A review of how dairy farmers can use and profit from genomic technologies
Jennie Pryce A C and Ben Hayes A BA Biosciences Research Division, Department of Primary Industries, 1 Park Drive, Bundoora, Vic. 3083, Australia.
B La Trobe University, Bundoora, Vic. 3086, Australia.
C Corresponding author. Email: jennie.pryce@dpi.vic.gov.au
Animal Production Science 52(3) 180-184 https://doi.org/10.1071/AN11172
Submitted: 9 August 2011 Accepted: 7 October 2011 Published: 6 March 2012
Journal Compilation © CSIRO Publishing 2012 Open Access CC BY-NC-ND
Abstract
New genomic technologies can help farmers to (1) achieve higher annual rates of genetic gain through using genomically tested bulls in their herds, (2) select for ‘difficult’ to measure traits, such as feed conversion efficiency, methane emissions and energy balance, (3) select the best heifers to become herd replacements, (4) sell pedigree heifers at a premium, (5) use mating plans to optimise rates of genetic gain while controlling inbreeding, (6) achieve certainty in parentage of individual cows and (7) avoid genetic defects that could arise from mating cows to bulls that are known carriers of genetic diseases that are the result of a single lethal mutation. The first use does not require genotyping females and could approximately double the net income per cow that arises due to genetic improvement, mainly through a reduction in generation interval. On the basis of current rates of genetic gain, the net profit from using genotyped bulls could be worth AU$20/cow per year and is permanent and cumulative. One of the most powerful uses of genomic selection is to select for economically important, yet difficult- or expensive-to-measure traits, such as residual feed intake or energy balance. Provided the accuracy of genomic breeding values is high enough (i.e. correlation between the true and estimated breeding values), these traits lend themselves well to genomic selection. For selecting replacement heifers, if genotyping costs are AU$50/cow, the net profit of genotyping 40 heifers to select the top 20 as replacements (per 100 cows) would be worth approximately AU$41 per cow. However, using parent average estimated breeding-value information is free and can already be used to select replacement heifers. So, genotyping costs would need to be very low to be more profitable than selecting on parent average estimated breeding value. However, extra value from genotyping can also be captured by using other strategies. For example, mating plans that use genomic relationships rather than pedigree relationships to capture inbreeding are superior in terms of reducing progeny inbreeding at a desired level of genetic gain, although pedigree does an adequate job. So, again, the benefits of genotyping are small (<AU$10). Ascertainment of pedigree is an additional use of genotyping and is potentially worth ~AU$30 per cow. Avoidance of genetic diseases and selling of pedigree heifers have a value that should be estimated case-by-case. Because genotyping costs continue to fall, it may become increasingly popular to capture the extra value from genotyping females.
Introduction
Genomic selection refers to selection decisions based on genomic breeding values (GEBV). The GEBVs are calculated as the sum of the effects of dense genetic markers that are approximately equally spaced across the entire genome, thereby potentially capturing most of the quantitative trait loci that contribute to variation in a trait.
The application of genomic selection to dairy cows has enabled breeding companies to redesign their breeding schemes (Pryce and Daetwyler 2012). More than two times the rate of genetic gain achieved through conventional progeny testing is feasible if bulls are used at a young age and large numbers of young bulls are screened (de Roos et al. 2011). However, other potential applications of genomics in dairy herds have been less well studied. It may be economically worthwhile for dairy farmers to invest in whole or partial herd low-density genotyping. Examples could include confirming parentage, selecting the best heifer calves born to keep as herd replacements, managing genetic defects and selling high-value pedigree stock.
The aim of the present review is to assess the potential value of genomic technology to dairy farmers.
Selection of sires
Under conventional progeny-testing schemes, farmers have a choice of purchasing semen from (1) young bulls that have entered progeny-testing (where the Australian breeding value, is a parent average with a reliability of ~30–35%), (2) 1st-proof sires with 50–100 daughters (typical reliabilities are 80–85%) and (3) proven 2nd-crop sires with 100s to 1000s of daughters and reliabilities of 90–99%. The reliability is the squared correlation between the true breeding value and the estimated breeding value. Therefore, a reliability of 99% is very close to the true breeding value. Reliabilities of milk-production traits evaluated using genomic selection and recently surveyed (April 2011) are currently ~60% (Pryce and Daetwyler 2012). For a trait with a heritability of 0.3, a reliability of 60% is approximately the same as proof based on phenotypes from 20 daughters.
Reducing risk from using young bulls with genomic estimated breeding values
The number of bulls selected for use (through artificial insemination) in a herd is primarily a function of the number of cows in that herd. A herd of 100 cows may choose a group of five proven bulls (1st proof). To obtain the same group or team reliabilities for genomically evaluated young bulls, more young bulls would need to be used. The reliability of a group of unrelated sires can be calculated as follows:
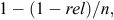
where rel is the individual reliability of bulls and n is the number of bulls in the group. The assumption that sires are unrelated is not strictly correct; in the majority of cases, many sires in a group will have common ancestry. Therefore, this is an upper limit of ‘group’ reliability. Using this equation, a group of 10 genomically tested sires with reliabilities of 60% have a collective reliability that is similar to the collective reliability of a group of five first-proof bulls with individual reliabilities of 80% (96% group reliability). Therefore, the 95% confidence interval around the mean of this example of 10 bulls with genomic evaluations and five with first proofs is about the same.
In addition to using a larger group of genomically tested bulls, compared with proven bulls to mitigate the risks associated with lower reliabilities of individual bulls, another strategy is using a mixture of proven and genomically tested bulls. However, as countries switch from progeny-testing to solely genomic selection (as is the case in France; Boichard et al. 2010), then bigger bull groups will probably become the accepted norm until the reliabilities achieved in genomic selection match the reliabilities of bulls that are progeny-tested (80–85%).
The new genomic era could revolutionise the way bulls are selected and used, but this does not mean that data collected on-farm (traditionally through progeny-testing) become less important. At the current marker densities (~50 000 single-nucleotide polymorphisms, SNPs), the prediction of GEBVs using associations between SNP data and phenotypes in a reference population of bulls needs to be continuously updated, otherwise the reliability of the GEBV erodes rapidly over generations due to the decay of genetic relationships between the reference and validation populations (e.g. Habier et al. 2007). Because fewer bulls are likely to be progeny-tested, it is likely that genotyped females will play an important role in updating future reference populations through providing phenotypes on important traits such as milk production, health, fertility, survival and type.
Selecting for difficult to measure traits
One of the most powerful uses of genomic selection is to select for economically important, yet difficult- or expensive-to-measure traits. Provided the accuracy of GEBVs is high enough (i.e. correlation between the true breeding value and GEBV), these traits lend themselves well to genomic selection.
Examples include traits related to feed intake, such as energy balance and residual feed intake and some measures of fertility, such as commencement of luteal activity. Residual feed intake (RFI) is the difference between the actual (measured) feed intake and the predicted feed requirements, based on liveweight, growth rate and level of milk production (for lactating cows). It is important because feed is a major component of on-farm costs. Recently, Pryce et al. (2012a) reported a cross-validation study of 2000 6-month-old heifers from Australia and New Zealand to predict the accuracy of RFI by using genomic selection and assessed using cross-validation. All heifers were genotyped with the Illumina high-density bovine SNP chip (Ilumina Inc., San Diego, CA, USA) and had detailed phenotypes on feed intake and liveweight, enabling RFI to be calculated. The average accuracy was 0.37 in Australian heifers and 0.31 in New Zealand heifers. Hayes et al. (2011) demonstrated that at this accuracy, including RFI in the Australian profit ranking (APR), and selection for the index, with DNA marker-derived breeding values for RFI, would improve the rate of annual gain for profitability by 3.8%.
Selection to improve ‘difficult’ traits is often hampered by the cost of measurement and, as a consequence, they are generally available only on small numbers of animals. Optimal strategies to collect the phenotypes and genotypes for these traits are required if they are to be incorporated in future breeding objectives. One way in which this can be done is to set up dedicated resource populations in which the phenotypes are collected. Alternatively, there may be opportunities for research organisations to combine data on genotypes and comparable phenotypes.
Genotyping females
With the availability of low-density SNP panels, there is an opportunity for farmers to genotype their own cows. But, is this economically worthwhile?
Selecting replacements
One use of genotyping is to identify the best heifers to become replacements. The benefit is the dollar advantage (i.e. APR) of the heifers that are selected using genomic selection, compared with the average APR of the whole group. APR is a selection index that measures net farm profit and includes milk, fat and protein yields, survival, fertility, somatic cell count, liveweight, temperament and milking speed. The standard deviation (s.d.) of APR (SD_APR) within the Holstein breed is AU$80.4 (Australian Dairy Herd Improvement Scheme 2010). It should be noted that within a herd, the SD_APR is likely to be lower than that across the whole population of dairy bulls.
The net profit advantage of a selected group of heifers can be calculated using an adaptation of the breeder’s equation (e.g. Falconer and Mackay 1996), as follows:
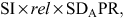
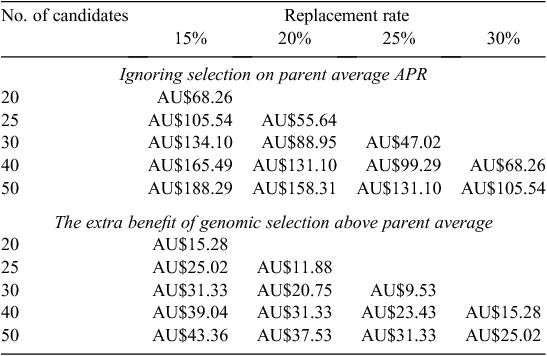
where SI is the selection intensity calculated from the proportion of heifers selected and rel is the individual reliability of bulls. To calculate SI, we assumed a range of replacement heifers (replacements) and heifers available for selection (candidates). The candidate heifers available ranged between 20 and 50 per 100 cows and the replacement rates varied between 15% and 30%. So, an example is selecting the best 15 heifers of 20 candidates. The cost of genotyping was assumed to be AU$5, AU$50 and AU$100. The cost of genotyping was spread across the number of replacements. Therefore, a herd that genotyped a large number of candidate heifers to select a low number of replacements had a large per-replacement genotyping cost.
Two scenarios were tested, one where there was no previous selection strategy (i.e. replacements were selected at random). The other scenario was where selection using genotyping was compared with selection using parent average APR. The only difference between parent average and genomic selection is that replacements would be selected with higher-reliability breeding values using genomic selection. Costs associated with rearing were not included in the calculations, because it was assumed that having a genomic test did not affect when surplus heifers were sold.
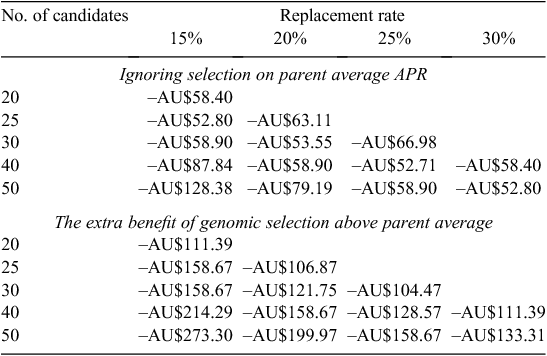
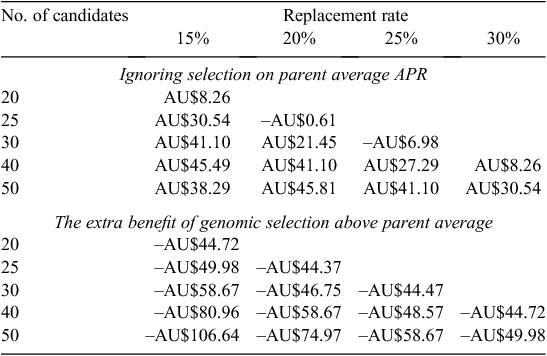
At current genotyping costs (AU$100/heifer), the dollar benefit of genotyping solely as a tool to select replacements was not worthwhile (Table 1). If the cost of genotyping was AU$50, then there was an advantage in genotyping (Table 2). However, this advantage was only in the scenario where selection was compared with no previous selection. If animals were chosen on the basis of their parent average estimated breeding value for APR, then the advantage of genotyping above that was not sufficient to make it profitable. If the cost of genotyping was AU$5, then genotyping became profitable in both scenarios (Table 3). The biggest advantage was when a large number of candidates were genotyped for relatively few replacements, i.e. genotyping 50 candidate heifers to select the best 15 generates AU$43 profit per replacement over selection on parent average. This very high selection intensity may be feasible if sexed semen and heifers to breed heifers are used in the breeding plan.
|
|
|
Marketing pedigree heifers
The situation is different where genotyping of females is used as a marketing tool, especially for pedigree heifers. Using genomics, the reliability of a heifer’s breeding values at birth can be as high as 60%, which is equivalent to a cow with three or four lactation records, and much higher than a heifer’s reliability without genomic selection (which is ~30%, depending on trait). An approximation of reliability can be calculated as a function of the number of records and heritability of a trait (e.g. Cameron 1997), as follows:
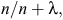
where n is the number of records and λ is 1 – h2/h2, where h2 is the heritability of the trait.
Genotyping young heifers will help alleviate concerns over preferential treatment, because the genomic part of the breeding value should be less biased than the part derived from pedigree relationships. This could see either positive implications for pedigree sale prices of genotyped heifers and/or embryos, or simply that genotyping of sale heifers becomes the norm.
Mating plans
Control of inbreeding levels in progeny can be implemented using mate allocation (Kinghorn 1998). Mate allocation can be considered independently of mate selection, if all candidates are to be mated; this is often the case for modern dairy herds. Poor fertility and the requirement to maintain compact calving patterns mean that often all available females are included in the mating plan. For large herds in particular, mating plans could help resolve the choice of sires to mate to cows in an optimal way. The idea is to maximise a specific breeding objective while constraining inbreeding in the progeny (Kinghorn 1998). Inbreeding affects profitability by adversely affecting traits related to fitness and production (Smith et al. 1998). On the basis of Australian data, Man (2004) calculated that the cost of inbreeding was 3.1 APR units per 1% increase in inbreeding. Smith et al. (1998) calculated that the cost of inbreeding over a cow’s lifetime was AU$24 in the USA, which Haile-Mariam et al. (2007) converted to an APR equivalent to $8.5 per 1% increase in inbreeding.
Traditionally pedigree relationships have been used to control inbreeding in mating plans. However, the genomic relationship matrix can be used instead (Sonesson et al. 2010; Pryce et al. 2012b). Sonesson et al. (2010) demonstrated that under genomic selection, controlling inbreeding using either pedigree or the genomic relationship matrix was effective. However, when inbreeding was controlled using pedigree relationships, the rate of genomic inbreeding was three times greater than when inbreeding was controlled using genomic relationships.
Pryce et al. (2012b) showed that controlling inbreeding using a genomic relationship matrix could reduce the rate of inbreeding by 1–2%, with very little loss in genetic gain in APR (less than AU$0.70). This reduction in inbreeding is worth between AU$4.96 and AU$9.71. However, pedigree did a reasonably good job as well, reducing inbreeding by around half the amount of genomically controlled inbreeding when both pedigree and genomically controlled inbreeding were assessed on the genomic scale. As pedigree is ‘free’, the value of controlling inbreeding using genomic relationships rather than pedigree relationships is small and by itself does not justify genotyping females.
For farmers using large groups of genomically tested sires, it may be difficult to manually work out which cow to mate to which bull, i.e. avoiding matings between relatives. This could mean that computerised mating plans become more common. However, it seems likely that, in the short term, only part of the herd is likely to be genotyped. One strategy is to replace part of the pedigree relationship matrix with genomic relationships (e.g. Legarra et al. 2009). So pedigree only is used for some of the relationships and genomic relationships are used between bulls and cows where available.
Parentage verification
Genomic tools to verify the paternity of calves are now available, with 100% certainty when more than 300 SNPs are genotyped on an animal and its sire (Hayes 2011). A calf can also be assigned to its dam provided the dam has also been genotyped. Using genotyping to resolve parentage may be particularly useful for herds with large numbers of calves being born over relatively short periods, where it is often logistically not possible to work out the sire and dam of a calf. The value of this is likely to be in reducing stress and reliance on staff around calving when a lot of calves are born over a short-period. Currently, Holstein Australia uses a service provided by the University of Queensland to verify parentage. The procedure uses 22 microsatellites and costs AU$36.30 (Matthew Shaffer, pers. comm., 2011). As with SNP data, both parents need to have microsatellites for full parentage verification.
Avoiding deleterious recessive alleles
Examples of single-gene diseases in dairy cattle include bovine leukocyte adhesion deficiency, which is a disease that affects the white blood cells and where affected calves are likely to suffer from bacterial infections. The mutation is believed to be a DNA-sequence change of A (adenine) to G (guanine) at position 383 of the CD18 gene (Shuster et al. 1992; Tajima et al. 1993). Complex vertebral malformation is a recessive deleterious mutation disease that results in stillborn or aborted fetuses. The mutation is in the SLC35A3 gene (Thomsen et al. 2006) at position 539 (guanine to adenine). Both of these diseases can be avoided by not mating known carriers to each other.
SNP data can be used to avoid mating known male carriers to females carrying SNP alleles that are in high linkage disequilibrium with genetic diseases that arise from single mutations, although it is better to have a genetic test for the actual mutation and these tests are available for both complex vertebral malformation and bovine leukocyte adhesion deficiency. Again, a computerised mating plan can be used to make sure that matings between carriers is avoided. In addition, genomic data should help us find future single-mutation genetic diseases that arise in dairy populations.
Conclusions
The benefits to a dairy herd of using semen from large groups of young bulls selected on the basis of GEBVs are large, with much higher rates of genetic gain being achievable, potentially doubling the net returns from genetic improvement. Conversely, the benefit of genotyping females is less certain at the commercial farm level. However, as genotyping costs reduce for low-density SNP chips, the prospect of using this technology on commercial dairy farms becomes increasingly attractive. Especially, if genotyping is used for several strategies, such as mating plans to control inbreeding, select the best replacements and parentage verification. The situation is different for pedigree breeders who may realise better sale prices from genotyped high genetic-merit heifers.
Acknowledgements
The Dairy Futures CRC (Melbourne, Australia) is gratefully acknowledged for funding this research. Michelle Axford (Australian Dairy Herd Improvement Scheme, Melbourne) is thanked for useful discussions in preparing this manuscript.
References
Australian Dairy Herd Improvement Scheme (2010) Ranges and means for bull ABV’s, August 2011. Available at http://www.adhis.com.au/v2/sitev2.nsf/(ContentByKey)/BreedingValuesBullABVs?open [Verified August 2010]Boichard D, Ducrocq V, Fritz S, Colleau JJ (2010) Where is dairy breeding going? A vision of the future. Interbull Bulletin 41, 63–68.
Cameron ND (1997) ‘Selection indices and prediction of genetic merit in animal breeding.’ (CAB International: Wallingford, UK)
de Roos APW, Schrooten C, Veerkamp RF, van Arendonk JAM (2011) Effects of genomic selection on genetic improvement, inbreeding, and merit of young versus proven bulls. Journal of Dairy Science 94, 1559–1567.
| Effects of genomic selection on genetic improvement, inbreeding, and merit of young versus proven bulls.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXmvFeit7k%3D&md5=8c3f3a6adf1cdc4396265db0f6da7d5bCAS |
Falconer DS, Mackay TFC (1996) ‘Introduction to quantitative genetics.’ Edn 4. (Longmans Green: Harlow, UK)
Habier D, Fernando RL, Dekkers JC (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177, 2389–2397.
Haile-Mariam ME, Bowman PJ, Goddard ME (2007) A practical approach for minimising inbreeding and maximising genetic gain in dairy cattle. Genetics, Selection, Evolution 39, 369–389.
| A practical approach for minimising inbreeding and maximising genetic gain in dairy cattle.Crossref | GoogleScholarGoogle Scholar |
Hayes BJ (2011) Technical note: efficient parentage assignment and pedigree reconstruction with dense single nucleotide polymorphism data. Journal of Dairy Science 94, 2114–2117.
| Technical note: efficient parentage assignment and pedigree reconstruction with dense single nucleotide polymorphism data.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXnvFChu7o%3D&md5=7bf654e1f2398730d72ae3a230ddd6bdCAS |
Hayes BJ, van der Werf JHJ, Pryce JE (2011) Economic benefit of genomic selection for residual feed intake (as a measure of feed conversion efficiency) in Australian dairy cattle. Recent Advances in Animal Nutrition 18, 31–36.
Kinghorn BP (1998) Managing genetic change under operational and cost constraints. In ‘36th national congress of the South African Association of Animal Science’, 5–8 April 1998. pp. 9–16. (University of Stellenbosch)
Legarra A, Aguillar I, Misztal I (2009) A relationship matrix including full pedigree and genomic information. Journal of Dairy Science 92, 4656–4663.
| A relationship matrix including full pedigree and genomic information.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVKqtr3E&md5=52dad86381f223dab5059bdac03e2e99CAS |
Man WYN (2004) Inbreeding in Australian Holstein Friesian cattle. PhD Thesis, University of Sydney.
Pryce JE, Daetwyler HD (2012) Designing dairy cattle breeding schemes under genomic selection: a review of international research. Animal Production Science 52, 107–114.
| Designing dairy cattle breeding schemes under genomic selection: a review of international research.Crossref | GoogleScholarGoogle Scholar |
Pryce JE, Arias J, Bowman PJ, Davis SR, Macdonald KA, Waghorn GC, Wales W, Williams YJ, Spelman RJ, Hayes BJ (2012a) Accuracy of genomic predictions of residual feed intake and 250 day bodyweight in growing heifers using 625 000 SNP markers. Journal of Dairy Science (in press).
Pryce JE, Hayes BJ, Goddard ME (2012b) Novel strategies to minimise progeny inbreeding while maximising genetic gain using genomic information. Journal of Dairy Science (in press).
Shuster DE, Kehrli ME, Ackermann MR, Gilbert RO (1992) Identification and prevalence of a genetic defect that causes leukocyte adhesion deficiency in Holstein cattle. Proceedings of the National Academy of Sciences, USA 89, 9225–9229.
| Identification and prevalence of a genetic defect that causes leukocyte adhesion deficiency in Holstein cattle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3sXkslSitLc%3D&md5=65de206e807b984f7b09bfa69c05af44CAS |
Smith LA, Cassell BG, Pearson RE (1998) The effects of inbreeding on lifetime performance of dairy cattle. Journal of Dairy Science 81, 2729–2737.
| The effects of inbreeding on lifetime performance of dairy cattle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXnt1Sms7Y%3D&md5=d4945357333202a4b18bac1298e774a4CAS |
Sonesson AK, Woolliams JA, Meuwissen THE (2010) Maximising genetic gain whilst controlling rates of genomic inbreeding using genomic optimum contribution selection. In ‘Proceedings of the 9th world congress on genetics applied to livestock production’.
Tajima M, Irie M, Kirisawa R, Hagiwara K, Kurosawa T, Takahashi K (1993) The detection of a mutation of CD18 gene in bovine leukocyte adhesion deficiency (BLAD). The Journal of Veterinary Medical Science 55, 145–146.
| The detection of a mutation of CD18 gene in bovine leukocyte adhesion deficiency (BLAD).Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK3s3htl2msw%3D%3D&md5=5ead4ddba34c8444c037a470b4e4e357CAS |
Thomsen B, Horn P, Panitz F, Bendixen E, Petersen AH, Holm L-E, Nielsen VH, Agerholm JS, Arnbjerg J, Bendixen C (2006) A missense mutation in the bovine SLC35A3 gene, encoding a UDP-N-acetylglucosamine transporter, causes complex vertebral malformation. Genome Research 16, 97–105.
| A missense mutation in the bovine SLC35A3 gene, encoding a UDP-N-acetylglucosamine transporter, causes complex vertebral malformation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhslChtLY%3D&md5=5f4123696d5fe851b56411ffc8aea8ecCAS |


