genetic diversity suggest that the populations of E. cordata comprise two differentiated regions. For differentiation to occur between such sizable groups of populations, a long separation with a lack of gene flow would be required. This suggests that in the past (probably before the last glacial epoch) the distribution of the two regions in the north-east and south-west were well separated (Links 4 and 5, Fig. 6). Surrounding the genetically diverse central core of each region are peripheral, often more divergent, populations of lower genetic diversity. Populations near the centre of a species geographical range generally exhibit higher genetic diversity and lower divergence than populations on the periphery (central–peripheral hypothesis; Brussard 1984; Eckert et al. 2008), which appears to hold for both the eastern and western cores of E. cordata. Indeed, Vucetich and Waite (2003) have shown that peripheral populations can experience up to 30 times greater genetic drift than central-core populations. Although peripheral populations have traditionally been studied in relation to a continuous central core (Eckert et al. 2008; Jones et al. 2013), in the case of E. cordata, the cores are not continuous but have become recently fragmented. This hypothesis is supported by the low differentiation between populations within the ‘phantom core’, which is suggestive of a more continuous distribution in the cores in the recent past.
Populations of low microsatellite diversity tend to separate the two central core areas and form a band through the central part of the E. cordata distribution, which includes E. morrisbyi (Fig. 4a). These populations are centred on the River Derwent valley and fringes of Storm Bay (even near sea level, e.g. Populations 10 and 23), and are likely the remnants of a more widespread distribution in this area during the various Quaternary glaciations. Lowered sea levels during the Quaternary glacials would have resulted in large expanses of low-altitude habitat suitable for eucalypt forest (Fig. 6; Kirkpatrick and Fowler 1998; McKinnon et al. 2004a; Jackson 2005). The affinities of the population at Taranna (14) on the Tasman Peninsula with western populations at Moogara (14) and Chimney Pot Hill (3) (Link 3, Fig. 6) rather than to geographically closer populations to the north provides support for this hypothesised past, more-or-less continuous distribution of E. cordata in the now-flooded Storm Bay. On the other hand, the genetic affinities between Penguin Island (10), Brown Mountain (1) and the Calverts Hill E. morrisbyi population (23) (Link 1, Fig. 6) provides evidence for an even older, more ancestral distribution in this area, which probably corresponds to an older glacial cycle. The antiquity of this link is suggested by the generally high differentiation of these populations from conspecific populations and the ancestral heteroblastic features in the two E. cordata populations. The genetic distinctiveness of the two isolated E. morrisbyi populations suggests long occupancy and isolation in this region, and the genetic affinities of the Meehan Range E. cordata population (8) and Risdon Hill E. morrisbyi (22) may reflect historical nuclear gene exchange between these two proximal populations early in the speciation process.
Chloroplast genome diversity was relatively low in many E. cordata populations, with most being fixed for a specific haplotype. This pattern was previously noted by McKinnon et al. (2004b) in a smaller scale study. However, the E. cordata population at Square Mountain (13) was an exception to this and had the highest chloroplast diversity. The unique chloroplast haplotypes found in Square Mountain have not been reported in any other eucalypt (McKinnon et al. 2001b; University of Tasmania DNA database 2014, unpubl. data, comprising 1780 samples from 44 species) and were not shared with other E. cordata populations. Chloroplast sharing with co-occurring eucalypts could explain the high haplotype diversity at Square Mountain (i.e. McKinnon et al. 2004b); however, the uniqueness of these chloroplast haplotypes to this E. cordata population argues against this diversity originating from hybridisation. The high haplotype diversity at Square Mountain contrasts with surrounding populations in the north-east, which tended to have high microsatellite diversity, yet are fixed for different chloroplast haplotypes.
The variation in the chloroplast genome in the north-eastern core populations likely reflects historical population processes associated with past population isolation or bottlenecks. Newly founded or expanded populations are expected to experience severe bottlenecks, which erode the genetic diversity (Excoffier et al. 2009). Because the chloroplast genome is maternally inherited in eucalypts (Byrne et al. 1993; McKinnon et al. 2001b), it is more susceptible to genetic drift than the nuclear genome (Petit et al. 1993). This pattern of low diversity of chloroplast haplotype in the north-eastern lineage could arise if the core had been established by founder events along an eastward expansion axis or re-established from multiple small remnants from a previous era during which chloroplast diversity had been eroded. Under this scenario, the greater pollen- than seed-mediated gene flow would allow nuclear genetic diversity to be rapidly re-established as populations expand, as demonstrated in several studies of isolated tree populations (Buschbom et al. 2011; Hampe et al. 2013). The high chloroplast diversity in the more western population at Square Mountain (13) suggests that it has not experienced a major bottleneck in the past and could be the source of the eastern expansion of E. cordata subspecies cordata in this north-eastern region (Link 4, Fig. 6).
In contrast to the north-eastern lineage, genetic diversity within the south-west lineage appears to have been shaped to some extent by recent, seed-mediated migration of populations across a mid-altitude plateau, south of the Mount Wellington Range (Fig. 6). Recent colonisation and maintenance of large populations is supported by the close microsatellite affinities and high diversity, and the sharing of a common chloroplast haplotype (JS43) among most populations in this core region. The low microsatellite diversity in two geographic outliers to this core, Cape Queen Elizabeth (4) and Mount Grosse (7), suggests that these populations have lost gene flow connectivity with this core. Although having greatest microsatellite affinities to the south-western lineage, the affinities of the low-altitude populations at Cape Queen Elizabeth (4) and Electrona (15) to the populations transgressing the Derwent Valley (Cluster 3, Fig. 4; Link 3, Fig. 6) may reflect their more ancestral, intermediate status.
The marked morphological differentiation between populations of the core of the south-western molecular lineage without major, putatively neutral microsatellite differentiation implies that selection has played a role in the evolution and maintenance of the subspecies quadrangulosa. This south-western core includes populations that are fixed for quadrangular branchlets and classify as subspecies quadrangulosa (e.g. Snug Plains, 21; Sherwood Hill, 20; Combes Hill, 19), populations that have round branchlets (e.g. O’Briens Hill, 9; Mount Grosse, 7) and are of subspecies cordata, and intermediates (Electrona, 15; Herringback (top), 16). This finding raises the question of the origin of the quadrangular branchlet phenotype. A possible scenario for the evolutionary origin of the quadrangular branchlets could be ancient hybridisation coupled with selection. Quadrangular branchlets have evolved several times within Maidenaria and they are most notable in juvenile E. globulus. Recent studies have found evidence of genome-wide introgression of adaptive traits in Helianthus (Yatabe et al. 2007; Vekemans 2010; Whitney et al. 2010) and Iris (reviewed in Arnold and Martin 2009), and some genomic regions of E. globulus could have been introgressed into an E. cordata-like hybrid. However, although introgression in the reverse direction (e.g. E. cordata markers into E. globulus) has been demonstrated, no evidence of nuclear introgression into E. cordata was evident (McKinnon et al. 2010). An alternative and perhaps more likely explanation for the quadrangular branchlets of subspecies quadrangulosa is that the trait has independently evolved within E. cordata as a direct or indirect response to directional selection for increased vegetative and reproductive organ size in the cooler, wetter and more productive environments occupied by this subspecies (Potts 1989) and may be of structural significance.
In conclusion, our genetic study of E. cordata showed that most populations maintain high levels of genetic diversity relative to other localised eucalypt species. This indicates that the species appears to be ‘genetically healthy’. The genetic clustering of individuals into five distinct groups provides a genetic framework for the management of the species. The preservation of populations in each of these groups as well as all of the relict populations will be important. Although most populations are well represented in reserves, the low number of individuals in some populations suggests that their ability to respond to stochastic environmental change may be limited, in particular, the small, relict populations at Penguin Island and Brown Mountain, which have preserved signals of the early speciation stage of E. cordata. The absence of marked genetic differentiation between many morphologically differentiated populations recognised as different E. cordata subspecies argues for recent evolution of the localised subspecies quadrangulosa through directional selection. The current distribution of subspecies quadrangulosa is more restricted than subspecies cordata, and although this subspecies is currently well reserved, the status and health of unreserved populations, including the intermediates, should be monitored to ensure long-term maintenance of the genetic diversity of this endemic species.
Acknowledgements
We thank the many public and private landholders for access to sample E. cordata populations on their land, including Parks and Wildlife Services Tasmania (TFL 14060), Forestry Tasmania, Norske Skog and the Australian Department of Defence. We also thank Sascha Wise and Joanne Harrison for their help with laboratory and field work. This project was supported by a Professor Newton Barbour Scholarship (PAH), the CRC for Forestry, and the Australian Research Council Discovery grants DP0986491, DP130104220 and DP120100501.
References
Arnold ML, Martin NH (2009) Adaptation by introgression. Journal of Biology 8, 82.1–82.3.| Adaptation by introgression.Crossref | GoogleScholarGoogle Scholar |
Bloomfield JA, Nevill P, Potts BM, Vaillancourt RE, Steane DA (2011) Molecular genetic variation in a widespread forest tree species Eucalyptus obliqua (Myrtaceae) on the island of Tasmania. Australian Journal of Botany 59, 226–237.
| Molecular genetic variation in a widespread forest tree species Eucalyptus obliqua (Myrtaceae) on the island of Tasmania.Crossref | GoogleScholarGoogle Scholar |
Brondani RPV, Brondani C, Tarchini R, Grattapaglia D (1998) Development, characterization and mapping of microsatellite markers in Eucalyptus grandis and E. urophylla. Theoretical and Applied Genetics 97, 816–827.
| Development, characterization and mapping of microsatellite markers in Eucalyptus grandis and E. urophylla.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXotVaht7o%3D&md5=37cef8a5ff1ad21a5b1896c6127dd08dCAS |
Brondani RPV, Williams ER, Brondani C, Grattapaglia D (2006) A microsatellite-based consensus linkage map for species of Eucalyptus and a novel set of 230 microsatellite markers for the genus. BMC Plant Biology 6,
| A microsatellite-based consensus linkage map for species of Eucalyptus and a novel set of 230 microsatellite markers for the genus.Crossref | GoogleScholarGoogle Scholar |
Brooker MIH (2000) A new classification of the genus Eucalyptus L’Her. (Myrtaceae). Australian Systematic Botany 13, 79–148.
| A new classification of the genus Eucalyptus L’Her. (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Brussard PF (1984) Geographic patterns and environmental gradients: the central–marginal model in Drosophila revisited. Annual Review of Ecology and Systematics 15, 25–64.
| Geographic patterns and environmental gradients: the central–marginal model in Drosophila revisited.Crossref | GoogleScholarGoogle Scholar |
Buschbom J, Yanbaev Y, Degen B (2011) Efficient long-distance gene flow into an isolated relict oak stand. The Journal of Heredity 102, 464–472.
| Efficient long-distance gene flow into an isolated relict oak stand.Crossref | GoogleScholarGoogle Scholar | 21525180PubMed |
Butcher PA, Otero A, McDonald MW, Moran GF (2002) Nuclear RFLP variation in Eucalyptus camaldulensis Dehnh. from northern Australia. Heredity 88, 402–412.
| Nuclear RFLP variation in Eucalyptus camaldulensis Dehnh. from northern Australia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XksFSrs70%3D&md5=daf02295e3582a23d6ac4bda5187a0beCAS | 11986878PubMed |
Butcher PA, Skinner AK, Gardiner CA (2005) Increased inbreeding and inter-species gene flow in remnant populations of the rare Eucalyptus benthamii. Conservation Genetics 6, 213–226.
| Increased inbreeding and inter-species gene flow in remnant populations of the rare Eucalyptus benthamii.Crossref | GoogleScholarGoogle Scholar |
Byrne M (2008) Phylogeny, diversity and evolution of eucalypts. In ‘Plant genome. Biodiversity and evolution. Vol. 1’. (Eds AK Sharma, A Sharma) pp. 303–346. (Science Publishers: Plymouth, UK)
Byrne M, Hopper SD (2008) Granite outcrops as ancient islands in old landscapes: evidence from the phylogeography and population genetics of Eucalyptus caesia (Myrtaceae) in Western Australia. Biological Journal of the Linnean Society. Linnean Society of London 93, 177–188.
| Granite outcrops as ancient islands in old landscapes: evidence from the phylogeography and population genetics of Eucalyptus caesia (Myrtaceae) in Western Australia.Crossref | GoogleScholarGoogle Scholar |
Byrne M, Moran GF, Tibbits WN (1993) Restriction map and maternal inheritance of chloroplast DNA in Eucalyptus nitens. The Journal of Heredity 84, 218–220.
Canestrelli D, Aloise G, Cecchetti S, Nascetti G (2010) Birth of a hotspot of intraspecific genetic diversity: notes from the underground. Molecular Ecology 19, 5432–5451.
| Birth of a hotspot of intraspecific genetic diversity: notes from the underground.Crossref | GoogleScholarGoogle Scholar | 21059127PubMed |
Chybicki IJ, Burczyk J (2009) Simultaneous estimation of null alleles and inbreeding coefficients. The Journal of Heredity 100, 106–113.
| Simultaneous estimation of null alleles and inbreeding coefficients.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVKmsA%3D%3D&md5=9c239e6f48faf049b2f6a61e64629a16CAS | 18936113PubMed |
Comps B, Gomory D, Letouzey J, Thiebaut B, Petit RJ (2001) Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics 157, 389–397.
Cremer KW (1977) Distance of seed dispersal in Eucalyptus estimated from seed weights. Australian Forest Research 7, 225–228.
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12, 13–15.
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4, 359–361.
| STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method.Crossref | GoogleScholarGoogle Scholar |
Eckert CG, Samis KE, Lougheed SC (2008) Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Molecular Ecology 17, 1170–1188.
| Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXksFShsbc%3D&md5=4d56d481228082e84adcec7cdff7ef25CAS | 18302683PubMed |
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24, 217–242.
| Population genetic consequences of small population size: implications for plant conservation.Crossref | GoogleScholarGoogle Scholar |
Ennos RA (1994) Estimating the relative rates of pollen and peed migration among plant-populations. Heredity 72, 250–259.
| Estimating the relative rates of pollen and peed migration among plant-populations.Crossref | GoogleScholarGoogle Scholar |
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14, 2611–2620.
| Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXmvF2qtrg%3D&md5=9768cb9d92ec62dae4e93a84977410f8CAS | 15969739PubMed |
Excoffier L, Foll M, Petit RJ (2009) Genetic consequences of range expansions. Annual Review of Ecology Evolution and Systematics 40, 481–501.
| Genetic consequences of range expansions.Crossref | GoogleScholarGoogle Scholar |
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587.
Field DL, Ayre DJ, Whelan RJ, Young AG (2009) Molecular and morphological evidence of natural interspecific hybridization between the uncommon Eucalyptus aggregata and the widespread E. rubida and E. viminalis. Conservation Genetics 10, 881–896.
| Molecular and morphological evidence of natural interspecific hybridization between the uncommon Eucalyptus aggregata and the widespread E. rubida and E. viminalis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXntlShurc%3D&md5=91f2d2a92ecad0d8e3e657a6f62227aaCAS |
Freeman JS, Jackson HD, Steane DA, McKinnon GE, Dutkowski GW, Potts BM, Vaillancourt RE (2001) Chloroplast DNA phylogeography of Eucalyptus globulus. Australian Journal of Botany 49, 585–596.
| Chloroplast DNA phylogeography of Eucalyptus globulus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXptVemu7w%3D&md5=000698299ba2aece477203fb156d7086CAS |
Goudet J (2002) ‘FSTAT, a program to estimate and test gene diversities and fixation indices. Version 2.9.3.’ (Department of Ecology & Evolution, UNIL: Lausanne, Switzerland). Available at http://www2.unil.ch/popgen/softwares/fstat.htm.[Verified 3 April 2014]
Hampe A, Pemonge MH, Petit RJ (2013) Efficient mitigation of founder effects during the establishment of a leading-edge oak population. Proceedings. Biological Sciences 280,
| Efficient mitigation of founder effects during the establishment of a leading-edge oak population.Crossref | GoogleScholarGoogle Scholar |
Hudson CJ, Freeman JS, Kullan ARK, Petroli CD, Sansaloni CP, Kilian A, Dettering F, Grattapaglia D, Potts BM, Myburg AA, Vaillancourt RE (2012) A reference linkage map for Eucalyptus. BMC Genomics 13, 240
| A reference linkage map for Eucalyptus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhslamu7zM&md5=d587321f71f6633511e37f4b06bd3d51CAS | 22702473PubMed |
Jackson WD (2005) Palaeohistory of vegetation change: the last 2 million years. In ‘Vegetation of Tasmania’. (Eds JB Reid, RS Hill, MJ Brown, MJ Hovenden) pp. 64–88. (Australian Biological Resources Study: Hobart)
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806.
| CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXpt1ahtbs%3D&md5=8ee17ac13abf31e1c79922b4db068da5CAS | 17485429PubMed |
Jones RC, McKinnon GE, Potts BM, Vaillancourt RE (2005) Genetic diversity and mating system of an endangered tree Eucalyptus morrisbyi. Australian Journal of Botany 53, 367–377.
| Genetic diversity and mating system of an endangered tree Eucalyptus morrisbyi.Crossref | GoogleScholarGoogle Scholar |
Jones RC, Steane DA, Lavery M, Vaillancourt RE, Potts BM (2013) Multiple evolutionary processes drive the patterns of genetic differentiation in a forest tree species complex. Ecology and Evolution 3, 1–17.
| Multiple evolutionary processes drive the patterns of genetic differentiation in a forest tree species complex.Crossref | GoogleScholarGoogle Scholar |
Kirkpatrick JB, Fowler M (1998) Locating likely glacial forest refugia in Tasmania using palynological and ecological information to test alternative climatic models. Biological Conservation 85, 171–182.
| Locating likely glacial forest refugia in Tasmania using palynological and ecological information to test alternative climatic models.Crossref | GoogleScholarGoogle Scholar |
Klier K, Leoschke MJ, Wendel JF (1991) Hybridisation and interogression in white and yellow ladyslipper orchards (Cypripedium candidum and C. pubescens). The Journal of Heredity 82, 305–319.
Larcombe M, Barbour RC, Vaillancourt RE, Potts BM (2014) Assessing the risk of exotic gene flow from Eucalyptus globulus plantations to native E. ovata forests. Forest Ecology and Management 312, 193–202.
| Assessing the risk of exotic gene flow from Eucalyptus globulus plantations to native E. ovata forests.Crossref | GoogleScholarGoogle Scholar |
Leimu R, Mutikainen P, Koricheva J, Fischer M (2006) How general are positive relationships between plant population size, fitness and genetic variation? Journal of Ecology 94, 942–952.
| How general are positive relationships between plant population size, fitness and genetic variation?Crossref | GoogleScholarGoogle Scholar |
Lewis PO, Zaykin D (2001) ‘Genetic data analysis: computer program for the analysis of allelic data. Version 1.1.’ Available at http://www.eeb.uconn.edu/people/plewis/software.php.[Verified 3 April 2014]
Maruyama T, Fuerst PA (1985) Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111, 675–689.
McKinnon GE, Vaillancourt RE, Jackson HD, Potts BM (2001a) Chloroplast sharing in the Tasmanian eucalypts. Evolution 55, 703–711.
| Chloroplast sharing in the Tasmanian eucalypts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXktleqtLs%3D&md5=2d9ad781175a1e23995a1b774aa8bd0bCAS | 11392388PubMed |
McKinnon GE, Vaillancourt RE, Tilyard PA, Potts BM (2001b) Maternal inheritance of the chloroplast genome in Eucalyptus globulus and interspecific hybrids. Genome 44, 831–835.
| Maternal inheritance of the chloroplast genome in Eucalyptus globulus and interspecific hybrids.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXos12ls78%3D&md5=acd2c803322f37c4dbe34be37dc99044CAS |
McKinnon GE, Jordan GJ, Vaillancourt RE, Steane DA, Potts BM (2004a) Glacial refugia and reticulate evolution: the case of the Tasmanian eucalypts. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 359, 275–284.
| Glacial refugia and reticulate evolution: the case of the Tasmanian eucalypts.Crossref | GoogleScholarGoogle Scholar | 15101583PubMed |
McKinnon GE, Vaillancourt RE, Steane DA, Potts BM (2004b) The rare silver gum, Eucalyptus cordata, is leaving its traces in the organellar gene pool of Eucalyptus globulus. Molecular Ecology 13, 3751–3762.
| The rare silver gum, Eucalyptus cordata, is leaving its traces in the organellar gene pool of Eucalyptus globulus.Crossref | GoogleScholarGoogle Scholar | 15548288PubMed |
McKinnon GE, Vaillancourt RE, Steane DA, Potts BM (2008) An AFLP marker approach to lower-level systematics in Eucalyptus (Myrtaceae). American Journal of Botany 95, 368–380.
| An AFLP marker approach to lower-level systematics in Eucalyptus (Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXktlyjs7Y%3D&md5=0eb70a7c44c6ee79b3bb4f96a51d319bCAS | 21632361PubMed |
McKinnon GE, Smith JJ, Potts BM (2010) Recurrent nuclear DNA introgression accompanies chloroplast DNA exchange between two eucalypt species. Molecular Ecology 19, 1367–1380.
| Recurrent nuclear DNA introgression accompanies chloroplast DNA exchange between two eucalypt species.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXlvVWhtrk%3D&md5=7c565fd5e2471b50431348cd63665b2dCAS | 20298471PubMed |
Nei M (1972) Genetic distance between populations. American Naturalist 106, 283
| Genetic distance between populations.Crossref | GoogleScholarGoogle Scholar |
Nei H (1987) ‘Molecular evolution genetics.’ (Columbia University Press: New York)
Nicolle D (2006) ‘Eucalypts of Victoria and Tasmania.’ (Bloomings Books: Melbourne)
Nicolle D, Potts BM, McKinnon GE (2008) Eucalyptus cordata subsp. quadrangulosa (Myrtaceae), a new taxon of restricted distribution from Southern Tasmania. In ‘Papers and proceedings of the Royal Society of Tasmania. No. 142, Vol. 2’. pp. 71–78. (The Royal Society of Tasmania: Hobart)
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539.
| GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVehtbjI&md5=039b7447ba51da3cb7b5b5ad3760bb54CAS | 22820204PubMed |
Petit RJ, Kremer A, Wagner DB (1993) Finite island model for organelle and nuclear genes in plants. Heredity 71, 630–641.
| Finite island model for organelle and nuclear genes in plants.Crossref | GoogleScholarGoogle Scholar |
Potts BM (1988) The distribution and type locality of Eucalyptus cordata Labill. – an historical account. In ‘Papers and proceedings of the Royal Society of Tasmania. No. 122, Vol. 2‘. pp. 31–38. (The Royal Society of Tasmania: Hobart)
Potts BM (1989) ‘Population variation and conservation status of a rare Tasmanian endemic, Eucalyptus cordata.’ (Tasmanian Forest Research Council: Hobart)
Potts BM, Wiltshire RJE (1997) Eucalypt genetics and genecology. In ‘Eucalypt ecology: individuals to ecosystems’. (Eds JE Williams, JCZ Woinarski) pp. 56–91. (Cambridge University Press: New York)
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
R Core Team (2012) ‘R: a language and environment for statistical computing. Version 2.15.1.’ (The R Foundation for Statistical Computing, Institute for Statistics and Mathematics: Vienna). Available at http://cran.r-project.org/.[Verified 3 April 2014]
Rathbone DA, McKinnon GE, Potts BM, Steane DA, Vaillancourt RE (2007) Microsatellite and cpDNA variation in island and mainland populations of a regionally rare eucalypt, Eucalyptus perriniana (Myrtaceae). Australian Journal of Botany 55, 513–520.
| Microsatellite and cpDNA variation in island and mainland populations of a regionally rare eucalypt, Eucalyptus perriniana (Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXpt1SltbY%3D&md5=0bbb12ba830fdcab3edf544d2a266f44CAS |
Rieseberg LH, Kim SC, Randell RA, Whitney KD, Gross BL, Lexer C, Clay K (2007) Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129, 149–165.
| Hybridization and the colonization of novel habitats by annual sunflowers.Crossref | GoogleScholarGoogle Scholar | 16955330PubMed |
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145, 1219–1228.
Sansaloni CP, Petroli CD, Jaccoud D, Carling J, Detering F, Grattapaglia D, Kilian A (2011) Diversity Arrays Technology (DArT) and next generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proceedings 5, 54–55.
| Diversity Arrays Technology (DArT) and next generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus.Crossref | GoogleScholarGoogle Scholar |
Slee AV, Brooker MIH, Duffy SM, West JG (2006) ‘EUCLID: eucalypts of Australia.’ 3rd edn (CSIRO Publishing: Melbourne)
Smith S, Hughes J, Wardell-Johnson G (2003) High population differentiation and extensive clonality in a rare mallee eucalypt: Eucalyptus curtisii – conservation genetics of a rare mallee eucalypt. Conservation Genetics 4, 289–300.
Steane DA, Vaillancourt RE, Russell J, Powell W, Marshall D, Potts BM (2001) Development and characterisation of microsatellite loci in Eucalyptus globulus (Myrtaceae). Silvae Genetica 50, 89–91.
Steane DA, Nicolle D, Sansaloni CP, Petroli CD, Carling J, Kilian A, Myburg AA, Grattapaglia D, Vaillancourt RE (2011) Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping. Molecular Phylogenetics and Evolution 59, 206–224.
| Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping.Crossref | GoogleScholarGoogle Scholar | 21310251PubMed |
Threatened Species Unit (2003) ‘Morrisby’s gum, Eucalyptus morrisbyi draft flora recovery plan, 2004–2008.’ (Department of Primary Industries, Water and Environment: Hobart)
Tyson M, Vaillancourt RE, Reid JB (1998) Determination of clone size and age in a mallee eucalypt using RAPDs. Australian Journal of Botany 46, 161–172.
| Determination of clone size and age in a mallee eucalypt using RAPDs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXksV2qurk%3D&md5=7728a74c3e2380866ac6091a2d470d35CAS |
Vekemans X (2010) What’s good for you may be good for me: evidence for adaptive introgression of multiple traits in wild sunflower. New Phytologist 187, 6–9.
| What’s good for you may be good for me: evidence for adaptive introgression of multiple traits in wild sunflower.Crossref | GoogleScholarGoogle Scholar | 20624229PubMed |
Vucetich JA, Waite TA (2003) Spatial patterns of demography and genetic processes across the species’ range: null hypotheses for landscape conservation genetics. Conservation Genetics 4, 639–645.
| Spatial patterns of demography and genetic processes across the species’ range: null hypotheses for landscape conservation genetics.Crossref | GoogleScholarGoogle Scholar |
Whitney KD, Randell RA, Rieseberg LH (2010) Adaptive introgression of abiotic tolerance traits in the sunflower Helianthus annuus. New Phytologist 187, 230–239.
| Adaptive introgression of abiotic tolerance traits in the sunflower Helianthus annuus.Crossref | GoogleScholarGoogle Scholar | 20345635PubMed |
Williams K, Potts M (1996) The natural distribution of Eucalyptus species in Tasmania. Tasforests 8, 39–149.
Wiltshire RJE, Potts BM, Reid JB (1991) Phenotypic affinities, variability and conservation status of a rare Tasmanian endemic, Eucalyptus morrisbyi R.G.Brett. In ‘Aspects of Tasmanian botany – a tribute to Winifred Curtis’. (Eds MR Banks, SJ Smith, AE Orchard, G Gantvilas) pp. 213–229. (Royal Society of Tasmania: Hobart)
Woodhams M, Steane DA, Jones RC, Nicolle D, Moulton V, Holland BR (2013) Novel distances for Dollo data. Systematic Biology 62, 62–77.
| Novel distances for Dollo data.Crossref | GoogleScholarGoogle Scholar | 22914977PubMed |
Yatabe Y, Kane NC, Scotti-Saintagne C, Rieseberg LH (2007) Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics 175, 1883–1893.
| Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmsFyls7w%3D&md5=2ee98812bbe15a684e663d250f0466dcCAS | 17277373PubMed |

|

|

|
Appendix 4. Plot of the ΔK method of Evanno et al. (2005) used to estimate the optimal number of K clusters using the STRUCTURE runs from K = 2 to K = 23


|



