Development of Angus SteerSELECT: a genomic-based tool to identify performance differences of Australian Angus steers during feedlot finishing: Phase 1 validation
Brad C. Hine A D , Christian J. Duff
A D , Christian J. Duff  B , Andrew Byrne B , Peter Parnell B , Laercio Porto-Neto C , Yutao Li C , Aaron B. Ingham C and Antonio Reverter
B , Andrew Byrne B , Peter Parnell B , Laercio Porto-Neto C , Yutao Li C , Aaron B. Ingham C and Antonio Reverter  C
C
A CSIRO Agriculture & Food, F.D. McMaster Laboratory, Chiswick, New England Highway, Armidale, NSW 2350, Australia.
B Angus Australia, 86 Glen Innes Road, Armidale, NSW 2350, Australia.
C CSIRO Agriculture & Food, Queensland Bioscience Precinct, 306 Carmody Road, St Lucia, Brisbane, Qld 4067, Australia.
D Corresponding author. Email: brad.hine@csiro.au
Animal Production Science - https://doi.org/10.1071/AN21051
Submitted: 4 February 2021 Accepted: 2 June 2021 Published online: 19 August 2021
Journal Compilation © CSIRO 2021 Open Access CC BY
Abstract
Context: Genomic-based technologies are allowing commercial beef producers to predict the genetic merit of individual animals of unknown pedigree with increased ease and accuracy. Genomic selection tools that can accurately predict the feedlot and carcass performance of steers have the potential to improve profitability for the beef supply chain.
Aims: To validate the ability of the Angus SteerSELECT genomic product to predict differences in performance of Australian Angus steers, in terms of carcass weight, marbling score, ossification score and carcass value, using a short-fed (100 days) or long-fed (270 days) finishing protocol at a commercial feedlot.
Methods: A reference population of 2763 Australian Angus steers was used to generate genomic prediction equations for three carcass traits, namely, carcass weight, marbling score and ossification. The accuracy and bias of genomic predictions of breeding values were then evaluated using a validation population of 522 Angus steers, either short- or long-fed at a commercial feedlot, by comparing breeding values to measured phenotypes. The potential economic benefits for feedlot operators when using Angus SteerSELECT were estimated on the basis of the ability of the tool to predict the carcass value of steers in the validation population.
Key results: The accuracy of genomic predictions of breeding values for carcass weight, marbling score and ossification score were 0.752, 0.723 and 0.734 respectively. When steers were ranked in quartiles for predicted carcass value, calculated using genomic predictions of breeding values for carcass weight and marbling score, the least-square mean carcass value for steers in each quartile, from bottom 25% predicted performers to top 25% predicted performers, were estimated at A$1794, A$1977, A$2021 and A$2148 for short-fed steers and A$3546, A$3780, A$3864 and A$4258 for long-fed steers. Differences in the carcass value least-squares mean between the bottom and top quartile were highly significant (P < 0.001) for both short-fed and long-fed steers.
Conclusions: Genomic prediction equations used in Angus SteerSELECT can predict differences in carcass weight, marbling score, ossification score and carcass value in both short-fed and long-fed Australian Angus steers.
Implications: Genomic selection tools that can predict differences in performance, in terms of growth and carcass characteristics, of commercial feedlot cattle have the potential to significantly increase profitability for the beef supply chain by improving the quality and consistency of the beef products they produce.
Keywords: beef cattle, feedlot performance, carcass, genomic predictions, accuracy.
Introduction
Genomic-based technologies are allowing commercial beef producers to predict the genetic merit of individual animals in their herds of unknown pedigree with increased ease and accuracy. Information on the predicted genetic merit of individual animals can be used by commercial producers to inform breeding selection decisions, such as identifying replacement breeding females, and to inform management decisions, matching animals to the most appropriate finishing path and end market destination.
Many variables contribute to the profitability of feedlot cattle including initial purchase price, feed costs, health-related costs, facility depreciation and maintenance costs, labour costs, transport costs, processing costs and carcass value at slaughter. Certain variables such as facility depreciation and maintenance, labour, transportation and processing can be estimated with some certainty when calculating expected returns, whereas other variable costs, such as initial purchase price, feed costs and carcass value at slaughter, are more difficult to estimate because they can fluctuate significantly with commodity markets. Feedlot operators often utilise forward contracts to lock in commodity prices for fixed time periods and provide some certainty around input costs and expected returns in fluctuating markets. The unexpected variation in performance of cattle during feedlot finishing can also have a major impact on profitability and is also difficult to predict without information on the genetic potential for animals to perform in the feedlot environment.
It is common practice for commercial beef producers in Australia to purchase sires from seed-stock producers to access registered bulls with known genetic backgrounds (Angus Australia 2019). Feedlot operators attempt to reduce the risk of animals underperforming at the feedlot or in the chiller by using a combination of decisions based on prior management history of cattle or by purchasing cattle from vendors with a known performance history or targeting new vendors who are known to manage their herds well and use sires of known genetic background. However, cattle from preferred vendors are not always available for purchase when required or in numbers required and genetic variation between animals is observed in all herds, including those with known genetic backgrounds. For example, in a cohort of long-fed steers (n = 71) included in the validation population for the present study (see details below), which were of a similar age and from the same herd, carcass weights varied from 393 kg to 515 kg and marble score varied from 340 to 800. If some of this between-animal variation could be predicted, feedlot operators could select animals with potential to perform better in their production system to improve financial returns. Therefore, a genomic-based tool that can predict the performance of steers could be used by feedlot operators or others in the beef-supply chain (e.g. breeders, backgrounders and brand owners) to better inform selection decisions, identify the most appropriate finishing path for animals once purchased and improve their ability to consistently meet market specifications. Such a test could be considered as a risk-management tool, aimed at reducing the risk of animals underperforming.
There are several genomic products currently available for commercial beef producers in Australia, including a test aimed at informing selection of replacement Angus females (HeiferSELECT, Angus Australia, https://www.angusaustralia.com.au/education/breeding-and-genetics/angus-heiferselect/) and a test aimed at predicting the genetic potential of crossbred beef heifers, steers and commercial bulls (Igenity® Beef, Neogen, https://www.neogen.com/igenity-beef/). The accuracy of genomic predictions is known to be affected by the heritability of the trait being predicted, the size and relevance of the reference population used to generate predictions and the relatedness of the animals being tested to those in the reference population. With this in mind, we have developed a genomic product specifically aimed at predicting differences in performance of Australian Angus steers during finishing under Australian feedlot conditions called Angus SteerSELECT. We report here the results of the initial Phase 1 validation of Angus SteerSELECT, in which genomic estimated breeding values (gEBVs) generated for steers in a validation population were compared with their measured phenotypes for three traits, namely, carcass weight, marbling score and ossification score, assessed at slaughter following steers undergoing either a short-fed (100 days) or long-fed (270 days) finishing protocol. We hypothesised that Angus SteerSELECT would be able to predict differences in carcass weight, marbling score, ossification score and carcass value of Australian Angus steers solely on the basis of their genomic profile and genetic predictions when either short- or long-fed, under standard Australian commercial feedlot conditions.
Materials and methods
Reference and validation population details
Phenotypes and matching genotypes were available for 3372 steers that were progeny of the Australian Angus Sire Benchmarking Program (ASBP), representing Years 1–7 of the program (described as Cohorts 1–7 respectively). The ASBP is a major initiative of Angus Australia with support from Meat & Livestock Australia (MLA) and industry partners that aims to generate progeny test data on contemporary Angus bulls, particularly for hard-to-measure traits such as feed efficiency, carcass measurements, meat quality attributes and female reproduction (https://www.angusaustralia.com.au/sire-benchmarking/about/general-information/). A subset of these steers (n = 2763), representing Cohorts 1–6, were used to form a reference population from which genomic prediction equations were generated for each trait. A further subset of steers (n = 522) representing Cohort 7 were used to form a validation population to compare gEBVs with measured phenotypes for selected traits. A small number of steers (n = 87) were excluded from the reference and validation populations because of restrictions imposed on having age at measurement recorded and a minimum number of animals permitted in fixed-effect categories when undertaking analyses. By design, there were minimal sire linkages across cohorts of the ASBP. For example, Cohort 7 steers were the progeny of 56 sires, of which only 14 had progeny in Cohort 6 and one had progeny in Cohort 5.
Phenotypic data collection
Steers included in the reference and validation populations were born on ASBP co-operator herd farms where they remained until post-weaning. Most steers were backgrounded on pasture at their property of origin until reaching feedlot entry weights and were then transported directly to the feedlot. A small number of steers were backgrounded on pasture at a location other than their property of origin and were then transported from that location to the feedlot once reaching feedlot entry weights. All steers (Cohorts 1–7) were initially transported to Tullimba feedlot (University of New England, Armidale, NSW, Australia) where their feed efficiency was assessed using the GrowSafe® System. Steers were fed at Tullimba for a minimum of 100 days. All steers in Cohorts 1–4 and a subset of steers from Cohorts 5–7 (n = 2682) were then transported to a commercial feedlot in northern NSW, Australia, and fed for a further minimum of 170 days before being slaughtered at a commercial abattoir (long-fed steers). A subset of steers from Cohorts 5–7 (n = 690) were slaughtered at a commercial abattoir directly following exit from the Tullimba feedlot (short-fed steers).
Three traits, namely, carcass weight (CWT), Meat Standards Australia (MSA) marbling score (MBL) and ossification score (OSS) were used in the current study to validate the ability of SteerSELECT to predict differences in carcass performance following feedlot finishing. These traits were selected as they can significantly influence carcass value. Phenotypes for CWT were collected on the day of slaughter as hot standard CWT. Phenotypes for MBL and OSS were collected the day following slaughter on chilled carcasses by experienced MSA accredited graders. MBL was assessed at the 12th to 13th rib of the carcass on the exposed rib eye, using the MSA grading system which ranges from 100 to 1190 in increments of 10 (https://www.mla.com.au/globalassets/mLa-corporate/marketing-beef-and-lamb/msa_tt_beefinfokit_jul13_lr.pdf). At the same time, from the same anatomical site, and by the same MSA grader, an AUSMEAT MBL was also assessed using the AUSMEAT scoring system, which ranges from 0 to 9 in increments of 1 (https://solutionstofeedback.mla.com.au/cattle/chiller-assessment/aus-meat-marbling/). The AUSMEAT MBL was used only as a parameter to estimate carcass values in the current study, as described below. Marbling was assessed according to the AUSMEAT requirements for chiller assessment when the temperature of the part of the carcass being assessed was below 12°C. Ossification of the cartilage within the vertebral spinous processes, a measure of physiological maturity of the carcass, was assessed across three areas of the backbone, being the sacral, lumbar and thoracic vertebrae, by using a scoring system that ranged from 100 to 590 in increments of 10 (https://www.mla.com.au/globalassets/mLa-corporate/marketing-beef-and-lamb/msa_tt_beefinfokit_jul13_lr.pdf). Carcass value (CVL) was estimated for all steers in the validation population by multiplying CWT by (A$5 + A$1 per kg of CWT for each AUSMEAT MBL). Therefore, a carcass with an AUSMEAT MBL of 0 was valued at A$5/kg CWT, whereas a carcass with an AUSMEAT MBL of 3 was valued at A$8/kg CWT.
Generation of genomic prediction equations
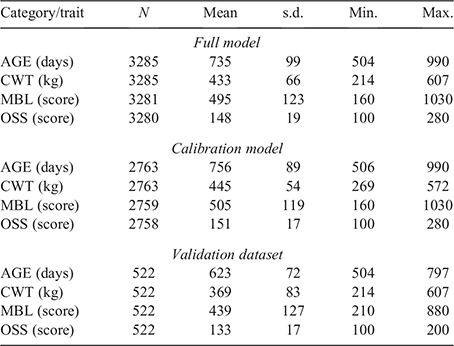
Genotypes for 45 364 autosomal single-nucleotide polymorphisms (SNPs) were available for all the animals included in the present study and were used to compute the genomic relationship matrix (G) following Method 1 of VanRaden (2008). Initially, a model using data from steers in both the reference and validation populations (Cohorts 1–7, defined hereafter as the ‘full model’) was used to obtain the most reliable set of fixed-effect solutions and genetic parameter estimates and to assist in the calibration when back-solving the SNP effects to generate the genomic prediction equations. The linear model contained the fixed effects of contemporary group (CG, 77 levels) and age of dam at birth of calf in years (6 levels, 2–7+ years) and the linear regression covariate of age at measurement in days. The CG was defined as a combination of cohort, property of origin, month of birth, management group and the date the phenotype was measured. Records without age at measurement or from steers in a CG with fewer than seven animals were excluded from analyses. After edits, the number of records available for analysis was 3285 for the full model, 2763 for the calibration model (Cohorts 1–6) and 522 in the validation dataset (Cohort 7). Additionally, the random additive polygenic and residual effects were fitted with assumed distributions N(0, G⊗Vg) and N(0, I⊗Ve) respectively, where G represents the genomic relationship matrix described earlier, Vg is the genetic variance matrix, I is an identity matrix, Ve is the residual variance matrix and ⊗ represents the Kronecker product.
Validation involved the following steps: (1) the above model (full model) was then re-run using a dataset in which the phenotypic data from steers in the reference population (Cohorts 1–6) were retained in the dataset and data from steers in the validation population (Cohort 7) were set as missing values (defined hereafter as the ‘calibration model’); (2) the calibration model was then used to generate gEBVs for steers in the validation population (Cohort 7) on the basis of their genomic relationship with steers in the reference population (Cohorts 1–6); and (3) the SNP effects, back-solved from the calibration model, were then further calibrated using results from the full model and the final gEBVs for steers in the validation population (Cohort 7) were generated on the basis of SNP effects and their genomic relationship with steers in the reference population (Cohorts 1–6). In summary, after solving the calibration model, where animals from the validation population contributed genotypes but not phenotypes, the SNP effects were back-solved and used to recompute the gEBVs for animals in the validation population. The optimality of these new recomputed gEBVs was assessed by comparing them with the gEBVs obtained using the full model, where animals from the validation population contributed both genotypes and phenotypes. All models were run using univariate analyses, one trait at a time, by using the Qxpak5 software (Pérez-Enciso and Misztal 2011).
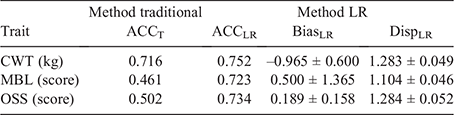
To validate the ability of Angus SteerSELECT to predict differences in performance of steers in the validation population, their raw phenotypic data for CWT, MBL and OSS were adjusted for fixed effects and covariates by using solutions from the full model. Traditional and Method LR (Legarra and Reverter 2018) approaches were used to estimate accuracy, bias and dispersion of gEBVs generated for steers in the validation population. Traditional accuracy (ACCT) was calculated as the correlation between gEBV and the adjusted phenotype divided by the square root of h2. Following Method LR approaches (Legarra and Reverter 2018), accuracy (ACCLR), bias (BiasLR) and dispersion (DispLR) were calculated by comparing the gEBVs for the validation population resulting from the full model with the gEBVs for the same individuals resulting from the calibration model. ACCT is grounded in theory. However, it does require knowledge of adjustment factors and can be affected dramatically when heritability is poorly estimated, which is possible when the selection process is inadequately described in the data and environmental trends are present. In contrast, the LR method obviates the need for adjustment factors and has been shown to perform optimally even if the model uses an incorrect heritability or a hidden trend exists in the data (Macedo et al. 2020).
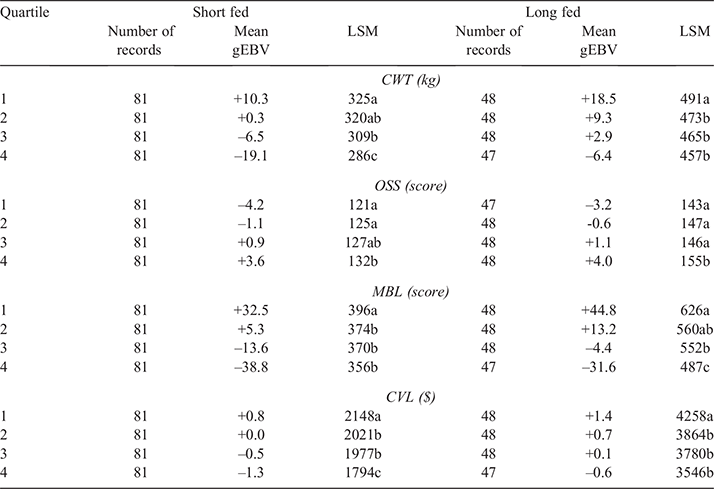
Steers in the validation population were then ranked on the basis of their gEBV for each of the traits CWT, MBL and OSS and assigned to quartiles, with the 25% of steers predicted to be the best performers for a given trait being assigned to quartile one, the next 25% to quartile two, the following 25% to quartile three and the 25% of steers predicted to be the worst performers to quartile four. Therefore, steers in quartile one were predicted to have the highest CWT and MBL and the lowest OSS. Short-fed (n = 324) and long-fed (n = 191) steers in the validation population were ranked independently. A small number of long-fed steers (n = 7) in the validation population were excluded from ranking because they were slaughtered at an abattoir different from where their herd mates were slaughtered. For CVL, the gEBVs for CWT and MBL for each individual steer in the validation population were standardised (by dividing values by the standard deviation of gEBVs from all steers in the validation population), and the average of these standardised gEBVs for CWT and MBL were used to rank animals for CVL. Steers were then assigned to quartiles as described above. Quartile measured phenotype least-square means (LSMs) were then generated for all traits by using a linear model, fitting the fixed effects of kill date and herd where significant, and the significance of differences among quartiles were analysed using R (R Core Team 2013). Multiple comparisons were evaluated using P-values adjusted using a Tukey correction.
Results and discussion
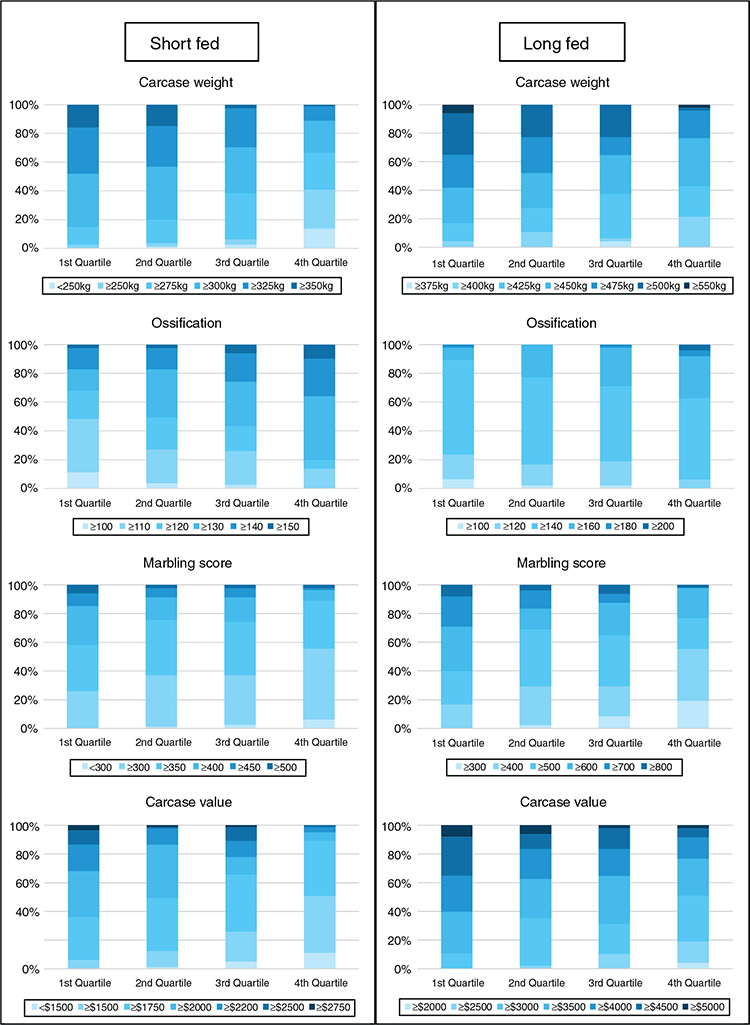
Results from the current study demonstrated that SteerSELECT was able to predict differences in CWT, MBL and OSS of both short-fed and long-fed Angus steers. Summary statistics for the traits analysed are presented in Table 1. The accuracy of CWT, MBL and OSS gEBVs for steers were 0.72, 0.46 and 0.50 respectively, when computed using the traditional method and 0.75, 0.72 and 0.73 when computed using Method LR (Table 2). The larger accuracy difference observed for MBL and OSS, than for CWT, using the different methods is not fully understood and will require further investigation. To demonstrate how differences in predicted gEBVs translated into differences observed at the phenotypic level, steers in the validation population were ranked on their gEBV for CWT, MBL and OSS and assigned to quartiles representing the predicted top 25% of performers for each trait in quartile one, followed by the next 25% and so on. The measured phenotype LSMs for steers in each quartile were then compared for each trait (Table 3). Short- and long-fed steers ranked in quartile one for CWT gEBV had carcasses that were, on average, 39 and 34 kg heavier respectively, than those ranked in quartile four. The mean MBL of short- and long-fed steers ranked in quartile one for MBL gEBV were 40 and 139 points higher respectively, than those ranked in quartile four. Genomic prediction equations were also able to successfully predict OSS, with steers ranked in quartile one for OSS gEBVs having mean OSS that were lower, 11 points in short-fed steers and 12 points in long-fed steers, than those of their counterparts ranked in quartile four. It should be noted that lower OSS would be targeted by feedlot operators. Differences in LSMs between quartiles one and four for all traits analysed, namely, CWT, MBL and OSS, were highly significant (P < 0.001) in both short-fed and long-fed steers (Table 3). The distribution of measured phenotype values for steers assigned to each quartile for CWT, MBL and OSS are shown in Fig. 1.
|
|
The concept of using SNP genotypes to predict the genetic merit of individual animals was first proposed by Meuwissen et al. (2001). This method allows breeding values to be estimated for unrelated animals in a population, by using information previously ignored by traditional pedigree-based methods (Clark et al. 2012). Three key factors have facilitated the accelerated genetic improvement of livestock through genomic selection, being (a) development of genomic selection methodology, (b) the discovery of large numbers of SNPs that can be used as genetic markers and (c) the improved cost-effectiveness of genotyping methods (Meuwissen et al. 2013, 2016). The implementation of low-density SNP panels to estimate the genetic merit of individual animals at the commercial level of the beef industry has the potential to inform decisions at all levels of the commercial production chain from breeding and management on farm, through to predicting carcass characteristics for individual animals at the point of slaughter (Miller 2010).
Successful adoption of genomic selection tools by commercial beef producers will be driven by the ability of genomic tests to accurately predict performance in commercial production environments. Several factors influence the accuracy of genomic predictions, including the number of animals represented in the reference population, the relatedness of animals in the reference and test populations, the heritability of the trait, the type of response variable used to estimate accuracies and the method used to cluster reference population data for validation (Boddhireddy et al. 2014). Where phenotypic data are available for animals in the validation population, adjusted phenotype measures are commonly used as the response variable to assess gEBV accuracies, whereas, when phenotypic data are not available, EBV and degressed EBV data are commonly used (Garrick et al. 2009).
The accuracy of genomic predictions of carcass-trait performance in beef cattle has been reported previously. Weber et al. (2012) reported gEBV accuracies of 0.35 (±0.10) for CWT and 0.23 (±0.06) for MBL in Angus cattle respectively. In more recent studies using Angus cattle, Bolormaa et al. (2013) reported gEBV accuracies of 0.16–0.18 for CWT and 0.1–0.21 for MBL (depending on the method used to generate genomic prediction equations) and Chen et al. (2015) reported gEBV accuracies of 0.35 (±0.02) for CWT and 0.37 (±0.03) for MBL. In each of these studies, a multibreed reference population was used to generate genomic prediction equations and accuracies were computed as the correlation between gEBVs and measured phenotypes. When there are limited numbers of animals of a target breed available for inclusion in the reference population, it is common to use multibreed reference populations to improve the accuracy of genomic predictions; however, improvements in accuracy are expected to be far greater when animals added to the reference population are of the same breed as those in the test population (Meuwissen et al. 2016). Using an Angus-specific reference population with a high EBV accuracy, Boddhireddy et al. (2014) reported CWT gEBV accuracies of 0.43 and 0.74 and MBL gEBV accuracies of 0.51 and 0.76 when EBVs or degressed EBVs respectively, were used as response variables. Accuracies associated with genomic predictions of carcass-trait performance have also been reported in other Bos taurus and Bos indicus beef cattle breeds. Mehrban et al. (2017) reported CWT and MBL gEBV accuracies of 0.32–0.4 and 0.25 respectively, in Hanwoo cattle. Similarly, a MBL gEBV accuracy of 0.32 has been reported in Nelore cattle (Magalhães et al. 2019).
The accuracy of CWT and MBL gEBVs generated in the current study were higher or, as a minimum, comparable with accuracies reported previously. As described above, this may be a reflection of (1) the high number of animals in the reference population, with accompanying high-quality, industry-relevant phenotypes used to generate genomic predictions underpinning Angus SteerSELECT and (2) the relatedness of steers in the reference and validation populations used in the validation. ACCT and ACCLR were both ~0.7 for CWT, while ACCT dropped to ~0.5 for MBL and OSS and ACCLR remained at ~0.7 for these two traits (Table 2). With a reference population of 4000 animals, a heritability of 0.3 and an effective population size of 100, a gEBV accuracy of ~0.45 is expected (Goddard and Hayes 2009). Similarly, with a reference population of 1743 Australian Angus cattle, Bolormaa et al. (2013) reported a gEBV accuracy of 0.26 averaged across 16 traits. The estimates of gEBV biases were all within two standard errors of zero (Table 2). However, there was evidence of gEBV over-dispersion, with Method-LR dispersion being >1 for the three traits (Table 2). This over-dispersion in the gEBV could be attributed to the Cohort 7 animals (validation population) being younger, lighter and with lower MBL and OSS than for animals in the calibration population (Table 1), and hence, prediction equations being based on heavier animals. Further research is needed to fully ascertain the reason for this over-dispersion.
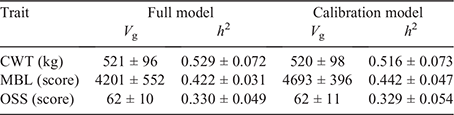
The heritability of traits influences the ability to predict genetic differences for those traits. In the current study, the genomic-based heritability of CWT, MBL and OSS was estimated at 0.53 ± 0.07, 0.42 ± 0.03 and 0.33 ± 0.05 respectively (Table 4). These estimates are higher than the heritability estimates of 0.37, 0.28 and 0.22 previously reported for CWT, MBL and OSS in Angus cattle (Boddhireddy et al. 2014; Jeyaruban et al. 2017), and also the results from the TransTasman Angus Cattle Evaluation (TACE), which estimated the heritability of CWT and intramuscular fat % (measured on carcasses) to be 0.41 and 0.32 respectively (https://www.angusaustralia.com.au/tace/resources/heritability-of-traits/).
|
For commercial beef producers to adopt genomic selection tools as part of routine management, the cost versus benefit of using such tools must be clearly demonstrated. To demonstrate the potential economic benefits for feedlot operators using SteerSELECT, the CVL of steers in the validation population was estimated. A CVL gEBV was calculated, by combining gEBVs for CWT and MBL, and steers were ranked into quartiles on the basis of their CVL gEBV, with the 25% of steers predicted to have the highest CVL being assigned to quartile one, the next 25% of steers to quartile two, and so on. The LSM of CVL for steers in quartiles one to four were A$2148, A$2021, A$1977 and A$1797 for short-fed steers and A$4258, A$3864, A$3780 and A$3546 for long-fed steers respectively, representing a difference in CVL between quartiles one and four of A$351 in short-fed steers and A$712 in long-fed steers. The distribution of estimated CVL for steers assigned to each quartile is shown in Fig. 1. Costs associated with the use of Angus SteerSELECT include labour costs to collect DNA samples from individual steers, genotyping costs and analysis costs. Clearly, the costs and benefits associated with identifying steers suitable for feedlot finishing and the most appropriate finishing path (e.g. short-fed or long-fed) will vary among those in the beef supply chain including feedlot operators. This will be specific to each production system, requiring specific cost–benefit analyses to be undertaken for each supply chain.
Use of genomic-based selection tools will provide beef cattle producers an opportunity to make genetic gains in novel, hard-to-measure traits such as feed efficiency and disease resistance (Miller 2010). We intend to include genomic predictions for a total of nine routinely measured and hard-to-measure traits in the final Angus SteerSELECT genomic selection tool, including traits related to growth (yearling weight), efficiency (average daily gain, dry matter intake) carcass characteristics (CWT, MBL, OSS, eye-muscle area, rib fat) and health (ImmuneDEX, Reverter et al. 2021). We are also developing weighted selection indexes based on combinations of relevant trait gEBVs, which are aimed at predicting the suitability of steers for different finishing protocols including short-fed, long-fed and grass-fed, with index values reported as part of Angus SteerSELECT. As the ability of Angus SteerSELECT to predict differences in performance of steers that are not purebred Angus, or that do not have a high Angus content, has not been evaluated, inclusion of a breed verification step in the analytical pipeline that generates predictions for the Angus SteerSELECT product is planned. This will allow DNA samples submitted for testing from crossbred animals or animals of a breed other than Angus to be identified, and predictions for these animals flagged as potentially of low accuracy.
Production losses due to disease are a major economic cost for the beef cattle feedlot industry in Australia (Lane et al. 2015). The Australian feedlot industry is actively investing in strategies to improve the health and welfare of cattle in their production systems and reduce their use of antibiotics to treat disease (Hine et al. 2019). Genomic selection tools have the potential to identify animals that are more resistant or susceptible to disease (Raszek et al. 2016); however, phenotypic data of sufficient quality and quantity to underpin the development of such tools has been lacking. We have developed methodology to assess immune competence, a proxy for general disease resistance, in beef cattle (Hine et al. 2019), and have used phenotypes to generate an index, termed ‘ImmuneDEX’, which can be used as a tool for the genetic improvement of immune competence in beef cattle (Reverter et al. 2021). Uniquely, genomic predictions for ImmuneDEX will be included as part of SteerSELECT, providing feedlot operators with information on the predicted ability of individual animals to resist disease for the first time.
Conclusions
Results from the present study suggest that the genomic predictions underpinning SteerSELECT can predict differences in CWT, MBL, OSS and CVL in both short-fed and long-fed Angus steers finished under commercial Australian feedlot conditions. This provides confidence that the commercialised Angus SteerSELECT product, based on a similar but larger reference population, will be able to accurately predict differences in performance, in terms of growth, efficiency, health and carcass characteristics, of Australian Angus steers. This will, in turn, provide businesses in the beef supply chain (e.g. breeders, backgrounders, feedlot operators, abattoirs and beef brand owners) with additional information on Angus animals to inform selection, purchasing and management decisions. Angus SteerSELECT will be especially beneficial to integrated beef supply chains that have the ability to target steers to different finishing paths, such as short-fed versus long-fed feeding programs, and/or with different brand specifications such as moderate versus high marbling requirements. Future studies will aim to validate the ability of Angus SteerSELECT to predict differences in performance of steers for additional traits (e.g. feed intake and immune competence), which will be included in the final product using the validation population described here. Studies will also be undertaken to assess the effectiveness of Angus SteerSELECT to predict differences in feedlot and carcass performance in commercial Angus steers that are independent of the ASBP and the Angus Australia reference population.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work was co-funded by Angus Australia and CSIRO. The authors acknowledge Angus Australia for facilitating access to progeny data from the Angus Sire Benchmarking Program. We also gratefully acknowledge cooperating herd owners, managers, and staff involved in the Angus Sire Benchmarking Program and co-funding for the program from Meat & Livestock Australia through the MLA Donor Co. (Project P.PSH.0528).
References
Angus Australia (2019) Australian beef breeding insights. Available at https://www.angusaustralia.com.au/australian-beef-breeding-insights/. [Verified 2 February 2021]Boddhireddy P, Kelly MJ, Northcutt S, Prayaga KC, Rumph J, DeNise S (2014) Genomic predictions in Angus cattle: comparisons of sample size, response variables, and clustering methods for cross-validation. Journal of Animal Science 92, 485–497.
| Genomic predictions in Angus cattle: comparisons of sample size, response variables, and clustering methods for cross-validation.Crossref | GoogleScholarGoogle Scholar | 24431338PubMed |
Bolormaa S, Pryce JE, Kemper K, Savin K, Hayes BJ, Barendse W, Zhang Y, Reich CM, Mason BA, Bunch RJ, Harrison BE, Reverter A, Herd RM, Tier B, Graser HU, Goddard ME (2013) Accuracy of prediction of genomic breeding values for residual feed intake, carcass and meat quality traits in Bos taurus, Bos indicus and composite beef cattle. Journal of Animal Science 91, 3088–3104.
| Accuracy of prediction of genomic breeding values for residual feed intake, carcass and meat quality traits in Bos taurus, Bos indicus and composite beef cattle.Crossref | GoogleScholarGoogle Scholar | 23658330PubMed |
Chen L, Vinsky M, Li C (2015) Accuracy of predicting genomic breeding values for carcass merit traits in Angus and Charolais beef cattle. Animal Genetics 46, 55–59.
| Accuracy of predicting genomic breeding values for carcass merit traits in Angus and Charolais beef cattle.Crossref | GoogleScholarGoogle Scholar | 25393962PubMed |
Clark SA, Hickey JM, Daetwyler HD, van der Werf JHJ (2012) The importance of information on relatives for the prediction of genomic breeding values and the implications for the makeup of reference data sets in livestock breeding schemes. Genetics, Selection, Evolution 44, 4
| The importance of information on relatives for the prediction of genomic breeding values and the implications for the makeup of reference data sets in livestock breeding schemes.Crossref | GoogleScholarGoogle Scholar | 22321529PubMed |
Garrick DJ, Taylor JF, Fernando RL (2009) Deregressing estimated breeding values and weighting information for genomic regression analyses. Genetics, Selection, Evolution 41, 55
| Deregressing estimated breeding values and weighting information for genomic regression analyses.Crossref | GoogleScholarGoogle Scholar | 20043827PubMed |
Goddard M, Hayes B (2009) Mapping genes for complex traits in domestic animals and their use in breeding programs. Nature Reviews Genetics 10, 381–391.
| Mapping genes for complex traits in domestic animals and their use in breeding programs.Crossref | GoogleScholarGoogle Scholar | 19448663PubMed |
Hine BC, Bell AM, Niemeyer DOD, Duff CJ, Butcher NM, Dominik S, Ingham AB, Colditz IG (2019) Immune competence traits assessed during the stress of weaning are heritable and favourably genetically correlated with temperament traits in Angus cattle. Journal of Animal Science 97, 4053–4065.
| Immune competence traits assessed during the stress of weaning are heritable and favourably genetically correlated with temperament traits in Angus cattle.Crossref | GoogleScholarGoogle Scholar | 31581299PubMed |
Jeyaruban MG, Johnston DJ, Walmsley BJ (2017) Genetic and phenotypic characterization of MSA index and its association with carcase and meat quality traits in Angus and Brahman cattle. In ‘Proceedings of the Association for the Advancement of Animal Breeding and Genetics’. (Ed. L Porton-Neto) pp. 313–316. (Townsville, Qld, Australia).
Lane J, Jubb T, Shephard R, Webb-Ware J, Fordyce G (2015) Priority list of endemic diseases for the red meat industries (MLA Project B.AHE.0010). Meat & Livestock Australia. Available at https://www.mla.com.au/research-and-development/search-rd-reports/final-report-details/Animal-Health-and-Biosecurity/Priority-list-of-endemic-diseases-for-the-red-meat-industries/2895 [Verified 1 February 2021]
Legarra A, Reverter A (2018) Semi-parametric estimates of population accuracy and bias of predictions of breeding values and future phenotypes using the LR method. Genetics, Selection, Evolution 50, 53
| Semi-parametric estimates of population accuracy and bias of predictions of breeding values and future phenotypes using the LR method.Crossref | GoogleScholarGoogle Scholar | 30400768PubMed |
Macedo FLL, Reverter A, Legarra A (2020) Behavior of the Linear Regression method to estimate bias and accuracies with correct and incorrect genetic evaluation models. Journal of Dairy Science 103, 529–544.
| Behavior of the Linear Regression method to estimate bias and accuracies with correct and incorrect genetic evaluation models.Crossref | GoogleScholarGoogle Scholar |
Magalhães AFB, Schenkel FS, Garcia DA, Gordo DGM, Tonussi RL, Espigolan R, de Oliveira Silva RM, Braz CU, Fernandes Júnior GA, Baldi F, Carvalheiro R, Boligon AA, de Oliveira HN, Chardulo LAL, de Albuquerque LG (2019) Genomic selection for meat quality traits in Nelore cattle. Meat Science 148, 32–37.
| Genomic selection for meat quality traits in Nelore cattle.Crossref | GoogleScholarGoogle Scholar | 30296711PubMed |
Mehrban H, Lee DH, Moradi MH, IlCho C, Naserkheil M, Ibáñez-Escriche N (2017) Predictive performance of genomic selection methods for carcass traits in Hanwoo beef cattle: impacts of the genetic architecture. Genetics, Selection, Evolution 49, 1
| Predictive performance of genomic selection methods for carcass traits in Hanwoo beef cattle: impacts of the genetic architecture.Crossref | GoogleScholarGoogle Scholar | 28093066PubMed |
Meuwissen TH, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829.
| Prediction of total genetic value using genome-wide dense marker maps.Crossref | GoogleScholarGoogle Scholar | 11290733PubMed |
Meuwissen T, Hayes B, Goddard M (2013) Accelerating improvement of livestock with genomic selection. Annual Review of Animal Biosciences 1, 221–237.
| Accelerating improvement of livestock with genomic selection.Crossref | GoogleScholarGoogle Scholar | 25387018PubMed |
Meuwissen T, Hayes B, Goddard M (2016) Genomic selection: a paradigm shift in animal breeding. Animal Frontiers 6, 6–14.
| Genomic selection: a paradigm shift in animal breeding.Crossref | GoogleScholarGoogle Scholar |
Miller S (2010) Genetic improvement of beef cattle through opportunities in genomics. Brazilian Journal of Animal Science 39, 247–255.
| Genetic improvement of beef cattle through opportunities in genomics.Crossref | GoogleScholarGoogle Scholar |
Pérez-Enciso M, Misztal I (2011) Qxpak.5: old mixed model solutions for new genomics problems. BMC Bioinformatics 12, 202
| Qxpak.5: old mixed model solutions for new genomics problems.Crossref | GoogleScholarGoogle Scholar | 21612630PubMed |
R Core Team (2013) ‘R: a language and environment for statistical computing.’ (R Foundation for Statistical Computing: Vienna, Austria)
Raszek MM, Guan LL, Plastow GS (2016) Use of Genomic Tools to Improve Cattle Health in the Context of Infectious Diseases. Frontiers in Genetics 7, 30
| Use of Genomic Tools to Improve Cattle Health in the Context of Infectious Diseases.Crossref | GoogleScholarGoogle Scholar | 27014337PubMed |
Reverter A, Hine BC, Porto-Neto L, Li Y, Duff CJ, Dominik S, Ingham AB (2021) ImmuneDEX: a strategy for the genetic improvement of immune competence in Australian Angus cattle. Journal of Animal Science 99, skaa384
| ImmuneDEX: a strategy for the genetic improvement of immune competence in Australian Angus cattle.Crossref | GoogleScholarGoogle Scholar | 33677583PubMed |
VanRaden PM (2008) Efficient methods to compute genomic predictions. Journal of Dairy Science 91, 4414–4423.
| Efficient methods to compute genomic predictions.Crossref | GoogleScholarGoogle Scholar | 18946147PubMed |
Weber KL, Thallman RM, Keele JW, Snelling WM, Bennett GL, Smith TPL, McDaneld TG, Allan MF, Van Eenennaam AL, Kuehn LA (2012) Accuracy of genomic breeding values in multibreed beef cattle populations derived from deregressed breeding values and phenotypes. Journal of Animal Science 90, 4177–4190.
| Accuracy of genomic breeding values in multibreed beef cattle populations derived from deregressed breeding values and phenotypes.Crossref | GoogleScholarGoogle Scholar | 22767091PubMed |