Microbiome analysis of the skin of sheep that are resistant or susceptible to breech flystrike
J. C. Greeff A B E , E. A. Paz
A B E , E. A. Paz  B , K. Munyard C , A. C. Schlink A , J. Smith D , L. J. E. Karlsson A , G. B. Martin B and D. Groth C
B , K. Munyard C , A. C. Schlink A , J. Smith D , L. J. E. Karlsson A , G. B. Martin B and D. Groth C
A Department of Primary Industries and Regional Development, Perth, 3 Baron Hay Court, South Perth, WA 6151, Australia.
B Institute of Agriculture, University of Western Australia, Crawley, 35 Stirling Highway, Crawley, WA 6009, Australia.
C Curtin Medical School, Curtin University, Perth, WA 6102, Australia.
D CSIRO, Livestock Industries, New England Highway, Armidale, NSW 2350, Australia.
E Corresponding author. Email: johan.greeff@dpird.wa.gov.au
Animal Production Science 61(18) 1774-1780 https://doi.org/10.1071/AN21063
Submitted: 8 February 2021 Accepted: 14 April 2021 Published: 7 July 2021
Journal Compilation © CSIRO 2021 Open Access CC BY
Abstract
Context: Breech strike is a serious disease for wool sheep. Skin wrinkle and dags are known predisposing factors for breech strike; however, a large part of the variation among sheep is unknown.
Aims: We studied the natural diversity and difference in microbial populations in the skin around the breech area in Merino sheep genetically resistant and susceptible to breech strike, by using 16S rRNA gene sequence analysis.
Methods: The sheep were from the breech strike flocks at the Mount Barker research station in Western Australia and from the CSIRO research station near Armidale in New South Wales. Skin samples were collected from the breech of all 2013-born progeny in both flocks before they were struck. Yearling ewes and rams were then naturally exposed to challenge by Lucilia cuprina blowflies. Breeding values for breech strike were estimated and used with phenotypic data to identify breech strike-resistant and -susceptible sheep. Skin samples of 78 unstruck and 73 struck sheep were selected, their microbiomes were analysed using 16S rRNA meta-barcoding, and operational taxonomic unit counts were analysed.
Results: The diversity analyses showed that the two flocks in the different environments had different microbiome profiles, but no difference was found between sexes or between breech strike-resistant and -susceptible sheep in either flock.
Conclusions: The results indicated that microbial differences on the skin of sheep are not associated with differences in susceptibility to breech strike.
Implications: Microbial differences do not offer opportunities to manage breech strike in Merino sheep.
Keywords: Merino sheep, breech strike, 16S rRNA gene, microbiome.
Introduction
Breech strike is a serious disease caused by the Australian sheep blowfly, Lucilia cuprina. Greeff et al. (2014, 2018a, 2018b, 2019) and Smith et al. (2009) showed that breech strike is a heritable trait, and that large differences exist among sires in the proportion of fly-struck sheep in both ewe and ram progeny groups. Faecal soiling (dags) and skin wrinkles of the breech explained most of the variation between struck and unstruck animals in a Mediterranean environment (Greeff et al. 2018a), whereas Smith et al. (2009) found that skin wrinkles were the most important predisposing factor for flystrike in a summer rainfall region. The presence of moisture is the critical factor in breech strike, skin-wrinkles and dags may change the micro-environment in the breech area, making it more favourable for blowfly larvae to develop (Seddon 1931). Greeff et al. (2018b) showed that sheep that were struck before hogget shearing at 16 months of age, are also more likely to be struck as adults under a management regime where the sheep were annually crutched to prevent the accumulation of dags. They showed that large differences exist among sire progeny groups for breech strike resistance that varied from only 3% of the progeny of the most resistant sire being struck, compared with 103% (some sheep were struck more than once) of the progeny of the most susceptible sire being struck under the same environmental conditions. These large differences among sire progeny groups contrast with the modest differences in visually detectable indicator traits such as dags, skin wrinkles and breech cover. They concluded that other, as yet unknown factors appear to contribute to differences among sheep in susceptibility to breech strike. Susceptibility to breech strike is also a repeatable trait in both winter- and summer-rainfall regions. In a winter-rainfall region, the correlation between breech strike from birth to weaning and breech strike from weaning to hogget shearing was 0.29 (Greeff et al. 2014), and between breech strike from birth to hogget age and breech strike in mature sheep, the correlation was 0.14 (Greeff et al. 2021). In a summer-rainfall region, Bird-Gardiner (2015) showed that the correlation between breech strike over 4 years was 0.34. These estimates are higher than the associated heritability, which indicates that other permanent environmental factors contribute to making sheep more resistant or susceptible to breech strike.
Greeff et al. (2013) showed that trained sniffer dogs could differentiate with a high degree of accuracy (>80%) between wool from resistant and wool from susceptible sheep that had not been struck by blowflies. This indicates that odour may play a role in attracting blowflies to sheep. Various studies (Emmens and Murray 1983; Eisemann 1985, 1995; Morris et al. 1997; Urech et al. 2004) have been conducted using mixtures of volatile compounds to determine blowfly attractants. Yan et al. (2019) used L. cuprina to compare the attractiveness of wool from unstruck breech strike-resistant and breech strike-susceptible sheep. They found that blowflies do show a preference for wool from specific sheep. Blowflies were more attracted to sheep whose wool contained more of the semiochemicals octanal and nonanal. These compounds are well known attractants for a variety of insects (https://www.pherobase.com/). However, the origin of these compounds is unknown and may originate from microbial populations on sheep, as Mulcock and Fraser (1958) indicated that the fleece is a suitable habitat for microorganisms and that all the requirements for microbial growth are present in the fleece.
Jackson et al. (2002) used culture-based studies to determine the bacterial composition in sheep fleece. However, this technique identifies less than 1% of all microbial species present in an environmental sample. The use of 16S rRNA analysis (Hugenholtz 2002) facilitates the characterisation of mixed microbial communities without culturing. Dixon et al. (2007) used 16S rRNA gene analysis to investigate the natural bacterial diversity on the skin of fleece rot-resistant and -susceptible sheep. They found that four operational taxonomic units (OTUs; groups of >97% sequence similarity) were present on susceptible, but absent from the resistant sheep. Hence, the current study was undertaken to determine whether the skin microbiome on the breech contributes to differences in susceptibility to blowflies in unstruck resistant and susceptible Merino sheep before the onset of the flystrike season, from both a summer- and winter-rainfall regions.
Materials and methods
The study was approved by the Animal Ethics Committee of the Department of Agriculture and Food Western Australia (Approval number AEC 17-4-09) and by the CSIRO Armidale animal Ethics Committee (Approval number 13/35).
Animals
Two Merino flocks were used in the present study. One flock was located in a summer-rainfall region (SR) at the CSIRO research station near Armidale, New South Wales (Smith et al. 2009), and the second flock was located at the Mount Barker research station of the Department of Agriculture and Food Western Australia in the winter-rainfall region (WR) of Western Australia (Greeff et al. 2014). The SR flock consisted of 439 sheep whereas the WR flock consisted of 937 sheep. The animals in both flocks were born in 2013.
Recording of breech strike
None of the sheep in this experiment was mulesed. The WR sheep were crutched in May, at ~10 months of age, 4 months before sampling in August, whereas the SR sheep were crutched at weaning in January, and skin-sampled in August. The sheep were then naturally exposed to blowflies. Daily inspections were undertaken to identify struck sheep. All blowfly strikes were recorded. Any struck sheep was clipped clean around the strike site, was treated with a short-acting insecticide (Extinosad®) at the manufacturers recommended rate, and was released back into the flock. A small proportion of sheep was struck more than once.
Estimating breeding values for breech strike
The breech strike records were analysed with best linear unbiased prediction mixed model methodology (Henderson 1984), using the ASRemL software package (Gilmour et al. 2015). All previous years’ breech strike records (from 2006 onward) were included in the analysis. Separate analyses were performed for each site. Full pedigrees were available on all sheep, and an animal model was fitted with year of birth, sex of the lamb, birth status (single or multiple) and age of the dam as fixed effects. Animal was fitted as a random effect. The sheep were ranked within sex and within site on their breeding value for breech strike.
Collection of skin samples
Skin samples were collected on all the sheep in both flocks before their first shearing and before the onset of the fly season. Both flocks were sampled in August 2014. No skin cleaning or clipping of the site was performed before collecting the skin sample. The site was anaesthetised with 0.5 mL lignocaine intradermally (2% Lignocaine, Troy Animal Health, Australia). A small (0.5–1.0 cm2) biopsy of skin was surgically removed with curved scissors from the designated breech area, ~2 cm to the right from the tip of the docked tail, at 11 months after birth on the lambs in the SR flock, and at 14–15 months of age in September 2014 on all the lambs in the WR flock. When an animal had excessive dags at sampling, care was taken to sample from a non-daggy area as close as possible to the designated sampling site, so as to ensure that the sample did not contain any dags. Some sheep in the SR flock were already struck by blowflies at sampling; however, their samples were not used in the study. None of the WR flock sheep was struck at, or before, sampling in spring. A separate sterilised pair of scissors was used on each lamb to prevent any cross-contamination. The wool on each skin sample was trimmed back to a length of ~3 mm. The sample was stored in RNAlater, and held at –20°C until processing.
Selection of sheep for 16S rRNA analysis
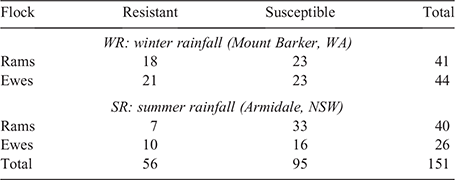
All the lambs were skin sampled in each flock. After the fly season was completed, the phenotypic breech strike data were analysed, as described above, to obtain breeding values on all the animals in both flocks. The breeding values from both sites were standardised by subtracting the flock mean breeding value from the breeding value of each animal in the flock, and by dividing the result with the standard deviation of the flock. This standardised value allowed for the identification of the most resistant and susceptible sheep in both locations. Any sheep with a high breeding value and that was not struck, and any sheep with a low breeding value and that was struck was excluded from genetic analysis. The rams and ewes with the highest (susceptible) and lowest (resistant) breeding values were then identified. Sixty-six sheep were selected from the SR flock, and 85 from the WR flock (Table 1). The skin samples from these 151 selected sheep were retrieved from the stored library of skin samples and sent to the Australian Genomic Research Facility (AGRF) for processing and sequencing.
|
DNA extraction and primary sequencing analysis
The extraction and isolation of DNA was undertaken by AGRF using the DNeasy PowerLyzer Powersoil Kit (QIAGEN 2017). Conserved primers were used for the 16S rRNA amplification (27F: AGAGTTTGATCMTGGCTCAG; and 519R: GWATTACCGCGGCKGCTG). The amplicon length was 300 bp.
The samples were sequenced using a paired-end protocol, and image analysis was performed in real time by the MiSeq Control Software v.2.6.1.1 and real-time analysis v.1.18.54. The Illumina bcl2fastq 2.17.4 pipeline was used to generate the de-multiplexed sequence files. The sequence data have been submitted to the NCBI Sequence Archive under the Bioproject number PRJNA657495.
Statistical analyses
16S rRNA analysis
Trimming of raw reads was undertaken using Sickle, setting the length-threshold at 200 bp and quality cut-off at 20 (https://github.com/najoshi/sickle). The first 10 bases of the forward and reverse reads were trimmed in the 16S rRNA analysis, and to ensure a minimum quality score of 30, the forward reads were truncated at 190 bases, and the reverse reads at 145 bases. Merging of paired-end sequences and clustering de novo into OTUs with a 97% similarity was performed using micca v.1.7.0 (Albanese et al. 2015). Taxonomic assignment was executed using the Bayesian Lowest Common Ancestor-based taxonomic classification method (Gao et al. 2017) on the basis of the NCBI 16S microbial database. A phylogenetic tree was built with FastTree, applying generalised time-reversible and CAT approximation (Price et al. 2010). To normalise the number of reads in each sample, we rarefied to a depth of 2732. Alpha and β diversities were estimated using QIIME v.1.9.1 (Caporaso et al. 2010). To determine the significance of the alpha metrics, a non-parametric two-sample Student’s t-test with Bonferroni correction was performed. A principal coordinates analysis was performed using unweighted UniFrac distance. The statistical significance of the distance matrix was tested using analysis of similarity with 1000 permutations. The linear discriminant analysis effect size algorithm was used to identify significant taxonomic differences.
Results
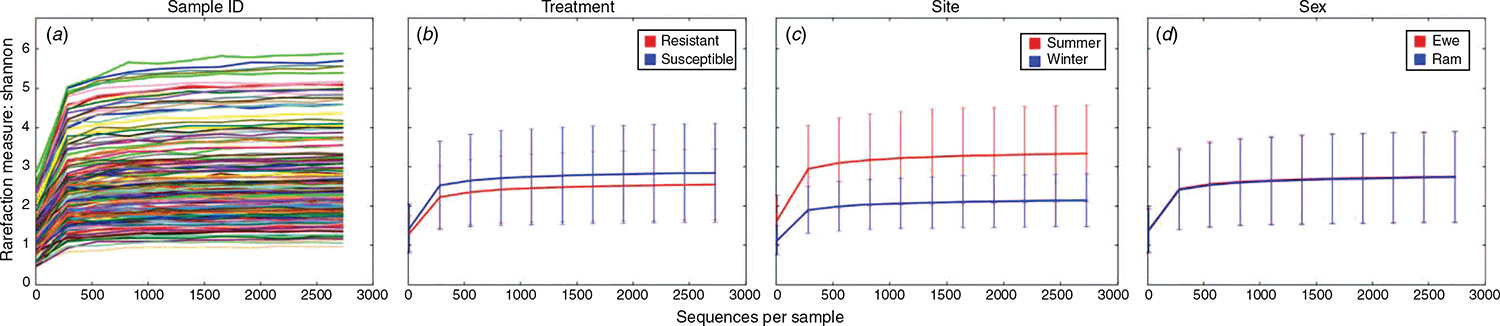
In total, 15935704 raw reads were obtained from all samples combined. After trimming, filtering and merging of paired-end reads, 4 654 408 tags were included in the workflow. In total, 3212 OTUs were clustered at 97% of similarity. The Shannon rarefaction curves reached a plateau and both groups had an acceptable sequence depth (Fig. 1). The rarefaction analysis shows that the observed OTUs reached a plateau at a sequencing depth of 2000 bases at both flocks. This resulted in 232 993 (16.5%) features in 151 (86.3%) subsamples of the total number of samples at this specified depth.
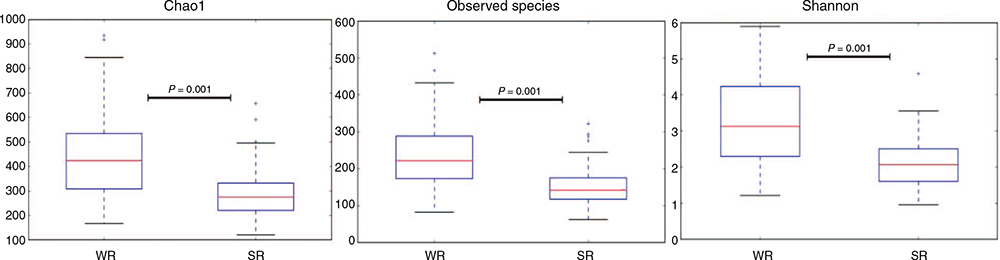
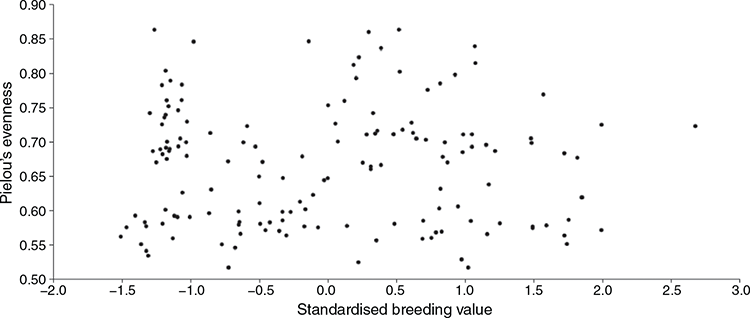
The Shannon diversity index plots are shown in Fig. 1 for the different samples, resistant or susceptible, location and sex, while the α diversity metrics, Chao1, observed species and Shannon analyses between the winter- and summer-rainfall regions are shown in Fig. 2. Higher microbial diversity was measured in the WR flock than in the SR flock. However, no significant differences were found between rams and ewes, or between resistant or susceptible animals within flocks. There was no correlation between microbial diversity and the individual standardised breeding value for breech strike for either flock (Fig. 3). The spearman test correlation between the standardised breeding value for breech strike and the 16S rRNA sequences was very low, that is 0.0044 (P-value = 0.9569).
|
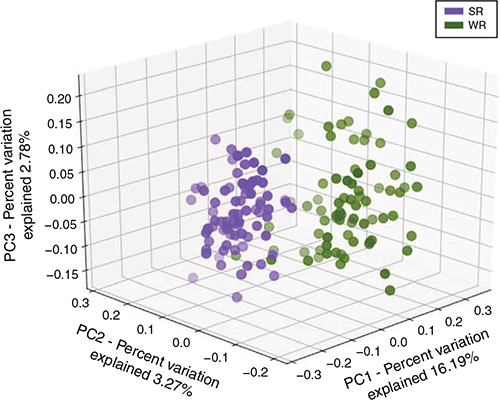
The principal coordinate analysis showed a distinct clustering of the samples by region (Fig. 4). Subsequent analysis of similarity found highly significant differences between the WR and SR regions (P = 0.00009; R2 = 0.724).
|
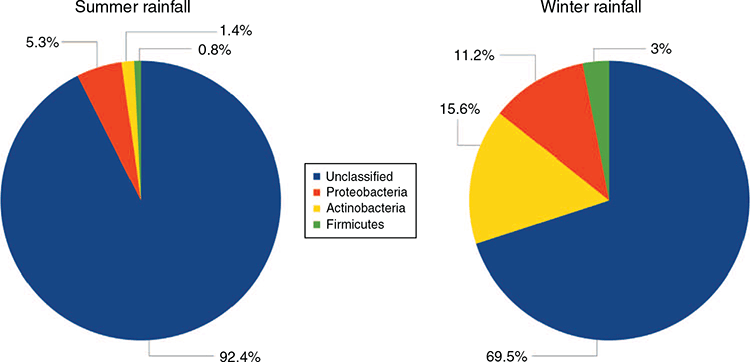
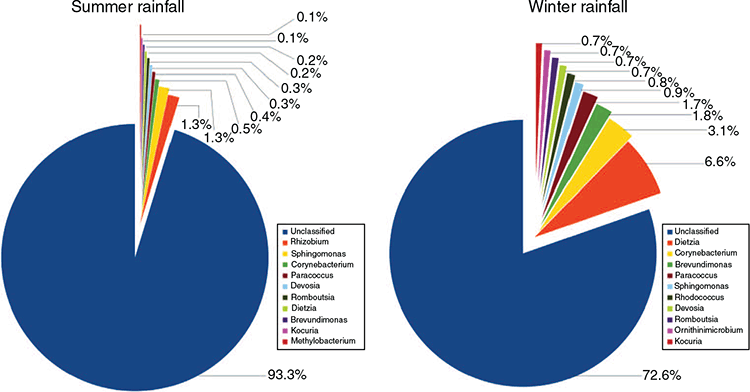
The microbial composition was evaluated at both phylum and genus levels. Proteobacteria were the dominant identifiable phylum in the SR flock (5.3%), followed by Actinobacteria (1.4%) and Firmicutes (0.8%). Whereas in the WR flock, Actinobacteria (15.6%), Proteobacteria (11.2%) and Firmicutes (3%) were the most abundant identifiable phyla (Fig. 5). However, most of OTUs in both flocks were unable to be classified at the phylum level (92.4% and 69.5%).
The top five most abundant genera in the study were Dietzia (3.4%), followed by Corynebacterium (1.8%), Sphingomonas (1.1%), Paracoccus (1%) and Brevundimonas (1%). In the SR flock, Rhizobium (1.3%) and Sphingomonas (1.3%) were the dominant taxa followed by Corynebacterium (0.5%). The genera most abundant in the WR flock were Dietzia (6.6%), Corynebacterium (3.1%) and Brevundimonas (1.8%; Fig. 6). In both flocks, a large proportion of the OTUs could not be classified to genus level (93.3% and 72.6%).
|
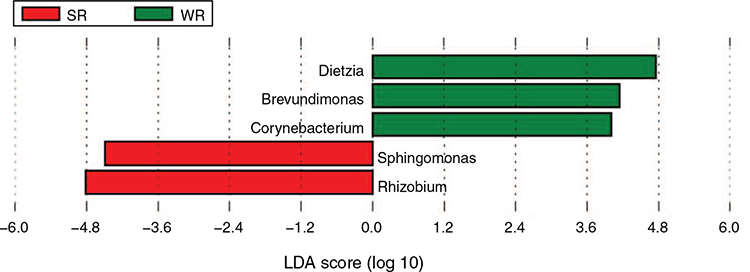
The linear discriminant analysis effect size analysis (Fig. 7) indicated that Rhizobium and Sphingomonas were significantly more abundant in the SR flock, whereas Dietzia, Brevundimonas and Corynebacterium were significantly more abundant in the WR flock.
|
Discussion
The present study was conducted to determine whether the microbial populations on the skin in the breech of sheep contribute to sheep being resistant or susceptible to breech strike. On the basis of the work of Greeff et al. (2014), it was speculated that unique odours that are more attractive or repulsive to blowflies, and/or create a more favourable environment for blowfly larvae in the breech, may be correlated with a sheep’s susceptibility or resistance to breech strike. The present study is the first study of this nature that has used proven genetically resistant and susceptible sheep. The only other related study was conducted by Dixon et al. (2007) who used sheep resistant and susceptible to fleece rot.
A large proportion of unknown OTUs was identified in the study and could not be classified even at the phylum level. In the summer-rainfall region, only 7.6% of the OTUs were associated with known microbial species, and 30.5% in the winter-rainfall region. Thus, sheep from a summer- and from a winter-rainfall region will host different microbial populations, related to that environment. In the summer-rainfall site, the genera Rhizobium and Sphingomonas were significantly more abundant, while at the winter-rainfall site, Brevundimonas and Dietzia were the most abundant. These four genera are mostly associated with plants and soils. Dixon et al. (2007) also found that 75% of the sequences in their study had not previously been identified in fleece-rot studies. However, contrary to our results, they did find that four OTUs were present on body strike-susceptible sheep, but absent from the body strike-resistant sheep. However, although they did find differences, they sequenced only two resistant and two susceptible sheep in their study.
Moisture is a key requirement for blowfly eggs and larvae to develop. Rams and ewes are anatomically very different and therefore ewes with urine stain may have more moist breeches that could be more conducive for blowfly larvae to develop. Greeff et al. (2018b) also showed that sheep with urine stain are more susceptible to breech strike. Therefore, these factors may indicate that rams and ewes could harbour different microbial species on their breeches. However, no significant (P > 0.05) difference was found between males and females that were managed separately in different paddocks in both the WR and SR flocks. These results indicate that the microbial species on ewes and rams were very similar across paddocks in both environments.
The present study has clearly shown that in a total of 3212 OTUs, no significant difference in microbial populations was able to be detected between sheep resistant and susceptible for breech strike. Thus, we conclude that the composition of microbial populations in the breech area of unstruck sheep before the blowfly season does not contribute to making sheep more or less susceptible to breech strike. Because large genetic differences exist between sire progeny groups for breech strike, the present study indicated that any difference between sheep is largely due to the animal itself, of which dags and breech wrinkle are the two most important predisposing factors. These two traits explain only up to 40% of the variation in breech strike (Greeff et al. 2018a), which indicates that more research is needed to determine why certain sheep are repeatedly struck over their lifetime, even in the absence of dags. Research should focus on the micro-environment in the breech area, to determine whether diurnal patterns of temperature and moisture differ between breech strike-resistant and breech strike-susceptible sheep.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank the Department of Agriculture and Food Western Australia for their support in conducting this experiment on the Department’s and CSIRO’s research stations, as well as wool producers and the Commonwealth government, through Australian Wool Innovation Ltd, for their generous financial and positive constructive contributions throughout the lifetime of this project. We also express our appreciation to Nicola Stanwyck, the technical officer involved with this experiment, for her diligence and dedication in recording the data on the animals in this flock.
References
Albanese D, Fontana P, De Fillippo C, Cavalieri D, Donati C (2015) MICCA: a complete and accurate software for taxonomic profiling of metagenomics data. Scientific Reports 5, 9743| MICCA: a complete and accurate software for taxonomic profiling of metagenomics data.Crossref | GoogleScholarGoogle Scholar | 25988396PubMed |
Bird-Gardiner T (2015) Genetic analysis of flystrike in sheep. Thesis for Research Masters of Rural Science of the University of New England, Armidale, NSW, Australia.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7, 335–336.
| QIIME allows analysis of high-throughput community sequencing data.Crossref | GoogleScholarGoogle Scholar | 20383131PubMed |
Dixon TJ, Mortimer SI, Norris BJ (2007) 16S RNA gene microbial analysis of the skin of fleece rot resistant and susceptible sheep. Australian Journal of Agricultural Research 58, 739–747.
| 16S RNA gene microbial analysis of the skin of fleece rot resistant and susceptible sheep.Crossref | GoogleScholarGoogle Scholar |
Eisemann CH (1985) A study of aspects of the host-finding behaviour of the Australian sheep blowfly, Lucilla cuprina (Wied.): and of the structure, distribution and behavioural significance of its antennal and palpal sensilla. PhD Dissertation. The University of Queensland, Brisbane, Qld, Australia.
Eisemann CH (1995) Orientation by gravid Australian sheep blowflies, Lucilia cuprina (Diptera: Calliphoridae), to fleece and synthetic chemical attractants in laboratory bioassays. Bulletin of Entomological Research 85, 473–477.
| Orientation by gravid Australian sheep blowflies, Lucilia cuprina (Diptera: Calliphoridae), to fleece and synthetic chemical attractants in laboratory bioassays.Crossref | GoogleScholarGoogle Scholar |
Emmens RL, Murray MD (1983) Bacterial odours as oviposition stimulants for Lucilia cuprina (Wiedemann) (Diptera: Calliphoridae), the Australian sheep blowfly. Bulletin of Entomological Research 73, 411–415.
| Bacterial odours as oviposition stimulants for Lucilia cuprina (Wiedemann) (Diptera: Calliphoridae), the Australian sheep blowfly.Crossref | GoogleScholarGoogle Scholar |
Gao X, Lin H, Revanna K, Dong Q (2017) A Bayesion taxonomic classification method for 16sRNA gene sequences with improved species-level accuracy. BMC Bioinformatics 10, 247
| A Bayesion taxonomic classification method for 16sRNA gene sequences with improved species-level accuracy.Crossref | GoogleScholarGoogle Scholar |
Gilmour AR, Gogel BJ, Cullis BR, Welham SJ, Thompson R (2015) ASReml User Guide Release 4.1 Functional Specification. VSN International Ltd, Hemel Hempstead, UK. Available at www.vsni.co.uk
Greeff JC, Biggs A, Grewar W, Crumblin P, Karlsson LJE, Schlink AC, Smith J (2013) Dogs can differentiate between odours from sheep that are resistant or susceptible to breech strike. Proceedings of the Association for the Advancement of Animal Breeding and Genetics 20, 397–400.
Greeff JC, Karlsson LJE, Schlink AC (2014) Identifying indicator traits for breech strike in Merino sheep in a Mediterranean environment. Animal Production Science 54, 125–140.
| Identifying indicator traits for breech strike in Merino sheep in a Mediterranean environment.Crossref | GoogleScholarGoogle Scholar |
Greeff JC, Karlsson LJE, Schlink AC, Gilmour AR (2018a) Factors explaining the incidence of breech strike in a Mediterranean environment in unmulesed and uncrutched Merino sheep. Animal Production Science 58, 1279–1288.
| Factors explaining the incidence of breech strike in a Mediterranean environment in unmulesed and uncrutched Merino sheep.Crossref | GoogleScholarGoogle Scholar |
Greeff JC, Schlink AC, Karlsson LJE (2018b) The impact of sire on the lifetime susceptibility of their progeny to breech strike in a Mediterranean environment. Animal Production Science 58, 1522–1530.
| The impact of sire on the lifetime susceptibility of their progeny to breech strike in a Mediterranean environment.Crossref | GoogleScholarGoogle Scholar |
Greeff JC, Karlsson LJE, Schlink AC (2019) Are breech strike, dags and breech wrinkle genetically the same trait in crutched, uncrutched and mulesed Merino sheep? Animal Production Science 59, 1777–1782.
| Are breech strike, dags and breech wrinkle genetically the same trait in crutched, uncrutched and mulesed Merino sheep?Crossref | GoogleScholarGoogle Scholar |
Greeff JC, Schlink AC, Karlsson LJE (2021) Genetic parameters of breech strike, neck wrinkles, dags and breech cover over the lifetime of crutched Merino ewes in a Mediterranean environment. Animal Production Science.
| Genetic parameters of breech strike, neck wrinkles, dags and breech cover over the lifetime of crutched Merino ewes in a Mediterranean environment.Crossref | GoogleScholarGoogle Scholar |
Henderson CR (1984) Applications of Linear Models in Animal Breeding. University of Guelph, Canada.
Hugenholtz P (2002) Exploring prokaryotic diversity in the genomic era. Genome Biology 3, 0003.1–0003.8.
Jackson TA, Pearson JF, Young SD, Armstrong S, O’Callaghan M (2002) Abundance and distribution of microbial populations in sheep fleece. New Zealand Journal of Agricultural Research 45, 49–55.
| Abundance and distribution of microbial populations in sheep fleece.Crossref | GoogleScholarGoogle Scholar |
Morris MC, Joyce MA, Heath AC, Rabel B, Delisle GW (1997) The responses of Lucilia cuprina to odours from sheep, offal and bacterial cultures. Medical and Veterinary Entomology 11, 58–64.
| The responses of Lucilia cuprina to odours from sheep, offal and bacterial cultures.Crossref | GoogleScholarGoogle Scholar | 9061678PubMed |
Mulcock AP, Fraser IEB (1958) Total counts of microorganisms in the fleece of two Corriedale fleece types. Australian Journal of Agricultural Research 9, 704–707.
| Total counts of microorganisms in the fleece of two Corriedale fleece types.Crossref | GoogleScholarGoogle Scholar |
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum likelihood trees for large alignments. PLoS One 5, e9490
| FastTree 2–approximately maximum likelihood trees for large alignments.Crossref | GoogleScholarGoogle Scholar | 20224823PubMed |
QIAGEN (2017) DNAeasy PowerLyzer Powersoil Kit Handbook, June 2017. Available at www.qiagen.com
Seddon HR (1931) Conditions which predispose sheep to blowfly attack. Agriculture Gazette of New South Wales 42, 581–594.
Smith JL, Brewer HG, Dyall T (2009) Heritability and phenotypic correlations for breech strike and breech strike resistance indicators in Merinos. Proceedings of the Association for the Advancement of Animal Breeding and Genetics 18, 334–337.
Urech R, Green PE, Rice MJ, Brown GW, Duncalfe F, Webb P (2004) Composition of chemical attractants affects trap catches of the Australian sheep blowfly, Lucilia cuprina, and other blowflies. Journal of Chemical Ecology 30, 851–866.
| Composition of chemical attractants affects trap catches of the Australian sheep blowfly, Lucilia cuprina, and other blowflies.Crossref | GoogleScholarGoogle Scholar | 15260228PubMed |
Yan G, Liu S, Schlink AC, Flematti GR, Brodie BS, Bohman B, Greeff JC, Vercoe PE, Hu J, Martin GB (2019) Volatiles from Merino fleece evoke antennal and behavioural responses in the Australian sheep blow fly Lucilia cuprina. Medical and Veterinary Entomology 33, 491–497.
| Volatiles from Merino fleece evoke antennal and behavioural responses in the Australian sheep blow fly Lucilia cuprina.Crossref | GoogleScholarGoogle Scholar | 31136024PubMed |