A preliminary study of the molecular epidemiology of Brachyspira hyodysenteriae isolates in Australia
P. K. Holyoake A C , T. La B , N. D. Phillips B and D. J. Hampson BA Holyoake Veterinary Consulting Pty Ltd, Strathfieldsaye, VIC 3551.
B Murdoch University, Murdoch, WA 6150.
C Corresponding author. Email: trishpigvet@icloud.com
Animal Production Science 55(12) 1531-1531 https://doi.org/10.1071/ANv55n12Ab076
Published: 11 November 2015
Swine dysentery (SD) is a mucohaemorrhagic colitis of grower-finisher pigs. Affected pigs have faeces ranging from soft, yellow-grey to mucoid, bloody diarrhoea. Swine dysentery is one of the most economically significant enteric diseases of pigs in Australia due to its effect on growth rate, feed efficiency, mortality and the associated medication control costs. The classical causative agent is a strongly β-haemolytic anaerobic intestinal spirochaete Brachyspira hyodysenteriae. Diagnosis of SD requires bacterial isolation and (or) identification using polymerase chain reaction (PCR). A number of PCR methods have been described for identifying B. hyodysenteriae. In this study, a multiplex PCR for B. hyodysenteriae, B. pilosicoli, L. intracellularis and Salmonella spp. including primers described by Elder et al. (1997) was compared with the PCR targeting NADH oxidase (nox) as described by La et al. (2006). Multi-locus sequence typing (MLST) was used to determine the relatedness of the B. hyodysenteriae isolates (La et al. 2009). The hypothesis was that isolates that were test-negative using the multiplex PCR but test-positive using the simple PCR were related but different to those positive on both PCR tests.
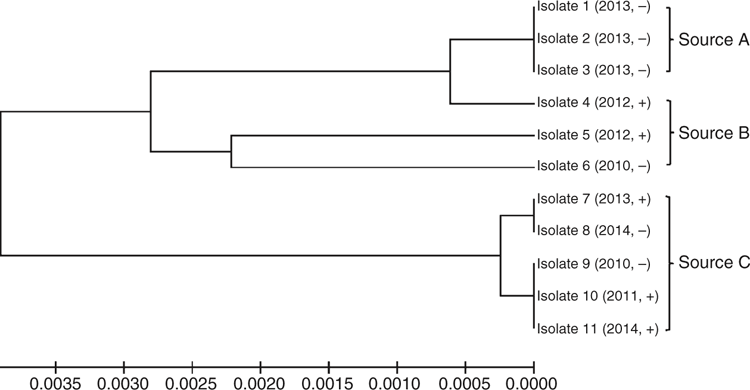
A total of 11 B. hyodysenteriae isolates from grower pigs from 11 farms having clinical signs of SD and collected over the period 2010–2014 was tested. Isolates were cultured to demonstrate pure cultures for MLST. The pigs originated from three genetic sources (Sources A, B and C). Isolates 1–3 were from three different farms supplied by Source A. Isolates 4, 5 and 6 were from three farms supplied by Source B. Isolates 7–11 were from five different farms supplied by Source C.
The multiplex PCR detected six (55%) of the nox PCR positive isolates. There was no clear relationship between the enteric PCR positive and negative isolates (Fig. 1). Isolates from different farms that obtained pigs from the same source generally were closely related, with isolates 1–3 (Source A) and isolates 7–11 (Source C) being identical or nearly identical, but different from those recovered elsewhere.
This study showed that there were no consistent strain-related patterns among multiplex PCR negative or positive isolates. Isolates from pigs from the same sources were similar in MLST, demonstrating that this method can reliably be used to map the movement of B. hyodysenteriae isolates between farms.
References
Elder RO, Duhamel GE, Mathiesen MR, Erickson ED, Gebhart CJ, Oberst RD (1997) Journal of Veterinary Diagnostic Investigation 9, 281–286.| Crossref | GoogleScholarGoogle Scholar |
La T, Collins AM, Phillips ND, Oksa A, Hampson DJ (2006) Letters in Applied Microbiology 42, 284–288.
| Crossref | GoogleScholarGoogle Scholar |
La T, Phillips ND, Harland BL, Wanchanthuek P, Bellgard MI, Hampson DJ (2009) Veterinary Microbiology 138, 330–338.
| Crossref | GoogleScholarGoogle Scholar |
Gene sequencing undertaken at Murdoch University was funded by the Pork CRC Limited Australia. We thank the Bendigo Pig Services Centre at the Department of Economic Development, Jobs, Transport and Resources for providing the isolates.