Genetic monitoring of southern hairy-nosed wombats over two decades reveals that individuals can live for at least 18 years in the same warrens
Faith M. Walker A B E , Jordyn R. Upton A B , Colin J. Sobek A B , David A. Taggart C and Matthew D. Gaughwin DA Bat Ecology and Genetics Laboratory, School of Forestry, Northern Arizona University, Flagstaff, AZ 86011, USA.
B Pathogen and Microbiome Institute, Northern Arizona University, Flagstaff, AZ 86011, USA.
C Department of Animal and Veterinary Science, University of Adelaide (Waite Campus), Glen Osmond, SA 5064, Australia.
D School of Public Health, University of Adelaide, Adelaide, SA 5005, Australia.
E Corresponding author. Email: faith.walker@nau.edu
Australian Mammalogy 43(1) 22-29 https://doi.org/10.1071/AM20012
Submitted: 4 February 2020 Accepted: 17 March 2020 Published: 28 April 2020
Journal Compilation © Australian Mammal Society 2021 Open Access CC BY-NC-ND
Abstract
Survival and growth rates are important demographic parameters to understand for long-term management of populations. Eighteen years have elapsed since non-invasive genetic methods were used to identify southern hairy-nosed wombats (Lasiorhinus latifrons), and determine space use and relatedness at Brookfield Conservation Park, South Australia. Because the species is long-lived (>30 years) and genetic methods can identify all or most wombats that use an area, it is possible to determine whether population size or warren use have changed and if any individuals are still alive. To this end, in April 2017 we collected hair from wombats from the same warrens as the earlier study using sticky tape suspended across burrows. We subjected DNA from selected hairs to 10 microsatellite loci and a Y-linked sex marker, and identified 76 wombats. Five wombats were detected 16–18 years before, and four of them were found in warrens that they had used previously. The number of tapes hit, wombats detected, and warrens used were greater than in April 2001 and similar to September 2001. This study illustrates that non-invasive sampling methods can be used to track free-ranging individuals in continuous habitat across decades, despite rapidly evolving genetic technology that can strand older datasets.
Additional keywords: legacy data, molecular analysis.
Introduction
Current rates of global environmental change are projected to influence the density and distribution of wildlife populations and to result in a rapid loss of biodiversity (Dirzo et al. 2014; Pimm et al. 2014; Newbold et al. 2016). The mechanisms and interactions of population size changes through time and density changes and movement in space can be elusive, and many approaches that are used to better understand these fundamental questions in ecology are cost prohibitive or invasive (Hebblewhite and Haydon 2010; Burgar et al. 2018). Non-invasive or minimally invasive genetic sampling, used alone or in concert with other methods, provides a powerful, efficient, and cost-effective approach to better understanding population processes (Walker et al. 2008a; Arandjelovic et al. 2010; Potter et al. 2012). Such methods commonly use remotely collected hair (Happe et al. 2020), faeces (Bradley et al. 2008), or feathers (Roy and Gregory 2019) as a DNA source.
Remote sampling and analytical methods for genetic monitoring were developed in the 1990s (Morin and Woodruff 1992; Taberlet et al. 1997; Sloane et al. 2000) and have been refined over the past 25 years (Carroll et al. 2018; Franklin et al. 2019). The early days of microsatellite DNA research used radioisotope incorporation during polymerase chain reaction (PCR) followed by autoradiography to visualise alleles, which was later replaced with capillary electrophoresis. More recently, high-throughput sequencing of microsatellite markers has shown great utility (De Barba et al. 2017). Each progression of technology leaves behind legacy datasets, as it is either not possible or not cost-effective to migrate ‘old’ data onto new platforms. Yet, for many studies using microsatellite DNA, incorporating these datasets into new genetic monitoring efforts will be extraordinarily valuable for insight into population changes over time.
One species with legacy microsatellite data is the southern hairy-nosed wombat (Lasiorhinus latifrons), a large (23–38 kg), long-lived (>30 years) herbivorous marsupial distributed in semiarid regions in southern Australia (Aitken 1971; Taggart and Temple-Smith 2008; Swinbourne et al. 2017). This species, which inhabits interconnected burrow systems called warrens, is problematic to study because it is nocturnal, shy, and difficult to capture. In the late 1990s and early 2000s, we deployed one of the largest hair trapping studies of the time with the aim of genetically identifying and tracking wombats via microsatellite DNA (Walker 2004). We found female-biased dispersal, male philopatry, and male kinship-based associations (Walker et al. 2008b), conservative space use with wombats using multiple warrens in a small area (<7.83 ha) (Walker et al. 2006), soil type as a driver of sociobiology (Walker et al. 2007), and increased population density and altered kin relationships in fragmented habitat (Walker et al. 2008a).
Our main study site for this work was at Brookfield Conservation Park in the Murraylands of South Australia, which has a large population of southern hairy-nosed wombats in relatively continuous habitat (McGregor and Wells 1998; Swinbourne et al. 2017). Since the time of our genetic studies, the Murraylands has experienced episodes of drought and is vulnerable to a changing climate (Marshall et al. 2018), and sarcoptic mange has cyclically impacted wombats (Ruykys et al. 2009). In order to better understand population size, survival, and warren use over a long period, we resampled our study area 18 years after the initial research. Specifically, we wished to determine: (1) how numbers and sex ratio of wombats compared with the earlier study, and (2) whether any of the same wombats from 1999–2001 could be detected and, if so, whether they used the same or different warrens. Such a study required navigating the disjunction between the original and currently used sequencing methods, while keeping laboratory costs low. This was achieved by applying a combination of recently developed (White et al. 2014) and original loci (Taylor et al. 1994; Alpers 1998; Beheregaray et al. 2000) along with positive control DNA of known genotype that had been used in our earlier work.
Materials and methods
This study was approved by the University of Adelaide Animal Ethics Committee (project no. S-2016-163) and South Australia’s Department of Environment, Water, and Natural Resources (permit no. Y26582-1). Sample export/import permits were acquired from the Australian Department of the Environment and Energy (permit no. PWS2018-AU-001967) and the US Fish and Wildlife Service (DEC control no. 2017141018). All genetic sampling and sequencing complied with relevant guidelines and regulations, and no wombats suffered injury or mortality as part of this study. Our non-invasive sample collection methods follow guidelines of the American Society of Mammalogists for animal care and use (Sikes 2016).
Study site and genetic sampling
Our field site was at Brookfield Conservation Park (−34.34618298, 139.48006), South Australia. This semiarid area has low, irregular rainfall (Fig. 1) and a large population of southern hairy-nosed wombats (McGregor and Wells 1998; Swinbourne et al. 2017). In April 2017, we collected hair samples from wombats in a 1.8 × 0.5 km area that was used in our hair-based genetic studies 16–18 years previously (Walker et al. 2006, 2007, 2008b). To do so, we suspended double-sided carpet tape (TESA Tape product 4970) between wooden garden stakes placed at either side of burrow entrances of 25 main study warrens. These were the same warrens, and were named the same, as in the previous studies, with the exception of warrens X and Alpha, which were abandoned, and Col, Suz, and PR, which were new and added to the study. GPS coordinates are provided in Supplementary Table S1. We taped all burrows at each warren except for those with no indication of recent use (spider webs or collapsed entrances). Tapes were suspended ~25 cm above the centre of burrow entrance floors. As wombats moved in and out of burrows during the night they left hairs behind on the tapes. Tapes were checked for hair each morning for five consecutive days, and were collected and replaced if hair was present. Five nights of taping detected all or nearly all wombats in the sampling area in our previous studies (Walker et al. 2006). We also taped seven peripheral warrens to the main study area, as was done in the early 2000s, in order to increase the number of wombats detected (Walker et al. 2006), and collected tapes with hair on Days 1 and 4. The total number of burrows taped across all warrens was 350, as in our previous work.

|
We performed same-day DNA extractions at the field camp using 5% Chelex solution (Sloane et al. 2000; Walker et al. 2006). Two DNA extractions were performed per hair tape, each from an individual hair follicle. We selected hair follicles based on size, and used hair from opposite ends of the tape when possible in order to maximise the chances that burrow sharing would be detected. DNA extractions were stored in a refrigerator for 10 days, after which they were transported to a −80°C freezer. For this study, we analysed the first four of the five days’ samples.
Microsatellite loci and PCR conditions
Our previous studies used radioisotope incorporation during PCR followed by autoradiography to visualise alleles, as it preceded capillary electrophoresis with fluorescently labelled primers that is now standard practice for microsatellite studies. Further, most of the early microsatellite loci developed for wombats (Taylor et al. 1994; Alpers 1998; Beheregaray et al. 2000) consist of dinucleotide repeats. This makes transferring to the newer technology problematic because they tend to suffer slippage during PCR, which appears as stutter peaks on chromatograms. Thus, in order to assign hairs to individual wombats and to recognise individuals from our previous studies, we used a combination of new tri- and tetranucleotide microsatellite loci developed by White et al. (2014) along with a subset of the original loci.
Loci for assigning hairs to wombats
Using the universal tail PCR labelling system of U’Ren et al. (2007), we tested the eight new primer sets of White et al. (2014), which were developed for northern hairy-nosed wombats (Lasiorhinus krefftii), the two older loci that they found to perform well with capillary electrophoresis (Ll68CA and Ll2), and the sexing marker we used in our earlier study (bSRY: Watson et al. 1998). We selected a panel of six loci that reliably amplified, were readily scored, and were polymorphic (6412, ELZRS, F3184, I85G2, DR470, Ll68CA). We PCR-amplified ELZRS, F3184, I85G2, DR470, 68CA, and bSRY as a multiplex or singleplexes (or in the case of bSRY, a duplex with DR470), and 6412 as a singleplex. The multiplex PCR contained 1X Mg-free PCR buffer (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA), 2 mm of MgCl2, 0.2 mm dNTPs, 0.2 µm fluorescently labeled forward primer, 0.2 µm unlabeled reverse primer, 0.4 U/µL Platinum Taq DNA polymerase (Invitrogen), 0.02 µg/µL Ultrapure non-acetylated Bovine Serum Albumin (BSA), and 5 µL of DNA template (mean 1.27 ng/µL) in a 15 µL reaction. PCRs for singleplexed loci contained 1X Mg-free PCR buffer (Invitrogen), 2 mm of MgCl2, 0.2 mm dNTPs, 0.05 µm unlabeled forward primer, 0.1 µm universal forward label (VIC, PET, 6-FAM, NED) (U’Ren et al. 2007), 0.1 µm unlabeled reverse primer, 0.1 U/µL Platinum Taq DNA polymerase (Invitrogen), 0.02 µg/µL BSA, and 5 µL of DNA template in a 15 µL reaction. PCRs for the sexing duplex of DR470 and bSRY contained the same quantities as the singleplexes aside from the primers and Taq polymerase: 0.1 µm of the smaller DR470 unlabeled forward primer, 0.2 µm universal forward label (PET) (U’Ren et al. 2007), 0.2 µm DR470 unlabeled reverse primer, 0.4 µm of the larger bSRY unlabeled forward primer, 0.8 µm universal forward label (6-FAM) (U’Ren et al. 2007), 0.8 µm bSRY unlabeled reverse primer, and 0.2 U/µL Platinum Taq DNA polymerase (Invitrogen). We performed thermal cycling on an Applied Biosystems SimpliAmp Thermal Cycler. Cycling conditions for all PCRs began with a denaturation step of 94°C for 5 min, followed by 45 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, then concluded with a final extension step of 72°C for 5 min.
After PCR amplification, product was diluted as follows before fragment analysis on an Applied Biosystems 3130 Genetic Analyzer: 1 : 50 for singleplexes, 1 : 100 for the duplex, and no dilution for the multiplex. We used GeneMarker software to score alleles, and program GeneAlEx 6.5 (Peakall and Smouse 2012) to match genotypes of wombats. DNA extractions with 100% allelic sharing across loci were treated as belonging to the same wombat. The power of these six loci for differentiating between individuals was high; the probability of two randomly selected wombats sharing a multilocus genotype was 4.6 × 10−8 (Waits et al. 2001).
Loci for identifying wombats from the early 2000s
To match wombats detected in 2017 with those from our 1999–2001 studies, we applied four loci (71CA, Lk23, Lla109, Ll2: Taylor et al. 1994; Alpers 1998; Beheregaray et al. 2000) to a representative sample from each wombat. These loci, along with Lla68CA above, were PCR amplified with positive control DNA of 10 Brookfield Conservation Park wombats from Taylor et al. (1994), as was done for our earlier studies (e.g. Walker et al. 2006). PCR reagents and conditions were as above for singleplexes. Any genotype with 100% allelic matching (at all five loci plus the sexing marker) with an individual from the 1999–2001 studies was considered to be the same individual. We repeated PCRs for all questionable matches using an additional representative sample from each wombat. The probability of identity for these five loci was 1.2 × 10−5.
Quality control
We used a suite of quality control measures to reduce and detect genotyping errors (Bonin et al. 2004; Waits and Paetkau 2005). As ~3671 PCR reactions were performed for this study, 11 single-locus genotypes can be expected to be false based on the 0.3% error rate of Sloane et al. (2000), who were the first to survey a hairy-nosed wombat population with these methods. Genotyping both hair samples from each tape assisted with scoring accuracy, since in our previous work 90.5% of these samples were from the same wombat. Further, we distinguished scoring errors, null alleles, and PCR artifacts by repeating PCRs for samples that were poorly amplifying (Taq polymerase increased to 0.2 U/µL), had novel alleles, and for multilocus genotypes differing at three or fewer loci. Samples that failed to amplify at four or more loci or that had high homozygosity were removed from the dataset. To facilitate scoring of questionable individuals, PCRs were repeated on the same PCR plate, placed on the Genetic Analyzer together, and scored side-by-side. Additionally, any putative individual wombats that were genetically similar and geographically more distant than wombats were detected to move in Walker et al. (2006) were also reamplified for all loci and compared side-by-side. As an additional conservative measure, after matching genotypes with wombats from the early 2000s, we included only unique genotypes that were detected at least twice, either on the same tape or different tapes.
Locus behaviour
We tested for deviations from Hardy–Weinberg equilibrium and for linkage disequilibrium between pairs of loci with program GENEPOP 4.7.3 (Rousset 2008). We tested for heterozygote or homozygote deficits in addition to the non-directional Hardy–Weinberg test, and we used a sequential Bonferroni procedure (Rice 1989) to adjust for multiple tests where required. We used program MICRO-CHECKER (Van Oosterhout et al. 2004) to test for the possibility of scoring errors, allelic dropout, and null alleles.
Results
Rate of attrition and samples analysed
Over the five sampling nights and 350 taped burrows, wombats had 1750 opportunities to donate hair to this study, 460 of which were taken (e.g. Fig. 2). Most tapes (95%) that were ‘hit’ by wombats had hair follicles, which indicates that tapes were set at an appropriate height to pluck hairs and that tape condition did not deteriorate between hits due to environmental variables (e.g. dew, dust). Of the hit tapes, 373 were from the main study warrens, which is similar to the number of hits in September 2001 (n = 370) but greater than in April 2001 (n = 172) (Walker et al. 2006). We analysed 663 DNA extractions (Days 1–4) of the 810 total extractions from five days of sampling. In our five sampling periods between 1999 and 2001, an average of three new wombats were detected on the fifth day (Walker et al. 2006), which suggests that analysis of the first four days allows detection of most wombats that use the area.

|
Wombat detection success and locus behaviour
We identified 76 distinct multilocus genotypes (Supplementary Table S2): 63 in the main study warrens and 13 in peripheral warrens. More females than males were detected (40 females and 36 males) (Fig. 3), but this difference was not statistically significant (P = 0.37; exact binomial test). The number of new wombats detected per night in the main study area declined to five and eight on Nights 3 and 4, respectively (Fig. 4). Individual wombats were detected up to 15 times (M = 4.79, s.d. = 2.92; Supplementary Table S2). There were 18 instances of within-night burrow sharing (two different wombats detected on the same hair tape), and 101 cases of both hair samples taken from a tape belonging to the same wombat. In 2001 we detected eight instances of within-night burrow sharing in April and 11 instances in September.

|

|
Mean expected heterozygosity for the 10 loci was 0.61, and mean number of alleles per locus was 5.4. All loci adhered to Hardy–Weinberg expectations for the probability test as well as tests for heterozygote or homozygote deficits. Likewise, no locus pairs were in linkage disequilibrium, and there was no evidence of null alleles for any locus.
Warren and burrow use
Warrens that were heavily used during this study were also heavily used in the early 2000s as well as when the authors first surveyed the study area in 1994 (Walker et al. 2006). In this study, two small warrens were new, and two small warrens had been abandoned. Wombats were detected to use 1–3 warrens (mean = 1.32, s.d. = 0.53). Up to 10 individuals used a single warren (mean = 4.11, s.d. = 2.63), and warrens with high numbers of individuals in 2000 and 2001 had similar numbers in this study (Table 1). In April 2017 our main study warrens were used by more wombats than in April 2001 and fewer wombats than in September 2001 (standardised warrens used and the number of sampling nights; Wilcoxon signed-ranks test: Z = 26, P < 0.021, Z = −28.5, P < 0.047, respectively). The number of wombats per active burrow, where an active burrow was defined as wombat hair on a tape, averaged 0.63 across all main study warrens (Table 1).
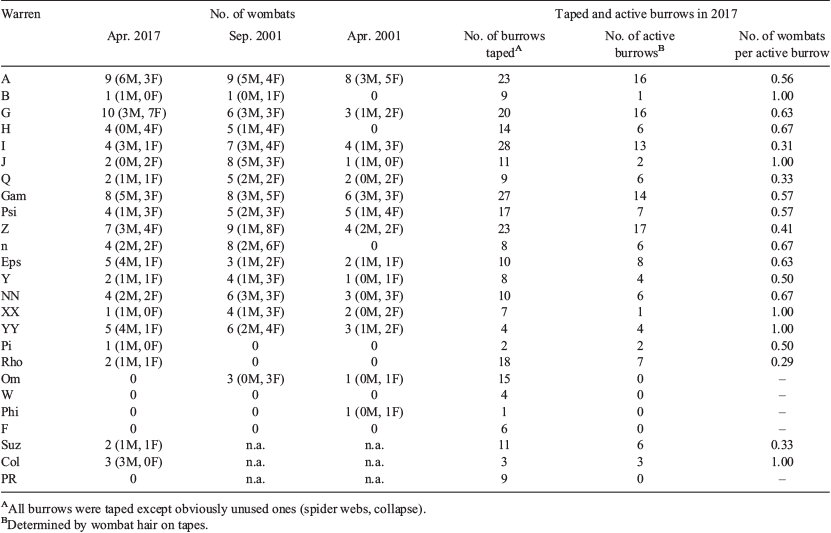
|
Wombats detected 16–18 years previously
Five wombats (6.6%) that were detected during five sampling periods in 1999–2001 were also detected during this study. Four of the five wombats were detected in a warren used by that individual 16–18 years previously. The exception was a female found in a warren 87 m from the cluster of warrens she was detected to use in 2001 (Table 2). There was a female sex bias in wombats detected over this timespan (four females, one male).
Discussion
This work is comparable to 347 captures of 76 individuals in a capture–mark–recapture study, and is the first study we are aware of to track the same individuals of a wild population occupying continuous habitat through nearly 20 years by using non-invasively collected samples. The five wombats that we detected again in this study were at least 16–18 years old, and likely older, since they were large enough to leave hair on a tape at the time of the original study. Species of this genus are known to be long-lived: two wild adult female southern hairy-nosed wombats in the Murraylands were recaptured 18 years after first capture (D. Taggart, pers. comm.), a captive female southern hairy-nosed wombat lived to be 33 years old (B. Cleaver, pers. comm.), and three wild northern hairy-nosed wombats (one male and two females) were determined to be at least 28 years old (Taylor 2012). Warren fidelity, which was documented previously (Gaughwin 1981; Walker et al. 2006), was particularly striking in that four of the five wombats were found in a warren they had used in 1999–2001. One of the long-lived wombats (female W30) was using the same burrow in the same warren as she used in 2001; in general, it was not possible to number the burrows to match the earlier study, but in some cases the burrows were recognisable.
The number of wombats detected and number of hit tapes was similar to those detected in September 2001, when the greatest number of wombats was detected in our earlier study. The numbers in April 2017, particularly given it was the non-breeding season, suggest that wombat population size in this area is similar to that in 2001, despite periods of drought in 2002 and 2008 (Fig. 1). There was higher than average rainfall in 2010 and 2011. Southern hairy-nosed wombat reproduction is linked to the plant growth index between July and September, which is correlated with rainfall (Gaughwin et al. 1998). Oogenesis and spermatogenesis are reduced or terminated when rainfall is low during these months (Gaughwin 1981). The sex ratio in this study was not significantly biased toward females, unlike in our previous work (Walker et al. 2006). Interestingly, there was a female sex bias in ‘old’ wombats: four of the five were female, and five of the six other long-lived hairy-nosed wombats (mentioned above) were also female. This may be related to greater mortality in male mammals, especially polygynous mammals (Clutton-Brock and Isvaran 2007).
We successfully migrated our early wombat data to what is now a traditional sequencing platform, and were able to do so cheaply (approximately US$14 500 for the 663 samples in this study, or US$22 per sample). We did not move our wombat data into the genomics era by high-throughput sequencing of microsatellite loci (De Barba et al. 2017) or SNP analysis (Kraus et al. 2015) due to the time and costs involved, but this would be a valuable future direction for hairy-nosed wombat research. For future hair censuses that use capillary electrophoresis, it is now possible to apply only the subset of loci from White et al. (2014), since they are discriminatory to the individual level even for our ‘old’ wombats. In order to keep hair censuses inexpensive, we recommend using the universal tail PCR labelling system of U’Ren et al. (2007) (the length of which should be subtracted from allele sizes in order to match with earlier work) and running singleplexes instead of multiplexing using fluorescently labelled primers. This is because we found that multiplexing had a high failure rate and required multiple PCR amplifications, and hence did not save time and was more costly overall. Our next step in analysis of these data is to examine parentage, pairwise relatedness, and relatedness of burrow sharers.
Conclusion
Genetic monitoring to estimate population demographics and track genetic diversity will likely become increasingly more important as wildlife populations decline due to anthropogenic changes. Tying in legacy microsatellite datasets has tremendous value for a long-term perspective of these parameters, as well as reidentifying individuals of long-lived species. Here, we show that it is possible to migrate between the earliest sequencing platform (involving radioisotopes) and a commonly used platform (capillary electrophoresis) in order to identify wombats that were at least 18 years old. The warren fidelity of these wombats over this time highlights the lack of vagility and hence vulnerability of this iconic species to a warming climate (Marshall et al. 2018). There is potential value in monitoring populations regularly to better estimate life-history parameters and to detect any effects of change in climate or disease. We recommend that studies using microsatellite markers bank DNA from each individual as well as high-quality DNA from a set of positive controls, so that high-throughput sequencing or other yet-to-be-developed methods can be used in the future.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
Funding was provided by the Australian Geographic Society, Schultz Foundation, Lirabenda Endowment Fund of the Field Naturalists Society of South Australia, and Northern Arizona University’s Interns2Scholars program. Thanks to Team Wombat’s J. Gladden, W. Noble, S. Romero, A. Gaughwin, and K. Judd for collecting hair samples, Bunnings Warehouse Mile End South Australia for donating garden stakes, and A. Taylor for positive control DNA. Thanks also to Conservation Volunteers Australia, South Australia, and to the Friends of Brookfield for providing logistical support.
References
Aitken, P. F. (1971). The distribution of the southern hairy nosed wombat (Lasiorhinus latifrons Owen). Part 1: Yorke Peninsula, Eyre Peninsula, The Gawler Ranges and Lake Harris. South Australian Naturalist 43, 93–104.Alpers, D. L. (1998). Genetic population structure of the southern hairy-nosed wombat (Lasiorhinus latifrons). Ph.D. Thesis, University of New South Wales, Sydney.
Arandjelovic, M., Head, J., Kuhl, H., Boesch, C., Robbins, M. M., Maisels, F., and Vigilant, L. (2010). Effective non-invasive genetic monitoring of multiple wild western gorilla groups. Biological Conservation 143, 1780–1791.
| Effective non-invasive genetic monitoring of multiple wild western gorilla groups.Crossref | GoogleScholarGoogle Scholar |
Beheregaray, L. B., Sunnucks, P., Alpers, D. L., Banks, S. C., and Taylor, A. C. (2000). A set of microsatellite loci for the hairy-nosed wombats (Lasiorhinus krefftii and L. latifrons). Conservation Genetics 1, 89–92.
| A set of microsatellite loci for the hairy-nosed wombats (Lasiorhinus krefftii and L. latifrons).Crossref | GoogleScholarGoogle Scholar |
Bonin, A., Bellemain, E., Bronken Eidesen, P., Pompanon, F., Brochmann, C., and Taberlet, P. (2004). How to track and assess genotyping errors in population genetics studies. Molecular Ecology 13, 3261–3273.
| How to track and assess genotyping errors in population genetics studies.Crossref | GoogleScholarGoogle Scholar | 15487987PubMed |
Bradley, B. J., Doran-Sheehy, D. M., and Vigilant, L. (2008). Genetic identification of elusive animals: re-evaluating tracking and nesting data for wild western gorillas. Journal of Zoology 275, 333–340.
| Genetic identification of elusive animals: re-evaluating tracking and nesting data for wild western gorillas.Crossref | GoogleScholarGoogle Scholar |
Burgar, J. M., Stewart, F. E. C., Volpe, J. P., Fisher, J. T., and Burton, A. C. (2018). Estimating density for species conservation: comparing camera trap spatial count models to genetic spatial capture–recapture models. Global Ecology and Conservation 15, e00411.
| Estimating density for species conservation: comparing camera trap spatial count models to genetic spatial capture–recapture models.Crossref | GoogleScholarGoogle Scholar |
Carroll, E. L., Bruford, M. W., DeWoody, J. A., Leroy, G., Strand, A., Waits, L., and Wang, J. (2018). Genetic and genomic monitoring with minimally invasive sampling methods. Evolutionary Applications 11, 1094–1119.
| Genetic and genomic monitoring with minimally invasive sampling methods.Crossref | GoogleScholarGoogle Scholar | 30026800PubMed |
Clutton-Brock, T. H., and Isvaran, K. (2007). Sex differences in ageing in natural populations of vertebrates. Proceedings of the Royal Society of London. Series B, Biological Sciences 274, 3097–3104.
| Sex differences in ageing in natural populations of vertebrates.Crossref | GoogleScholarGoogle Scholar |
De Barba, M., Miquel, C., Lobréaux, S., Quenette, P. Y., Swenson, J. E., and Taberlet, P. (2017). High-throughput microsatellite genotyping in ecology: improved accuracy, efficiency, standardization and success with low-quantity and degraded DNA. Molecular Ecology Resources 17, 492–507.
| High-throughput microsatellite genotyping in ecology: improved accuracy, efficiency, standardization and success with low-quantity and degraded DNA.Crossref | GoogleScholarGoogle Scholar | 27505280PubMed |
Dirzo, R., Young, H. S., Galetti, M., Ceballos, G., Isaac, N. J. B., and Collen, B. (2014). Defaunation in the Anthropocene. Science 345, 401–406.
| Defaunation in the Anthropocene.Crossref | GoogleScholarGoogle Scholar | 25061202PubMed |
Franklin, T. W., McKelvey, K. S., Golding, J. D., Mason, D. H., Dysthe, J. C., Pilgrim, K. L., Squires, J. R., Aubry, K. B., Long, R. A., Greaves, S. E., Raley, C. M., Jackson, S., MacKay, P., Lisbon, J., Sauder, J. D., Pruss, M. T., Heffington, D., and Schwartz, M. K. (2019). Using environmental DNA methods to improve winter surveys for rare carnivores: DNA from snow and improved noninvasive techniques. Biological Conservation 229, 50–58.
| Using environmental DNA methods to improve winter surveys for rare carnivores: DNA from snow and improved noninvasive techniques.Crossref | GoogleScholarGoogle Scholar |
Gaughwin, M. D. (1981). Socio-ecology of the hairy-nosed wombat (Lasiorhinus latifrons) in the Blanche Town region of South Australia. Ph.D. Thesis, University of Adelaide.
Gaughwin, M. D., Breed, W. G., and Wells, R. T. (1998). Seasonal reproduction in a population of southern hairy-nosed wombats Lasiorhinus latifrons in the Blanchetown region of South Australia. In ‘Wombats’. (Eds R. T. Wells, and P. A. Pridmore.) pp. 109–112. (Surrey Beatty: Sydney.)
Happe, P. J., Jenkins, K. J., McCaffery, R. M., Lewis, J. C., Pilgrim, K. L., and Schwartz, M. K. (2020). Occupancy patterns in a reintroduced fisher population during reestablishment. The Journal of Wildlife Management 84, 344–358.
| Occupancy patterns in a reintroduced fisher population during reestablishment.Crossref | GoogleScholarGoogle Scholar |
Hebblewhite, M., and Haydon, D. T. (2010). Distinguishing technology from biology: a critical review of the use of GPS telemetry data in ecology. Proceedings of the Royal Society of London. Series B, Biological Sciences 365, 2303–2312.
| Distinguishing technology from biology: a critical review of the use of GPS telemetry data in ecology.Crossref | GoogleScholarGoogle Scholar |
Kraus, R. H. S., vonHoldt, B., Cocchiararo, B., Harms, V., Bayerl, H., Kühn, R., Förster, D. W., Fickel, J., Roos, C., and Nowak, C. (2015). A single-nucleotide polymorphism-based approach for rapid and cost-effective genetic wolf monitoring in Europe based on noninvasively collected samples. Molecular Ecology Resources 15, 295–305.
| A single-nucleotide polymorphism-based approach for rapid and cost-effective genetic wolf monitoring in Europe based on noninvasively collected samples.Crossref | GoogleScholarGoogle Scholar |
Marshall, V. M., Taggart, D. A., and Ostendorf, B. (2018). Scale-dependent habitat analysis and implications for climate change risk for the southern hairy-nosed wombat. Australian Mammalogy 40, 162–172.
| Scale-dependent habitat analysis and implications for climate change risk for the southern hairy-nosed wombat.Crossref | GoogleScholarGoogle Scholar |
McGregor, F., and Wells, R. T. (1998). Population status of the southern hairy-nosed wombat Lasiorhinus latifrons in the Murraylands of South Australia. In ‘Wombats’. (Eds R. T. Wells, and P. A. Pridmore.) pp. 228–242. (Surrey Beatty: Sydney.)
Morin, P., and Woodruff, D. (1992). Paternity exclusion using multiple hypervariable microsatellite loci amplified from nuclear DNA of hair cells. In ‘Paternity in Primates: Genetic Tests and Theories’. (Eds R. Martin, A. Dixson, and E. Wickings.) pp. 63–81. (Karger: Basel.)
Newbold, T., Hudson, L. N., Arnell, A. P., Contu, S., De Palma, A., Ferrier, S., Hill, S. L. L., Hoskins, A. J., Lysenko, I., Phillips, H. R. P., Burton, V. J., Chng, C. W. T., Emerson, S., Gao, D., Pask-Hale, G., Hutton, J., Jung, M., Sanchez-Ortiz, K., Simmons, B. I., Whitmee, S., Zhang, H., Scharlemann, J. P. W., and Purvis, A. (2016). Has land use pushed terrestrial biodiversity beyond the planetary boundary? A global assessment. Science 353, 288–291.
| Has land use pushed terrestrial biodiversity beyond the planetary boundary? A global assessment.Crossref | GoogleScholarGoogle Scholar | 27418509PubMed |
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28, 2537–2539.
| GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update.Crossref | GoogleScholarGoogle Scholar | 22820204PubMed |
Pimm, S. L., Jenkins, C. N., Abell, R., Brooks, T. M., Gittleman, J. L., Joppa, L. N., Raven, P. H., Roberts, C. M., and Sexton, J. O. (2014). The biodiversity of species and their rates of extinction, distribution, and protection. Science 344, 1246752.
| The biodiversity of species and their rates of extinction, distribution, and protection.Crossref | GoogleScholarGoogle Scholar | 24876501PubMed |
Potter, S., Eldridge, M. D. B., Cooper, S. J. B., Paplinska, J. Z., and Taggart, D. A. (2012). Habitat connectivity, more than species’ biology, influences genetic differentiation in a habitat specialist, the short-eared rock-wallaby (Petrogale brachyotis). Conservation Genetics 13, 937–952.
| Habitat connectivity, more than species’ biology, influences genetic differentiation in a habitat specialist, the short-eared rock-wallaby (Petrogale brachyotis).Crossref | GoogleScholarGoogle Scholar |
Rice, W. R. (1989). Analyzing tables of statistical tests. Evolution 43, 223–225.
| Analyzing tables of statistical tests.Crossref | GoogleScholarGoogle Scholar | 28568501PubMed |
Rousset, F. (2008). genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources 8, 103–106.
| genepop’007: a complete re-implementation of the genepop software for Windows and Linux.Crossref | GoogleScholarGoogle Scholar | 21585727PubMed |
Roy, C. L., and Gregory, A. J. (2019). Landscape and population genetics reveal long distance sharp-tailed grouse (Tympanuchus phasianellus) movements and a recent bottleneck in Minnesota. Conservation Genetics 20, 259–273.
| Landscape and population genetics reveal long distance sharp-tailed grouse (Tympanuchus phasianellus) movements and a recent bottleneck in Minnesota.Crossref | GoogleScholarGoogle Scholar |
Ruykys, L., Taggart, D. A., Breed, W. G., and Schultz, D. (2009). Sarcoptic mange in southern hairy-nosed wombats (Lasiorhinus latifrons): distribution and prevalence in the Murraylands of South Australia. Australian Journal of Zoology 57, 129–138.
| Sarcoptic mange in southern hairy-nosed wombats (Lasiorhinus latifrons): distribution and prevalence in the Murraylands of South Australia.Crossref | GoogleScholarGoogle Scholar |
Sikes, R. S. (2016). Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. Journal of Mammalogy 97, 663–688.
| Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education.Crossref | GoogleScholarGoogle Scholar | 29692469PubMed |
Sloane, M. A., Sunnucks, P., Alpers, D., Beheregaray, L. B., and Taylor, A. C. (2000). Highly reliable genetic identification of individual northern hairy-nosed wombats from single remotely collected hairs: a feasible censusing method. Molecular Ecology 9, 1233–1240.
| Highly reliable genetic identification of individual northern hairy-nosed wombats from single remotely collected hairs: a feasible censusing method.Crossref | GoogleScholarGoogle Scholar | 10972763PubMed |
Swinbourne, M., Taggart, D., Peacock, D., and Ostendorf, B. (2017). Historical changes in the distribution of hairy-nosed wombats (Lasiorhinus spp.): a review. Australian Mammalogy 39, 1–16.
| Historical changes in the distribution of hairy-nosed wombats (Lasiorhinus spp.): a review.Crossref | GoogleScholarGoogle Scholar |
Taberlet, P., Camarra, J. J., Griffin, S., Uhres, E., Hanotte, O., Waits, L. P., Dubois-Paganon, C., Burke, T., and Bouvet, J. (1997). Noninvasive genetic tracking of the endangered Pyrenean brown bear population. Molecular Ecology 6, 869–876.
| Noninvasive genetic tracking of the endangered Pyrenean brown bear population.Crossref | GoogleScholarGoogle Scholar | 9301075PubMed |
Taggart, D. A., and Temple-Smith, P. D. M. (2008). Southern hairy-nosed wombat. In ‘The Mammals of Australia’. (Eds S. Van Dyck, and R. Strahan.) pp. 204–206. (New Holland Publishers: Sydney.)
Taylor, A. C. (2012). Northern hairy-nosed wombat hair census final report. Internal report to the Queensland Department of Environment and Heritage Protection. Monash University, Melbourne.
Taylor, A. C., Sherwin, W. B., and Wayne, R. K. (1994). Genetic variation of microsatellite loci in a bottlenecked species: the northern hairy-nosed wombat Lasiorhinus krefftii. Molecular Ecology 3, 277–290.
| Genetic variation of microsatellite loci in a bottlenecked species: the northern hairy-nosed wombat Lasiorhinus krefftii.Crossref | GoogleScholarGoogle Scholar | 7921355PubMed |
U’Ren, J. M., Schupp, J. M., Pearson, T., Hornstra, H., Clark Friedman, C. L., Smith, K. L., Leadem Daugherty, R. R., Rhoton, S. D., Leadem, B., Georgia, S., Cardon, M., Huynh, L. Y., DeShazer, D., Harvey, S. P., Robison, R., Gal, D., Mayo, M. J., Wagner, D., Currie, B. J., and Keim, P. (2007). Tandem repeat regions within the Burkholderia pseudomallei genome and their application for high resolution genotyping. BMC Microbiology 7, 23–43.
| Tandem repeat regions within the Burkholderia pseudomallei genome and their application for high resolution genotyping.Crossref | GoogleScholarGoogle Scholar | 17397553PubMed |
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4, 535–538.
| micro-checker: software for identifying and correcting genotyping errors in microsatellite data.Crossref | GoogleScholarGoogle Scholar |
Waits, L. P., and Paetkau, D. (2005). Noninvasive genetic sampling tools for wildlife biologists: a review of applications and recommendations for accurate data collection. Journal of Wildlife Management 69, 1419–1433.
| Noninvasive genetic sampling tools for wildlife biologists: a review of applications and recommendations for accurate data collection.Crossref | GoogleScholarGoogle Scholar |
Waits, L. P., Luikart, G., and Taberlet, P. (2001). Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Molecular Ecology 10, 249–256.
| Estimating the probability of identity among genotypes in natural populations: cautions and guidelines.Crossref | GoogleScholarGoogle Scholar | 11251803PubMed |
Walker, F. M. (2004). Sociobiology inferred from relatedness structure via remotely-collected DNA in southern hairy-nosed wombats, Lasiorhinus latifrons. Ph.D. Thesis, Monash University, Melbourne.
Walker, F. M., Sunnucks, P., and Taylor, A. C. (2006). Genotyping of “captured” hairs reveals burrow-use and ranging behavior of southern hairy-nosed wombats. Journal of Mammalogy 87, 690–699.
| Genotyping of “captured” hairs reveals burrow-use and ranging behavior of southern hairy-nosed wombats.Crossref | GoogleScholarGoogle Scholar |
Walker, F. M., Taylor, A. C., and Sunnucks, P. (2007). Does soil type drive social organization in southern hairy-nosed wombats? Molecular Ecology 16, 199–208.
| Does soil type drive social organization in southern hairy-nosed wombats?Crossref | GoogleScholarGoogle Scholar | 17181731PubMed |
Walker, F. M., Sunnucks, P., and Taylor, A. C. (2008a). Evidence for habitat fragmentation altering within-population processes in wombats. Molecular Ecology 17, 1674–1684.
| Evidence for habitat fragmentation altering within-population processes in wombats.Crossref | GoogleScholarGoogle Scholar | 18386311PubMed |
Walker, F. M., Taylor, A. C., and Sunnucks, P. (2008b). Female dispersal and male kinship-based association in southern hairy-nosed wombats (Lasiorhinus latifrons). Molecular Ecology 17, 1361–1374.
| Female dispersal and male kinship-based association in southern hairy-nosed wombats (Lasiorhinus latifrons).Crossref | GoogleScholarGoogle Scholar | 18302694PubMed |
Watson, C. M., Margan, S. H., and Johnston, P. G. (1998). Sex-chromosome elimination in the bandicoot Isoodon macrourus using Y-linked markers. Cytogenetic and Genome Research 81, 54–59.
| Sex-chromosome elimination in the bandicoot Isoodon macrourus using Y-linked markers.Crossref | GoogleScholarGoogle Scholar |
White, L. C., Horsup, A., Taylor, A. C., and Austin, J. J. (2014). Improving genetic monitoring of the northern hairy-nosed wombat (Lasiorhinus krefftii). Australian Journal of Zoology 62, 246–250.
| Improving genetic monitoring of the northern hairy-nosed wombat (Lasiorhinus krefftii).Crossref | GoogleScholarGoogle Scholar |



