Exploring genomic approaches to fast-track genetic gains in breechstrike resistance in Merino sheep
S. Dominik A D , A. Reverter
A D , A. Reverter  B , L. R. Porto-Neto B , J. C. Greeff
B , L. R. Porto-Neto B , J. C. Greeff  C and J. L. Smith A
C and J. L. Smith A
A CSIRO Agriculture and Food, FD McMasters Laboratories, 9308 New England Highway, Armidale, NSW 2350, Australia.
B CSIRO Agriculture and Food, Queensland Bioprecinct, 306 Carmody Road, St Lucia, Qld 4067, Australia.
C Department of Primary Industries and Regional Development, 3 Baron-Hay Court, South Perth, WA 6151, Australia.
D Corresponding author. Email: Sonja.dominik@csiro.au
Animal Production Science - https://doi.org/10.1071/AN21124
Submitted: 5 March 2021 Accepted: 9 July 2021 Published online: 30 August 2021
Journal Compilation © CSIRO 2021 Open Access CC BY-NC-ND
Abstract
Context: Breech flystrike is a costly trait to measure. Industry investment into genetic solutions for breech flystrike has resulted in the availability of estimated breeding values for indicator traits, such as breech wrinkle, breech cover and dag. However, selection is based on indicator traits rather than breech flystrike itself, and genetic gains could be enhanced through genomic selection approaches.
Aim: This study investigated whether genomic approaches based on major genes, such as marker-assisted selection, or genomic selection based on genomic breeding values, would be the most efficient application of genomic information to enhance genetic gains for breech flystrike resistance.
Methods: The analysis comprised 1535 sheep of the Merino Breeding for Breech Flystrike Resistance Resource flocks from New South Wales and Western Australia with high density genotypes (actual and imputed). A genome-wide association study was conducted on breech flystrike and its indicator traits, namely, breech wrinkle, dag and breech cover. The study also estimated genomic breeding values and their accuracy.
Key results: The SNP associations found in this study did not point to the existence of few genes with major effects on breech flystrike resistance or its indicator traits. Throughout the genome, associations of small effect were found, which enabled the estimation of genomic breeding values. However, these were of low accuracy, as expected for the size of the dataset.
Conclusion: Genomic prediction of breeding values for breech flystrike resistance is a feasible tool for applying genomic technology in the Merino industry.
Implications: A reference population of appropriate size needs to be established for this difficult-to-measure trait, and a dispersed reference population could be an effective option.
Keywords: flystrike, Merino sheep, mulesing, genomic selection.
Introduction
Resistance to breech flystrike has long been a priority research area for Australian Wool Innovation. Australian sheep breeding values for indicator traits of breech flystrike resistance are available through Sheep Genetics (Brown et al. 2010) and provide industry with tools to improve breech flystrike through indirect selection on indicator traits. Substantial genetic gains that reduce breech flystrike incidence to as low as 0.1 strikes per sheep per year have been predicted over a 10–20 year period with the use of indicator traits in selection index scenarios (Brien et al. 2021). Genetic gain, particularly in wool-focussed Merino sheep breeding programs, could be fast-tracked with the application of genomic selection (van der Werf 2009). It has been shown that the major benefit for industry will be realised from applying genomic technology to traits that are difficult or expensive to measure on live animals, such as disease traits, including breech flystrike, that cause a compromise in production and welfare or reproduction traits measured later in life (van der Werf 2009).
Breech flystrike resistance is a difficult and expensive trait to measure (Smith et al. 2009), and would qualify as a potentially good candidate for genomic approaches, such as genomic or marker-assisted selection (Meuwissen et al. 2001; Dekkers 2004).
Release 43 of the Sheep Quantitative Trait Loci (QTL) database, SheepQTLdb, in 2020 (Hu et al. 2013) reported 3562 major genes for 270 traits in sheep. However, only a small number of the reported major genes, such as a test for polledness, have been targeted with direct selection in Australian sheep breeding programs. No QTL associated with breech flystrike resistance or its indicator traits have been reported to date.
Genomic selection has been adopted at varying speed across the various livestock species. Application in the dairy industry has been revolutionary, driven by the benefit that young bulls can be selected without the need for progeny testing, reducing the generation interval and resulting in double the annual genetic gain (Boichard et al. 2016). The main limiting factor to the development of genomic selection tools is the generation of a reference population of a sufficient size due to the cost of phenotyping, in particular for traits of low heritability or those that are difficult to measure (Boichard et al. 2016). However, genomic selection does provide significant opportunities; for example, if the accuracy of a selection index with genomic estimated breeding values (GEBV) are at least as high as the square root of the heritability, genetic gains in sheep breeding programs could be increased by up to 40% (van der Werf 2009).
This study hypothesised that due to a lack of major effects of a few genes, a reference population and the estimation of GEBV would be an applicable genomic approach to fast-track genetic improvement for breech flystrike resistance in Merino sheep. Analysis of genomic data forms the basis for recommended pathways of commercial application of genomic technologies, such as marker-assisted selection or genomic selection (Dekkers 2004; Goddard 2009; Daetwyler et al. 2010; Zeng et al. 2012). Here, genome-wide association studies were conducted for breech flystrike resistance and its indicator traits to investigate whether genes of major effect exist, providing potential for marker-assisted selection approaches, and to explore the estimation of GEBV for application in genomic selection. An initial report of the experimental work, described below, has appeared in report for Australian Wool Innovation (2019a).
Materials and methods
Genotype data and quality control
The genotype and phenotype data for analysis contained two datasets, on the basis of the genotyping platform used. Dataset 1 included animals of the Breech Flystrike Resource flocks in New South Wales (NSW) and Western Australia (WA) (Smith et al. 2009; Greeff et al. 2014) born between 2005 and 2011. Dataset 2 included animals from the same flocks born between 2011 and 2014. The data were subsets of the Breech Flystrike Resource flocks and were selected to have a good representation of sires across phenotypes and fixed effects.
Dataset 1 comprised a total of 959 DNA samples from individual sheep of the Breech Flystrike Resource flocks in WA (n = 266) and NSW (n = 675), including 18 internal control samples. All samples were genotyped using the Illumina Ovine HD Beadchip with 606 006 single-nucleotide polymorphisms (SNPs).
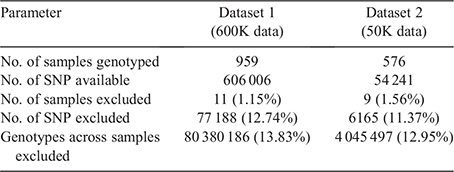
The genotypic data were submitted into a quality control (QC) process using snpQC (Gondro et al. 2014). Table 1 summarises the excluded number of SNPs and samples. In summary, quality control excluded 77 188 SNPs (12.74%) and 11 samples (1.15%), with 528 818 SNPs and 948 samples remaining in the dataset. Some SNPs failed multiple criteria. Flagged samples and SNPs were not included in the analysis, including those on the sex chromosomes X and Y and unmapped chromosomes (Chromosome 0).
|
Dataset 2 included a total of 576 DNA samples from sheep of the Breech Flystrike Resource flocks in NSW (n = 288) and WA (n = 288). All samples were genotyped with the Ovine 50K Illumina SNP chip. Quality control using snpQC (Gondro et al. 2014) excluded nine samples that did not pass the filtering criteria (1.56%). From the 54 241 SNPs, 6165 were excluded (11.37%). Of the total of 31 242 816 genotypes, 4 045 497 were excluded (12.95%; Table 1). Flagged samples and SNPs were removed from the analysis, including those on the sex chromosomes X and Y and unmapped chromosomes (Chromosome 0).
Phasing and imputation
To combine the two datasets, the 50K dataset was first prepared using PLINK (Purcell et al. 2007) and phased using the software Eagle ver. 2.3.5 (Loh et al. 2016). The 600K genotype data were also phased and missing genotypes imputed using Minimac3 (Das et al. 2016). In the next step, the 50K genotypes were imputed up to 600K density using Minimac3 (Das et al. 2016). The process of phasing and imputation with Eagle and Minimac3 has been described in Al-Mamun et al. (2017). As a final step, the two datasets were combined.
Phenotype data
The phenotype records of Dataset 1 were collected on 926 animals, with a full set of trait records and fixed effects. Dataset 2 contained phenotypes collected from 567 animals after genotype QC, of which 554 have a full set of phenotype records. Overall, the combined phenotype datasets contained 1480 animals comprising 949 from the NSW flock and 531 from the WA flock.
Traits for analysis included breech flystrike (STRIKE), coded as ‘struck’ or ‘not struck’ and its indicator traits, namely breech cover (BCOV), dag (DAG) and breech wrinkle (BWRK). Sheep at both sites were scored in five categories for these three traits as described in the ‘Visual Sheep Scores’ (Australian Wool Innovation 2019b). The traits DAG and BWRK had low mean scores and low variability in NSW and WA flocks respectively (Smith et al. 2009; Greeff et al. 2014). Therefore, to have adequate representation of phenotypic categories, all indicator traits were regrouped for both flocks to low, medium or high level of trait expression for analysis. All traits were categorised as, low (trait score ≤2.5), moderate (>2.5 trait score ≤4.0) and high (trait score >4.0) for both flocks. For the NSW flock all DAG scores and for WA flock all breech wrinkle scores fell into the low category (trait scores ≤2.5). Fixed effects included site (NSW or WA), sex (male or female), drop (2005–2009 or 2011–2014) and mules status (MULES, mulesed or unmulesed). Mules status was confounded with year because neither of the resource flocks was mulesed after 2011. In Dataset 2, all sheep in the WA flock were 2014 drop. Both breech flystrike phenotypes were well represented in the data, with 704 sheep struck and 768 not struck across all year drops.
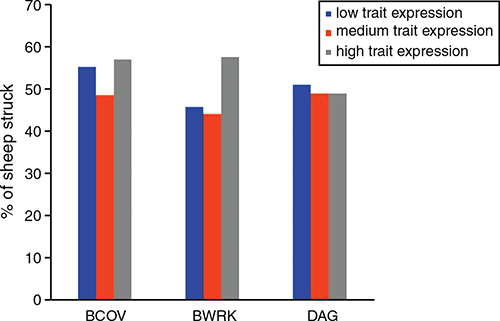
Animals selected from the larger population for genotyping were chosen to have even representation of both breech flystrike conditions (struck or not) within different expression levels for BCOV, BWRK and DAG, aiming at 50%. Breech wrinkle and DAG are site-specific indicator traits, with lower DAG variation for the NSW site and low variation in BWRK for the WA site. Figure 1 demonstrates that in the data that combine NSW and WA within each of the high, medium and low expression levels, both struck and not struck animals were well represented, with ~44% to 56% of animals in each expression level having been struck (Fig. 1).
|
Genomic analysis
The software GCTA (Yang et al. 2011) was used to generate the genomic relationship matrix (GRM) for the genome-wide association study (GWAS) and the estimation of GEBV. The GWAS was conducted using GCTA–MLMA for mixed linear model based association analysis to explore associations of SNPs with STRIKE, BCOV, BWRK and DAG. The mixed model fitted site, drop, sex and mules status as fixed effects, individual SNPs as a covariate for testing additive effect and the GRM as a random effect to take account of polygenic effects. The same fixed and random effects were also fitted in the mixed model to estimate the GEBV of individual animals by using GCTA–GREML module of GCTA (Yang et al. 2011). Any SNPs that exceeded the genome-wide significance threshold (P < 1e–5) were compared against the most recent annotation of the ovine genome (Ensembl Version 3.1.94; ftp://ftp.ensembl.org/pub/release-94/gff3/ovis_aries/Ovis_aries.Oar_v3.1.94.chr.gff3.gz) for identification of the closest genes. For this step, closest feature in bedops (Neph et al. 2012) was used.
Accuracy of GEBV
The accuracies of the genomic predictions (rGEBV) were calculated according to a function (Eqn 1) of the accuracy of genomic prediction as a predictor of effects captured by markers (rQhat) and the proportion of genetic variance captured by markers (q2); (Dekkers 2007; Goddard 2009).

Both components are dependent on the effective population size (Ne), the number of independent chromosome segments (Me, here calculated according to Goddard et al. (2011).

with L as the average chromosome length in Morgan (L = 1), Ne as the effective population size (Ne = 400) and k as the number of chromosomes (k = 26), the numbers of animals in the reference population (n = 1000–10 000), the heritability of the trait (h2 = 0.1–0.5) and the number of markers (nm = 500 000).
The effective population size (Ne) was evaluated by the proportional contribution of the WA and NSW flocks and their respective Ne. Heritabilities varied from h2 = 0.1 to h2 = 0.5 to capture the trait heritabilities for breech flystrike and its indicator traits as estimated for the WA and NSW flocks (Smith et al. 2009; Greeff et al. 2014). The sheep genome has a length of 2587 Mb (https://www.ncbi.nlm.nih.gov/genome?term = ovis%20aries), which translates to the average length of 1 Morgan for the 26 chromosomes of the sheep genome.
Results
Genome-wide association study
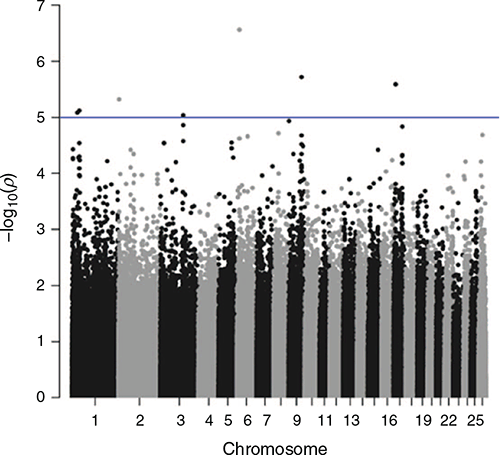
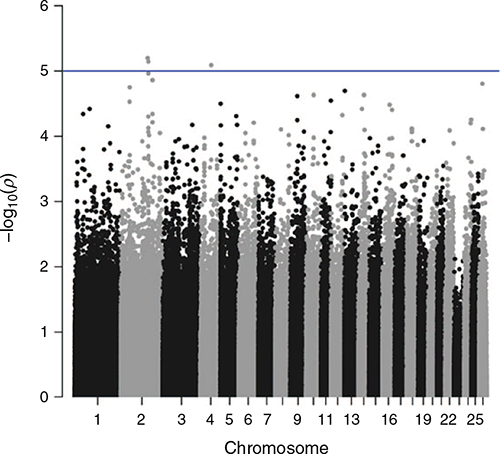
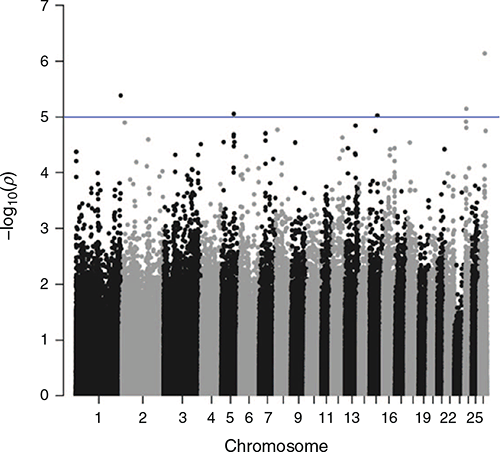
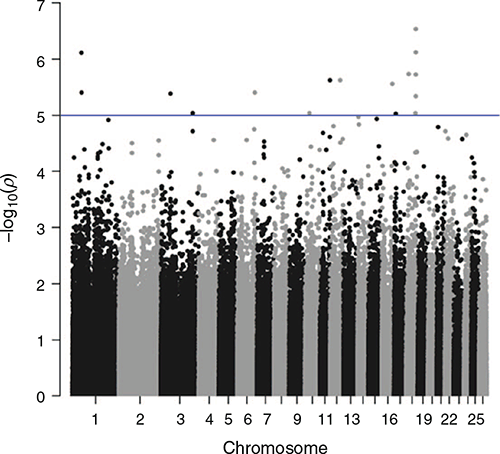
Genome-wide association studies were undertaken for STRIKE, BCOV, DAG and BWRK. Results are shown in Figs 2–5. The graphs show the significance level as –log(P-values) on the y-axis. The genome-wide significance threshold is drawn at –log10(1e–5).
|
|
|
|
No traits showed any outstanding significant peaks formed by multiple SNPs. In total, across the four traits, 32 SNPs were found to exceed the chromosome-wide significance level. All of these SNPs were checked for the closest genes up or downstream of their position on the chromosome.
For STRIKE, two of the SNPs just above the chromosome-wide significance threshold on Chromosome 3 are in proximity of the gene Transcription Factor CP2 (TFCP2) or α-globin transcription factor. None of the other genes in proximity of the two SNPs had a function that appeared relevant to the breech flystrike resistance traits.
The only cluster of several significant SNPs that exceeded the chromosome-wide significance threshold was four SNPs on Chromosome 18 for DAG (56 159 080–56 407 757 bp). The analysis of the closest genes did not yield anything of particular interest. However, GOLGA5 has been positioned at 56 405 660–56 406 553 bp on Chromosome 18, but was not picked up as the closest feature to any of the SNPs.
Several other genes were found to be close to significant SNPs, but they did not appear to be of relevance to breech flystrike or any of the indicator traits.
Genomic breeding values (GEBV)
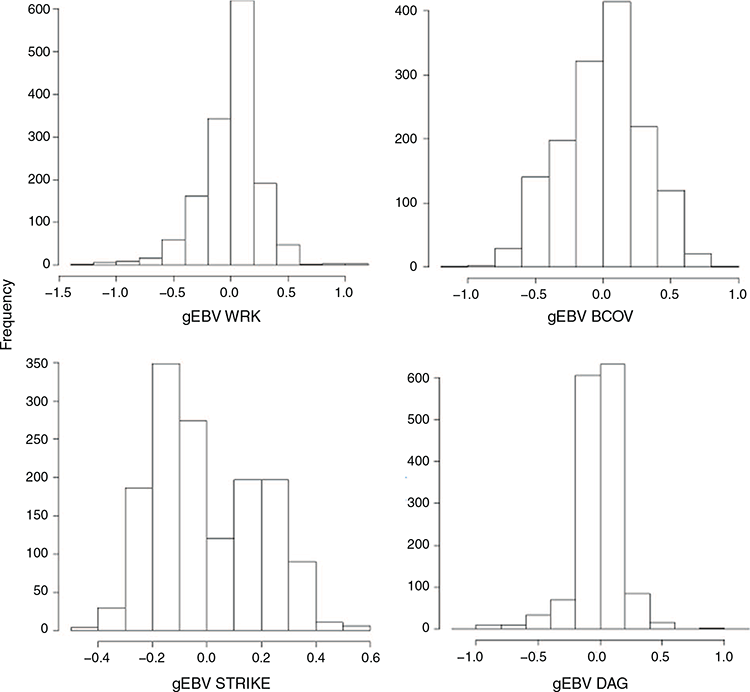
Genomic breeding values were estimated for all four traits using a genomic relationship matrix. The distribution of GEBV is shown in Fig. 6. The distribution for breech flystrike has two peaks, indicating the differentiation of GEBV for high and low breech flystrike resistance for this trait. It was expected that we would observe the same pattern for the indicator traits due to underlying genetic correlations; however, the GEBV for the other three traits displayed normal distributions. A possible reason is that the samples for genotyping across the two sites were specifically selected to have a good representation of struck versus not struck sheep across sire groups.
|
Accuracy of the genomic predictions (rGEBV)
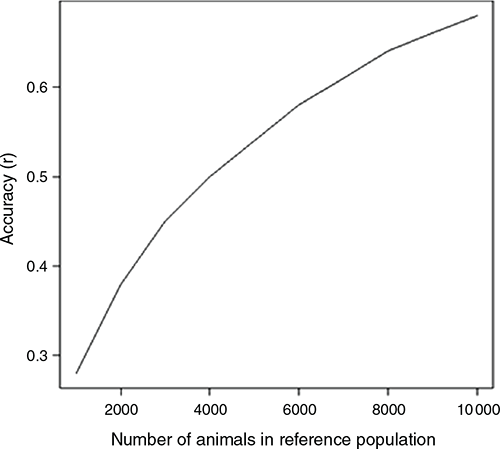
The accuracy of the genomic prediction, plotted for different numbers of animals in a reference population for a general trait with a heritability of h2 = 0.3, is presented in Fig. 7. With the ~1500 genotyped animals from the Breech Flystrike Resource flocks that currently form the reference population, an accuracy of rGEBV = of 0.33 was achievable. The accuracy of GEBV is expected to double with ~7500 animals in the reference population (Fig. 7). Increasing the number of animals in the reference population to 10 000 has limited benefits, with the accuracy being only 0.05 higher than with 7500 animals. The rGEBV for traits with varying heritability and based on a reference population of 1500 animals demonstrated that, for example, a trait with h2 = 0.1, such as DAG in the NSW flock, is expected to have an accuracy of 0.20 (Table 2). A larger reference population is required if the trait of interest has a low heritability.
|

|
Discussion
Results from genomic analyses help inform the most efficient approaches for the application of genomic information in breeding programs. Applications could be based on major genes, such as marker-assisted selection, or based on genomic breeding values that draw on the variation explained by all SNPs in genomic selection. The present study provided proof for the hypothesis that due to a lack of major effects of a few genes, a reference population and the estimation of GEBV would be an applicable genomic approach to fast-track genetic improvement for breech flystrike resistance in Merino sheep.
As a first step, the existence of SNPs that were significantly associated with STRIKE, BCOV, WRK or DAG was explored in a genome-wide association study. All four traits did not display any outstanding peaks formed by multiple SNPs that would indicate strong evidence for a major gene. In preparation for analysis, BCOV, WRK and DAG were amalgamated into three categories to achieve a reasonable number of records per category. This could have led to a loss of information and could have affected the results. Another potential underestimation of the SNP effects could have been due to the confounding that occurs when the GRM and SNP effects are both fitted in the model, which is a known pitfall of mixed model GWAS (Yang et al. 2014). Two regions with SNPs of significance on Chromosomes 3 and 18 have been located in proximity to potentially interesting genes for STRIKE (TFCP2) and DAG (GOLGA5). It can be only hypothesised that it has a functional relationship to breech flystrike, but some other members of the TFCP2/Grainyhead family are involved in cutaneous wound healing (Mace et al. 2005; Ting et al. 2005; Caddy et al. 2010). In addition, in humans, the α-globin transcription factor is involved in rare disorders called alpha-thalassemia, which is a blood disorder that reduces the production of haemoglobin (https://rarediseases.info.nih.gov/diseases/621/alpha-thalassemia). In 5–29% of people, it is associated with abnormalities of the immune system. In Altamurana sheep, a breed from southern Italy, polymorphisms in the α-globin gene have been found to associate with phenotypic variation in red blood cells similar to the human condition (Pieragostini et al. 2003). The authors discuss the link between blood feeding parasites and variation in red blood cells.
GOLGA5 codes for a protein family, the golgins, that are located in the golgi apparatus and are involved in vesicle tethering and docking. The Golgi apparatus is involved in the assembly of peptides, such as antimicrobial peptides that are excreted from the gastric mucosa (Bulet et al. 2004), that are involved in the first line of defence to internal parasites in the intestine. There could be an indirect effect on breech flystrike if GOLGA5 expression influences the formation of dags in sheep.
Both SNPs do not display peaks formed through a high level of significance of a cluster of several SNPs, which would be expected of a chromosome region or gene family that harbours a major gene. However, it is possible that some variation was masked, e.g. DAG and BCOV did not display a lot of variation in the NSW and WA environment respectively, and further investigation of these two regions is warranted. Therefore, splitting the data by site or choosing extreme animals, e.g. on the basis of lifetime strike, combined with a signature of selection approaches that do not require large number of animals, might be considered in future analyses.
The study suggested that a growing number of records in the reference population, a GEBV for breech flystrike could provide a tool for direct selection to improve breech flystrike resistance. Advantages of a GEBV are that all animals can obtain objective information without having to phenotype the actual incidence of flystrike, which is time consuming and requires long-term monitoring. The current genomic resource of the Breech Flystrike Resource flocks provides an excellent base for a reference population for the estimation of GEBV to be generated. Ideally, the rGEBV needs to be increased from those reported here to provide a reliable selection tool for industry. So as to build on the current reference population, the cost of phenotyping further animals has to be taken into consideration as incidence of breech flystrike is expensive to measure. Therefore, generation of a specific reference population for this trait would be costly and impractical to run. An advantage would be that the resource could be used for genomic predictions of other traits as well. Rather than a single reference population, a dispersed reference population based on opportunistic sampling in high-quality recorded stud or research flocks with reliable phenotypes would be advantageous. However, to include unstandardised or commercial phenotypes in a reference population, it would need to be confirmed that breech flystrike breeding values are highly correlated across sites, indicating no genotype × environment interactions. Other options of adding more commercial phenotypes could be explored, such as collecting ‘pooled’ phenotypes from commercial sheep operations. To achieve sufficient accuracy of genomic breeding values for reliable selection for breech flystrike, it is recommended that a future reference population should have at least 7500 phenotypes, which would require phenotyping of a further 6000 animals with accompanying genotypes, which could be added from other existing resources, including the Merino Lifetime Productivity project, and the Meat and Livestock Australia Resource flock, among others.
Conclusions
Genomic selection can provide substantial benefit to Merino breeding programs that focus on wool traits in their breeding objective and the present study suggests that there is a large number of genes involved in the expression of breech flystrike, which can be captured in a GEBV as a tool to fast track direct selection on the trait. The Merino industry is in a position to build on the existing genomic breech flystrike resource to create reliable selection tools to fast-track genetic progress in breech flystrike. Considering the cost to measure breech flystrike, it is recommended to establish a dispersed reference population for breech flystrike resistance of at least 7500 animals on the basis of the existing research flock animals with phenotypes and genotypes and/or suitable commercial animals.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgement
This research was funded by Australian Wool Innovation (AWI). AWI is grateful for its funding, which is primarily provided by Australian woolgrowers through a wool levy and by the Australian Government which provides a matching contribution for eligible R&D activities.
References
Al-Mamun HA, Bernardes PA, Lim D, Park B, Gondro C (2017) A guide to imputation of low density single nucleotide polymorphism data up to sequence level. Journal of Animal Breeding and Genomics 1, 59–68.| A guide to imputation of low density single nucleotide polymorphism data up to sequence level.Crossref | GoogleScholarGoogle Scholar |
Australian Wool Innovation (2019a) Genotyping of breech flystrike resource: update. Final report ON-00515. Available at https://www.wool.com/globalassets/wool/sheep/welfare/breech-flystrike/breeding-for-breech-strike-resistance/link-collection-1/final-report.pdf [Verified 23 June 2021]
Australian Wool Innovation (2019b) Visual Sheep Scores. Producer Version 3. Available at https://www.wool.com/globalassets/wool/sheep/welfare/breech-flystrike/breeding-for-breech-strike-resistance/visual-sheep-scores-producer-version-2019.pdf.
Boichard D, Ducrocq V, Croiseau P, Fritz S (2016) Genomic selection in domestic animals: principles, applications and perspectives. Comptes Rendus Biologies 339, 274–277.
| Genomic selection in domestic animals: principles, applications and perspectives.Crossref | GoogleScholarGoogle Scholar | 27185591PubMed |
Brien FD, Walkom SF, Swan AA, Brown DJ (2021) Substantial genetic gains in reducing breech flystrike and in improving productivity traits are achievable in Merino sheep by using index selection. Animal Production Science 60, 345–362.
| Substantial genetic gains in reducing breech flystrike and in improving productivity traits are achievable in Merino sheep by using index selection.Crossref | GoogleScholarGoogle Scholar |
Brown DJ, Swan AA, Gill JS (2010) Within- and across-flock genetic relationships for breech fly strike resistance indicator traits. Animal Production Science 50, 1060–1068.
| Within- and across-flock genetic relationships for breech fly strike resistance indicator traits.Crossref | GoogleScholarGoogle Scholar |
Bulet P, Stocklin R, Menin L (2004) Anti-microbial peptides: from invertebrates to vertebrates. Immunological Reviews 198, 169–184.
| Anti-microbial peptides: from invertebrates to vertebrates.Crossref | GoogleScholarGoogle Scholar | 15199962PubMed |
Caddy J, Wilanowski T, Darido C, Dworkin S, Ting SB, Zhao Q, Rank G, Auden A, Srivastava S, Papenfuss TA, Murdoch JN, Humbert PO, Boulos N, Weber T, Zuo J, Cunningham JM, Jane SM (2010) Epidermal Wound Repair Is Regulated by the Planar Cell Polarity Signaling Pathway. Developmental Cell 19, 138–147.
| Epidermal Wound Repair Is Regulated by the Planar Cell Polarity Signaling Pathway.Crossref | GoogleScholarGoogle Scholar | 20643356PubMed |
Daetwyler HD, Pong-Wong R, Villanueva B, Woolliams JA (2010) The Impact of Genetic Architecture on Genome-Wide Evaluation Methods. Genetics 185, 1021
| The Impact of Genetic Architecture on Genome-Wide Evaluation Methods.Crossref | GoogleScholarGoogle Scholar | 20407128PubMed |
Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwon A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C (2016) Next-generation genotype imputation service and methods. Nature Genetics 48, 1284–1287.
| Next-generation genotype imputation service and methods.Crossref | GoogleScholarGoogle Scholar | 27571263PubMed |
Dekkers JCM (2004) Commercial application of marker- and gene-assisted selection in livestock: strategies and lessons. Journal of Animal Science 82, E313–E328.
Dekkers JCM (2007) Prediction of response to marker-assisted and genomic selection using selection index theory. Journal of Animal Breeding and Genetics 124, 331
| Prediction of response to marker-assisted and genomic selection using selection index theory.Crossref | GoogleScholarGoogle Scholar |
Goddard M (2009) Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136, 245
| Genomic selection: prediction of accuracy and maximisation of long term response.Crossref | GoogleScholarGoogle Scholar | 18704696PubMed |
Goddard ME, Hayes BJ, Meuwissen THE (2011) Using the genomic relationship matrix to predict the accuracy of genomic selection. Journal of Animal Breeding and Genetics 128, 409
| Using the genomic relationship matrix to predict the accuracy of genomic selection.Crossref | GoogleScholarGoogle Scholar | 22059574PubMed |
Gondro C, Porto-Neto LR, Lee SH (2014) snpqc: an R pipeline for quality control of Illumina SNP genotyping array data. Animal Genetics 45, 758
| snpqc: an R pipeline for quality control of Illumina SNP genotyping array data.Crossref | GoogleScholarGoogle Scholar | 25040453PubMed |
Greeff JC, Karlsson LJE, Schlink AC (2014) Identifying indicator traits for breech strike in Merino sheep in a Mediterranean environment. Animal Production Science 54, 125
| Identifying indicator traits for breech strike in Merino sheep in a Mediterranean environment.Crossref | GoogleScholarGoogle Scholar |
Hu Z-L, Park CA, Wu X-L, Reecy JM (2013) Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Research 41, D871
| Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era.Crossref | GoogleScholarGoogle Scholar | 23180796PubMed |
Loh P-R, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane HK, Schoenherr S, Forer L, McCarthy S, Abecasis GR, Durbin R, Price AL (2016) Reference-based phasing using the Haplotype Reference Consortium panel. Nature Genetics 48, 1443
| Reference-based phasing using the Haplotype Reference Consortium panel.Crossref | GoogleScholarGoogle Scholar | 27694958PubMed |
Mace KA, Pearson JC, McGinnis W (2005) An Epidermal Barrier Wound Repair Pathway in Drosophila Is Mediated by grainy head. Science 308, 381–385.
| An Epidermal Barrier Wound Repair Pathway in Drosophila Is Mediated by grainy head.Crossref | GoogleScholarGoogle Scholar | 15831751PubMed |
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829.
| Prediction of total genetic value using genome-wide dense marker maps.Crossref | GoogleScholarGoogle Scholar |
Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, Sandstrom R, Humbert R, Stamatoyannopoulos JA (2012) BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919
| BEDOPS: high-performance genomic feature operations.Crossref | GoogleScholarGoogle Scholar | 22576172PubMed |
Pieragostini E, Petazzi F, Di Luccia A (2003) The relationship between the presence of extra α-globin genes and blood cell traits in Altamurana sheep. Genetic Selection Evolution 35, S121–S133.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a Tool Set for Whole-genome Association and Population-based Linkage Analyses. American Journal of Human Genetics 81, 559
| PLINK: a Tool Set for Whole-genome Association and Population-based Linkage Analyses.Crossref | GoogleScholarGoogle Scholar | 17701901PubMed |
Smith J, Brewer HG, Dyall T (2009) Heritability and phenotypic corrrelations for breech strike and breech strike resistance indicators in Merinos. In ‘Proceedings of the 18th Conference of the Association for the Advancement of Animal Breeding and Genetics’. (Eds. A Safari, B Pattie, B Restall) pp. 334–337. (University of Adelaide, Roseworthy)
Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM (2005) A Homolog of Drosophila grainy head Is Essential for Epidermal Integrity in Mice. Science 308, 411–413.
| A Homolog of Drosophila grainy head Is Essential for Epidermal Integrity in Mice.Crossref | GoogleScholarGoogle Scholar | 15831758PubMed |
van der Werf JHJ (2009) Potential benefit of genomic selection in sheep. In ‘Proceedings of the 18th Conference of the Association for the Advancement of Animal Breeding and Genetics’. (Eds. A Safari, B Pattie, B Restall) pp. 38–41. (University of Adelaide, Roseworthy)
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a Tool for Genome-wide Complex Trait Analysis. American Journal of Human Genetics 88, 76
| GCTA: a Tool for Genome-wide Complex Trait Analysis.Crossref | GoogleScholarGoogle Scholar | 21167468PubMed |
Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL (2014) Advantages and pitfalls in the application of mixed-model association methods. Nature Genetics 46, 100
| Advantages and pitfalls in the application of mixed-model association methods.Crossref | GoogleScholarGoogle Scholar | 24473328PubMed |
Zeng J, Pszczola M, Wolc A, Strabel T, Fernando RH, Garrick DJ, Dekkers JCM (2012) Genomic breeding value prediction and QTL mapping of QTLMAS2011 data using Bayesian and GBLUP methods. BMC Proceedings 6, S7
| Genomic breeding value prediction and QTL mapping of QTLMAS2011 data using Bayesian and GBLUP methods.Crossref | GoogleScholarGoogle Scholar | 22640755PubMed |


