Genotyping of rams based on melatonin receptor 1A gene polymorphisms: a tool in sire selection?
Victoria Peña-Delgado A , Agustí Noya
A , Agustí Noya  A , Melissa Carvajal-Serna
A , Melissa Carvajal-Serna  A , Francisco Canto
A , Francisco Canto  A , María Carmen Sánchez B , Eva Letosa B , Antonio Vicente B , Ignacio Morato B , Ángel Macías C , José Alfonso Abecia
A , María Carmen Sánchez B , Eva Letosa B , Antonio Vicente B , Ignacio Morato B , Ángel Macías C , José Alfonso Abecia  A , Adriana Casao
A , Adriana Casao  A and Rosaura Pérez-Pe
A and Rosaura Pérez-Pe  A *
A *
A
B
C
Abstract
Several polymorphisms in the melatonin receptor 1A gene (MTNR1A) have been related to reproductive performance in ovine.
To investigate the effect of the RsaI and MnlI polymorphisms on ram seminal quality.
Eighteen Rasa Aragonesa rams were genotyped for the RsaI (C/C, C/T, T/T) and MnlI (G/G, G/A, A/A) allelic variants of the MTNR1A gene. Individual ejaculates were analysed once a month throughout the whole year. Sperm motility, morphology, membrane integrity, levels of reactive oxygen species (ROS), phosphatidylserine (PS) inversion, DNA fragmentation and capacitation status were assessed. The effect of the season and polymorphisms on seminal quality was evaluated by mixed ANOVA.
Both polymorphisms had an effect on membrane integrity and viable spermatozoa with low levels of ROS and without PS translocation, and RsaI also on motile and DNA-intact spermatozoa. An interaction between both polymorphisms was found, pointing to a negative effect on seminal quality of carrying the T or A allele in homozygosity. Differences were higher in the reproductive than in the non-reproductive season.
Mutations substituting C by T and G by A at RsaI and MnlI polymorphic sites, respectively, in the MTNR1A gene in rams could decrease the seminal quality.
Genotyping of rams based on melatonin receptor 1A could be a powerful tool in sire selection.
Keywords: genotyping, MTNR1A gene, melatonin receptor, MnlI, polymorphisms, ram, RsaI, seasonality, sire selection, sperm quality.
Introduction
The ovine species has a very marked seasonal reproduction when located in temperate regions, with a breeding season from early autumn to late winter, and a non-reproductive season from late winter to late summer (Delgadillo et al. 2020). This reproductive seasonality is governed by photoperiod and melatonin secretion (Chemineau et al. 2008). This hormone, involved in multiple physiological processes, can exert its actions directly, by crossing the plasma membrane, or by binding to its specific receptors (Zhao et al. 2019). In mammals, two high-affinity melatonin receptors have been identified (Dubocovich and Markowska 2005). These receptors, called MT1 and MT2, belong to the guanine nucleotide-binding protein (G-protein)-coupled receptors and share a common structure consisting of seven transmembrane domains linked by alternating intracellular and extracellular loops (Dubocovich et al. 2003). The MT1 receptor, encoded by the MTNR1A (melatonin receptor 1A) gene, appears to be the one involved in the control of seasonality in several species, including the sheep (Weaver et al. 1996; Martínez-Royo et al. 2012).
The MTNR1A gene is located on chromosome 26 in the ovine species, and its structure consists of Exon I, which encodes the first transmembrane domain and the first intracellular loop; an intron of 8 kb length; and Exon II, which encodes the remaining part of the receptor (Barrett et al. 1997). Several polymorphic sites related to certain reproductive traits have been found in Exon II (Pelletier et al. 2000; Notter et al. 2003). Among them, two single nucleotide polymorphisms (SNPs) classically called RsaI (g.17355458 C > T) and MnlI (g. 17355452 G > A) (Messer et al. 1997) have been associated with the reproductive seasonality and performance in ewes of different Mediterranean breeds (Carcangiu et al. 2011; Luridiana et al. 2020; Starič et al. 2020; Cosso et al. 2021; Arjoune et al. 2023), including Rasa Aragonesa (Martínez-Royo et al. 2012; Calvo et al. 2018). Rasa Aragonesa is a local Spanish breed with a short seasonal anoestrus (Forcada et al. 1992). However, in contrast to ewes, there are hardly any studies on the effect of MTNR1A polymorphisms in rams. Previous research by our group has shown that the T/T or the G/G genotype for the RsaI and MnlI polymorphisms, respectively, has a positive effect on the sexual performance of young and adult Rasa Aragonesa rams (Abecia et al. 2020). Specifically, autumn-born ram lambs that carried the T/T or the G/G genotype were considerably younger at first mounting, while adult T/T and G/G rams exhibited a more intense reproductive behaviour than rams without these genotypes (Abecia et al. 2020) although it was not associated with differences in social dominance (Abecia et al. 2022). Moreover, the polymorphisms of the MTNR1A gene of rams seem to affect field fertility after artificial insemination (AI) depending on season (Abecia et al. 2023), although the reason for these differences remains unknown.
Melatonin can influence ovine reproduction through mechanisms other than photoperiod translation. It can also be synthesised in ram testes (Martínez-Marcos et al. 2019) and is present in seminal plasma (Casao et al. 2010). Moreover, both melatonin receptors, MT1 and MT2, have been found in the ram reproductive tract (González-Arto et al. 2017) and the plasma membrane of ovine spermatozoa (Casao et al. 2012; González-Arto et al. 2016). But until now, it is unknown whether carrying different genotypes for MT1 in rams affects the functionality of that receptor and therefore the interaction with melatonin, both at the level of the tissues of the male reproductive tract and the sperm level.
Therefore, the aim of this study was to examine the effect of the RsaI and MnlI MTNR1A gene polymorphisms on ram seminal quality during the reproductive and non-reproductive seasons.
Materials and methods
Unless otherwise stated, all reagents were purchased from Sigma-Aldrich (St Louis, MO, USA).
Animals
Eighteen Rasa Aragonesa rams of proven fertility were used in this study. The rams belonged to the Asociación Nacional de Criadores de Ganado Ovino Selecto de Raza Rasa Aragonesa (ANGRA), were housed in the facilities of the Centro de Selección y Reproducción Animal (CENSYRA, Centro de Transferencia Agroalimentaria, Movera, Zaragoza, Spain) and were used routinely for field AI in commercial farms.
All animal procedures were performed in accordance with the Spanish Animal Protection Regulation 348/2000 which conforms to European Union Regulation 98/58/CE. Approval from the Ethics Committee of the University of Zaragoza was not a prerequisite for this study since we worked with semen or blood samples.
Genotyping
Blood samples were collected from each ram from the jugular vein using sterile vacuum tubes (BD Vacutainer Systems, Belliver Industrial Estate, Plymouth, UK) with ethylenediamine tetraacetic acid (EDTA) as an anticoagulant. The blood samples were aliquoted in aliquots of 200 μL and stored at −20°C until analysis. DNA was extracted from an aliquot of whole blood using a commercial DNA extraction kit (NucleoSpin Blood Mini Kit, Macherey–Nagel, Dueren, Germany). After extraction, both DNA quantity and purity were assessed using a Nanodrop1000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA). The amount of DNA obtained was quantified by measuring the absorbance at 260 nm, and purity was determined by evaluating the A260/A280 and A260/A230 ratios.
To amplify the region of the Exon II where the RsaI and MnlI polymorphisms are located, we used the set of primers (forward: TCCCTCTGCTACGTGTTCCT; reverse: GTTTGTTGTCCGGTTTCACC) following Calvo et al. (2018). The fragments were amplified by PCR in a final volume of 50 μL of a reaction mix that contained 5.0 μL of 10 × Reaction Buffer MgCl2 FREE (Biotools, B&M Labs, Madrid, Spain), 1.0 μL of 10 pM of each primer, 1.0 μL of 200 nM dNTPs, 2.0 μL of 50 mM MgCl2 solution (Biotools, B&M Labs, Madrid, Spain) and 1 U of Pfu DNA polymerase (Biotools, B&M Labs, Madrid, Spain). When the DNA concentration obtained was lower than 20 ng/μL, an EmeraldAmp® GT PCR Master Mix (Takara Bio, Kusatsu, Japan) was used, following the manufacturer’s indications. The PCR conditions were the following: initial denaturation step at 94°C for 3 min; 35 cycles of 94°C for 1 min, the annealing temperature (58°C for the pair of primers above) for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 10 min. The PCR products were confirmed by electrophoresis in 1 × TBE buffer on a 1.7% (w/v) agarose gel containing 5 μL of SYBR™ Safe DNA Gel Stain (Invitrogen, Waltham, MA, USA) parallel with a 100 bp marker (GeneRuler 100 bp DNA Ladder, Thermo Scientific, Waltham, MA, USA) at a constant voltage of 110 V for 40 min. The bands obtained were then displayed using an ultraviolet light trans-illuminator (BioRad Gel Doc XR Imaging System, Hercules, CA, USA).
All PCR products were purified and sequenced in both directions by a commercial sequencing service (STAB VIDA, Lda., Caparica, Portugal). The Blast algorithm (www.ncbi.nlm.nih.gov/blast/) was used to compare the sequences obtained with the genome version Oar_rambouillet_v1.0 (GenBank assembly accession number: GCF_002742125.1). Nucleotide sequence alignments were carried out using the BioEdit Sequence Alignment Editor software (www.mbio.ncsu.edu/BioEdit/BioEdit). Finally, the sequences were analysed with FinchTV software ver. 1.4. (finchtv.software.informer.com/1.4/).
Semen collection and transport
The rams were in a regular semen collection regime, of at least one weekly collections since they are the sires used to carry out inseminations on the farms of the association’s members. Ram ejaculates were collected individually using an artificial vagina by the staff of the center where the animals were housed. After collection, a routine semen assessment was performed in the semen collection facilities, which included the volume of the ejaculate (measured in the graduated collection tube); concentration measured with a spectrophotometer (AccuRead, IMV Technologies, L’Aigle, France); mass motility scored from 0 to 5 and assessed by optical microscopy at 100× magnification; and the proportion of motile spermatozoa (evaluated with a CASA system, AI Station, Sperm Analysis Technologies S.L, Buñol, Valencia, Spain). Thereafter, semen was diluted to 1.6 × 109 spermatozoa/mL with INRA 96 (Imv Technologies, L’Aigle, France), put into 0.5-mL straws and refrigerated at 15°C for transport. Once a month throughout a whole year (from September to August), refrigerated semen samples from the 18 rams were sent to the facilities of the University of Zaragoza. Once the samples arrived at the laboratory, an extensive semen quality analysis was performed, which included sperm motility, morphology, membrane integrity, intracellular levels of reactive oxygen species (ROS), phosphatidylserine (PS) inversion, DNA fragmentation and capacitation status.
Sperm motility analysis
Motility parameters (percentages of motile (TM) and progressive motile (PM) spermatozoa) were measured using a computer-assisted CASA system (ISAS 1.0.4; Proiser SL, Valencia, Spain) as described previously (Gimeno-Martos et al. 2019). Sperm motility was recorded with a video camera (Basler A312f, Basler AG, Ahrensburg, Germany) mounted on a microscope (Nikon Eclipse 50i, Nikon Instruments Inc., Tokyo, Japan) equipped with a 10× negative-phase contrast lens. Samples were diluted 1:15 in a medium with the following composition: 0.25 M sucrose, 100 mM EGTA, 0.5 mM sodium phosphate, 50 mM glucose, 100 mM HEPES and 20 mM KOH. A drop of 8 μL of each diluted sample was placed between a pre-warmed slide and coverslip and kept at 37°C in a heated slide holder during analysis. Five videos at 25 frames/s for 1 s were recorded for each sample. The CASA system settings for the analysis were the following:
Particle area (μm2): 3 (min)−70 (max); VCL (μm/s): 10 > slow > 45 > medium > 75 > rapid; Progressive (% STR): 80; Connectivity: 12.
Sperm morphology evaluation
Semen samples were diluted 1:6 in the aforementioned medium before being mixed (10 μL) with 5 μL eosin and 5 μL nigrosine (Ax et al. 2000). After mixing, 20 μL of the stained sample were placed on a slide and spread with the help of another one. Smears were air-dried and then observed by bright-field microscopy using a Nikon Eclipse E-400 microscope (Kanagawa, Japan). At least 200 spermatozoa were analysed for each sample at 1000× magnification to determine the percentage of normal morphology cells. Sperm cells that showed a detached head, bent tail, coiled tail or proximal or distal droplet were considered abnormal (Ax et al. 2000).
Flow cytometry analyses
Flow cytometry measurements were performed using a Beckman Coulter FC 500 flow cytometer (Beckman Coulter Inc., Fullerton, CA, USA) equipped with two lasers of excitation (argon-ion laser, 488 nm; and solid-state laser, 633 nm), five filters of absorbance (FL1-525, FL2-575, FL3-610, FL4-675 and FL5-755; ±5 nm each bandpass filter) and the CXP software. A flow rate stabilised at 200–300 cells/s was used, and a minimum of 20,000 events were recorded in all experiments. The sperm population was gated for further analysis based on its specific forward (FS) and side scatter (SS) properties to exclude other non-sperm events.
Cell membrane integrity (viability) was evaluated following a modification of the procedure described by Harrison and Vickers (1990). Semen samples were diluted 1:75 to reach a final volume of 500 μL and then stained with 3 μL of 10 μM carboxyfluorescein diacetate (CFDA) and 3 μL of 1.5 mM propidium iodide (PI) and fixed with 3 μL formaldehyde (0.5% (v/v) in water). Samples were examined by flow cytometry following incubation (15 min, 37°C, in the dark). The FS log, SS log, FL1 log (CFDA), FL4 log (PI) and the proportion of viable spermatozoa (CFDA+/PI−) were the evaluated parameters.
Diluted semen samples (1:75, 500 μL final volume) were stained with 5 μL of H2DCFDA (20 mM) and 3 μL of 1.5 mM PI and fixed with 3 μL formaldehyde (0.5% (v/v) in water) (Guthrie and Welch 2006). After incubation (15 min, 37°C in darkness), samples were analysed by flow cytometry. The monitored parameters were FS log, SS log, FL1 log (H2DCFDA), and FL4 log (PI) and the percentage of viable spermatozoa with low ROS was evaluated.
FITC-Annexin V (Thermo Fisher Scientific, Waltham, MA, USA) was used to detect PS translocation and combined with PI to differentiate between membrane-intact and damaged cells, with or without PS translocation (Peña et al. 2003). For this analysis, aliquots of 350 μL (cells diluted 1:75 in Annexin-binding buffer, Invitrogen, Waltham, MA, USA) were stained with 2 μL FITC-Annexin V and 3 μL 1.5 mM PI. After incubation at 37°C in darkness for 10 min, samples were evaluated by flow cytometry. The monitored parameters were FS log, SS log, FL1 log (FITC-Annexin V) and FL4 log (PI). The percentage of viable spermatozoa without PS translocation was considered.
The presence of apoptosis-like DNA strand breaks in sperm samples was evaluated by the TUNEL (terminal transferase-mediated dUDP nick end-labelling) assay using the In Situ Cell Death Detection Kit with fluorescein labelled dUTP (Roche, Mannheim, Germany) (Li and Darzynkiewicz 1995). Previously, semen samples were diluted 1:6 and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature (RT) for 1 h. After two washes with 100 μL of PBS at 600g for 10 min at RT, samples were permeabilised with 0.1% Triton X-100 in 0.1% sodium citrate. Then, the reaction was performed by incubating the pellet obtained with 50 μL of labelling solution that contained the TdT enzyme and dUTP, for 1 h at 37°C in the dark. Two subsequent washes with PBS at 600g for 10 min at RT were performed to stop the reaction, and then samples were assessed by flow cytometry. The monitored parameters were FS log, SS log and FL1 (TUNEL). TUNEL-negative spermatozoa were considered DNA-intact cells.
Capacitation status
The capacitation status was determined using chlortetracycline (CTC) staining (Ward and Storey 1984). A CTC solution (750 μM) was prepared on the day of each experiment in a buffer containing 20 mM Tris, 130 mM NaCl and 5 μM cysteine (pH 7.8), and then passed through a 0.22-μm filter. For the analysis, 20 μL of each diluted sample (dilution 1:6) was mixed with 20 μL of CTC solution and fixed with 5 μL of 1.25% (w/v) paraformaldehyde in 0.5 M Tris–HCl (pH 7.8). Samples were incubated at 4°C in the dark for at least 30 min. After incubation, a 4-μL aliquot of the stained sample was placed on a glass slide and mixed with 2 μL of 0.22 M triethylenediamine (DABCO) in glycerol : PBS (9:1, v/v) at RT and semi-darkness. Samples were then covered with 24 × 48-mm coverslips, sealed with nail polish, and stored in the dark at −20°C. Samples were examined using a Nikon Eclipse E-400 microscope (Kanagawa, Japan) under epifluorescence illumination using a V-2A filter to evaluate CTC patterns, and 200 spermatozoa were scored per slide. Sperm classification followed Gillan et al. (1997) as follows: non-capacitated spermatozoa (NC, with even yellow fluorescence over the head, with or without a bright equatorial band); capacitated cells (C, with fluorescence on the acrosome) and acrosome-reacted cells (R, without fluorescence on the head and with or without a bright equatorial band).
Statistical analysis
Data are shown as mean ± s.e.m. The effect of the season (reproductive season: from September to February; non-reproductive season: from March to August) and RsaI and MnlI polymorphism on seminal quality was evaluated by mixed ANOVA. First, possible outliers were identified by the ROUT method (Motulsky and Brown 2006). The normality of the data was assessed by the Kolmogorov–Smirnov test, and then values were normalised by arcsine transformation. The homogeneity of variances was assessed by Levene’s test. For the mixed ANOVA, the season was considered the ‘within-subjects’ factor, whereas the RsaI and MnlI genotypes were the ‘between-subjects’ factors. For post hoc analyses, Tukey’s multiple comparisons test or Dunn’s method was used when homoscedasticity and heteroscedasticity were detected, respectively. Statistical analyses were performed with SPSS Statistics ver. 26 (IBM Analytic, Armonk, NY, USA) and GraphPad Prism ver. 8 (La Jolla, CA, USA).
Results
Genotypes of the rams used in the study
The result of ram genotyping is shown in Table 1. For the RsaI polymorphism, nine rams were C/C, five rams were C/T and four were T/T. For the MnlI polymorphism, seven of them were G/G, 7 G/A and 4 A/A. Three genotypes (T/T*G/A, C/T*A/A and T/T*A/A) were not present.
| RsaI polymorphism | Total | |||||
|---|---|---|---|---|---|---|
| C/C | C/T | T/T | ||||
| MnlI polymorphism | G/G | 1 (5.6%) | 2 (11.1%) | 4 (22.2%) | 7 (38.9%) | |
| G/A | 4 (22.2%) | 3 (16.7%) | 0 (0.0%) | 7 (38.9%) | ||
| A/A | 4 (22.2%) | 0 (0.0%) | 0 (0.0%) | 4 (22.2%) | ||
| Total | 9 (50%) | 5 (27.8%) | 4 (22.2%) | 18 | ||
| Allele frequency | C = 0.63 | T = 0.36 | G = 0.58 | A = 0.41 | ||
Interaction between RsaI, MnlI and season
A summary of the results of mixed ANOVA is shown in Table 2. No statistically significant three-way interaction between RsaI, MnlI and season, or two-way interaction between season and RsaI or MnlI, was found in any seminal quality parameters analysed. Nevertheless, we found a significant two-way interaction between RsaI and MnlI for total motility, cell membrane integrity (viability) and viable spermatozoa with low ROS. There was also a main effect of the RsaI genotype for total motility, viability, viable spermatozoa with low ROS, viable spermatozoa without PS translocation and DNA intact spermatozoa, and the MnlI genotype for viability, viable spermatozoa with low ROS and viable spermatozoa without PS translocation.
| Parameter | Mixed ANOVA results | |||||||
|---|---|---|---|---|---|---|---|---|
| RsaI | MnlI | Season | RsaI × season | MnlI × season | RsaI × MnlI | RsaI × MnlI × season | ||
| Total motility | P = 0.002 | n.s. | P = 0.041 | n.s. | n.s. | P = 0.018 | n.s. | |
| Progressive motility | n.s. | n.s. | P = 0.036 | n.s. | n.s. | n.s. | n.s. | |
| Membrane integrity (viability) | P < 0.001 | P < 0.001 | P = 0.030 | n.s. | n.s. | P = 0.008 | n.s. | |
| Viable sperm with low ROS levels | P < 0.001 | P < 0.001 | P = 0.019 | n.s. | n.s. | P = 0.010 | n.s. | |
| Viable sperm without PS translocation | P < 0.001 | P < 0.001 | n.s. | n.s. | n.s. | n.s. | n.s. | |
| Non-capacitated sperm | n.s. | n.s. | P = 0.001 | n.s. | n.s. | n.s. | n.s. | |
| Capacitated sperm | n.s. | n.s. | P = 0.001 | n.s. | n.s. | n.s. | n.s. | |
| Acrosome reacted sperm | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | |
| DNA intact sperm | P = 0.048 | n.s. | P < 0.001 | n.s. | n.s. | n.s. | n.s. | |
| Normal morphology | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | |
n.s., non significant.
We likewise detected a seasonal effect on total and progressive motility, viability, viable spermatozoa with low ROS, capacitation status and DNA fragmentation. Due to this seasonal effect on most of the studied parameters, further results were separated between reproductive and non-reproductive seasons. Only significant results (i.e. total motility, viability, viable spermatozoa with low ROS, viable spermatozoa without PS translocation and DNA fragmentation) are described below, and non-significant values (progressive motility, capacitation status and morphology) are shown in the supplementary material (Tables S1, S2 and S3).
Effect of RsaI polymorphism on sperm quality
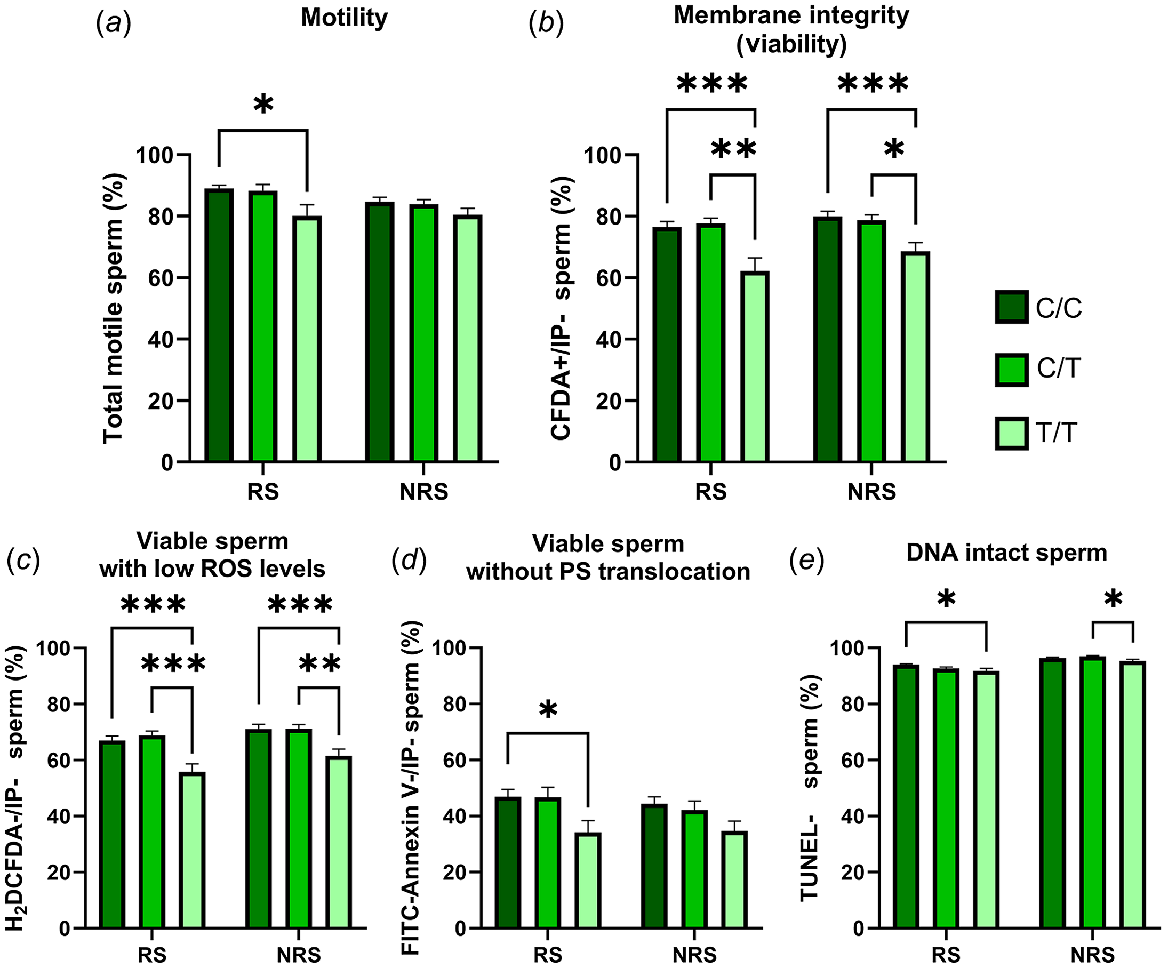
The seminal quality of T/T rams was lower than the other genotypes. Thus, these males presented a lower percentage of motile, membrane intact, viable with low ROS or without PS translocation and DNA intact spermatozoa than C/C or C/T rams. However, these differences were higher in reproductive than in the non-reproductive season and were not detected for total motility or the percentage of spermatozoa without PS translocation during the non-reproductive season (Fig. 1). The other two genotypes, C/C and C/T, did not show any statistical differences between them.
Percentage of total motile (a), viable (b), viable with low ROS levels (c), viable without PS translocation (d) and DNA intact (e) spermatozoa in semen samples from animals carrying different RsaI genotypes (C/C = nine rams; C/T = five rams; and T/T = four rams) during the reproductive season (RS) and the non-reproductive season (NRS). Data are shown as mean ± s.e.m. (n = number of rams of each genotype × 6 semen samples per season). *P < 0.05; **P < 0.01 and ***P < 0.001.
Effect of MnlI polymorphism on sperm quality
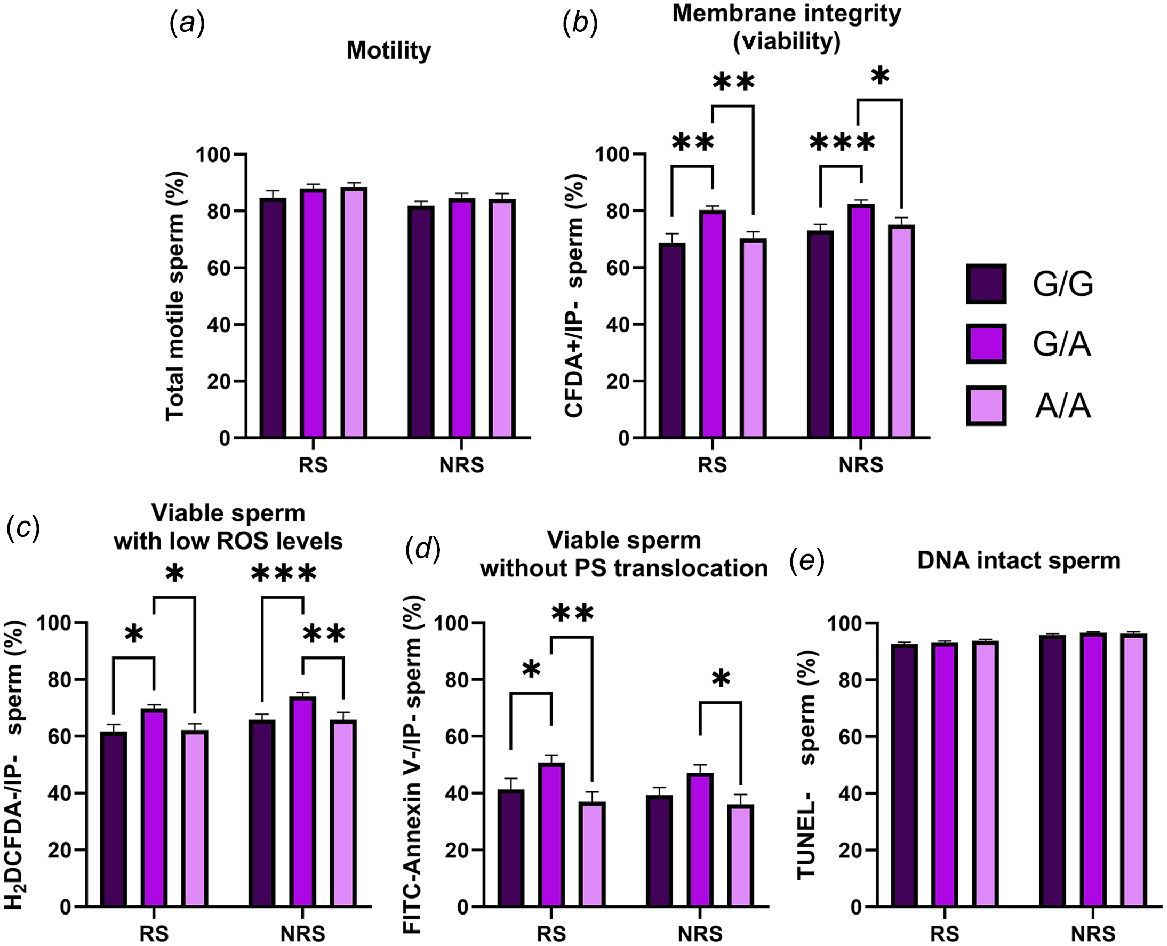
When the effect of the MnlI polymorphism was evaluated, we found a higher percentage of viable spermatozoa, viable sperm with low ROS and viable cells without PS translocation in samples from G/A rams (Fig. 2). These differences were also higher in the reproductive season but for viable spermatozoa with low ROS. No differences were found for total motility and DNA integrity in any season.
Percentage of total motile (a), viable (b), viable with low ROS levels (c), viable without PS translocation (d) and DNA intact (e) spermatozoa in semen samples from animals carrying different MnlI genotypes (G/G = seven rams; G/A = seven rams; and A/A = four rams) during the reproductive season (RS) and the non-reproductive season (NRS). Data are shown as mean ± s.e.m. (n = number of rams of each genotype × 6 semen samples per season). *P < 0.05; **P < 0.01 and ***P < 0.001.
Effect of the combination of RsaI and MnlI polymorphisms on sperm quality
Since mixed ANOVA analysis had detected a significant interaction between RsaI and MnlI polymorphisms in total motility, viability and viable spermatozoa with low ROS (Table 2) and a significance of P = 0.062 in viable sperm without PS translocation, it was essential to study the effect of the combination of both polymorphisms on seminal quality.
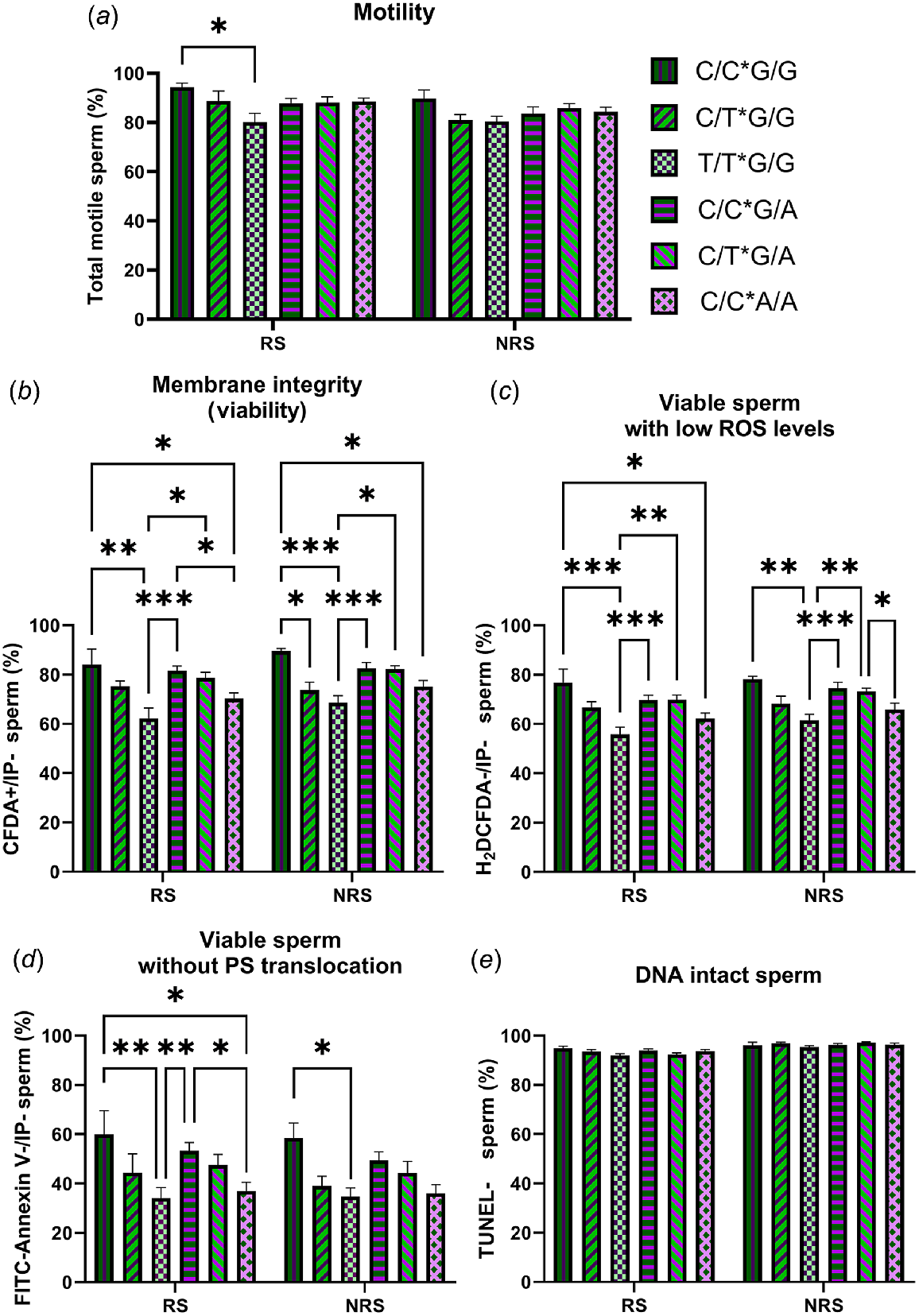
The results showed that, within the G/G males, those that also carried the T/T genotype for RsaI had worse semen quality than the G/G*C/C and the G/G*C/T (Fig. 3). The same was observed, albeit to a lesser extent, in males that carried the A/A genotype for MnlI within the C/C males for RsaI.
Percentage of total motile (a), viable (b), viable with low ROS levels (c), viable without PS translocation (d) and DNA intact (e) spermatozoa in semen samples from animals carrying different combinations of RsaI and MnlI polimorphysms (C/C*G/G = one ram; C/T*G/G = two rams; T/T*G/G = four rams; C/C*G/A = four rams; C/T*G/A = three rams; and C/C*A/A = four rams) during the reproductive season (RS) and the non-reproductive season (NRS). Data are shown as mean ± s.e.m. (n = number of rams of each genotype × 6 semen samples per season). *P < 0.05; **P < 0.01 and ***P < 0.001.
Given that no differences in seminal quality had been observed between males that were not T/T or A/A, in order to avoid the problem of the small number of animals with some combinations of alleles, data were grouped to compare the effect on seminal quality of carrying the lowest frequency alleles in homozygosity (T/T or A/A).
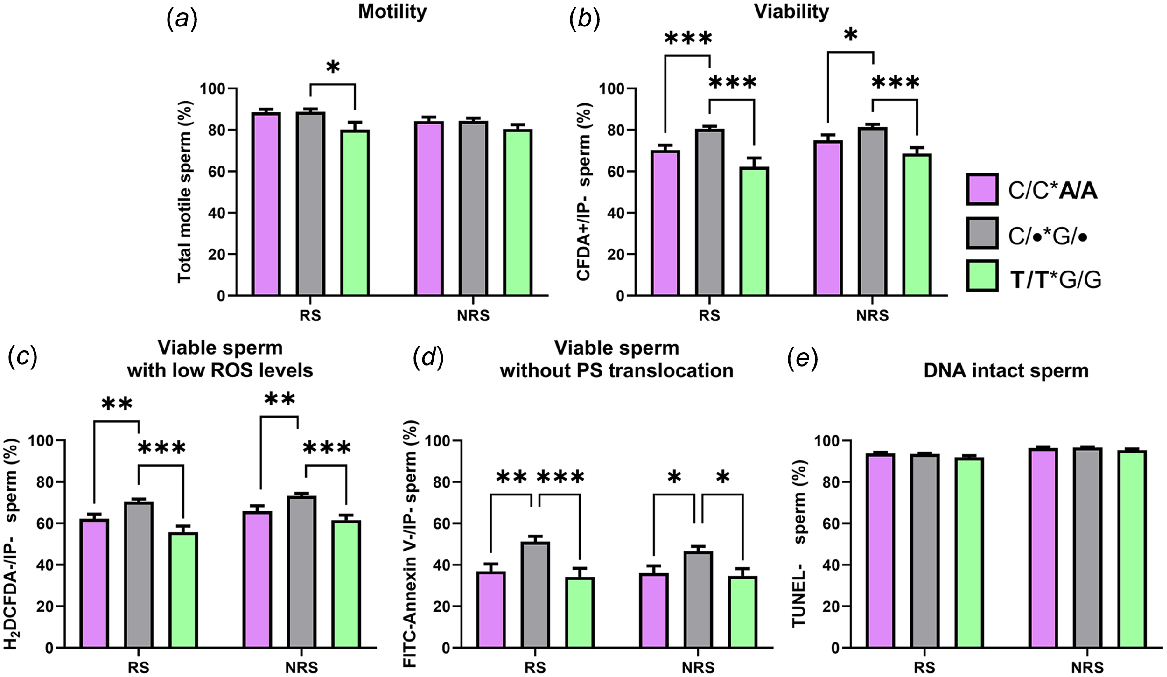
The results revealed that rams carrying the T/T or A/A genotype, regardless of their genotype for the other polymorphism, presented lower seminal quality, especially for viability, viable sperm with low ROS and viable cells without PS translocation (Fig. 4). Moreover, differences were more significant for T/T rams than A/A rams and during the reproductive season. T/T rams also showed differences in motility. No differences were found for DNA-intact spermatozoa.
Percentage of total motile (a), viable (b), viable with low ROS levels (c), viable without PS translocation (d) and DNA intact (e) spermatozoa in semen samples from animals carrying the C/C*A/A genotype (n = four rams); the T/T*G/G genotype (n = four rams) and the other possibilities of genotypes (C/•*G/•, n = 10 rams) during the reproductive season (RS) and the non-reproductive season (NRS). Data are shown as mean ± s.e.m. (n = number of rams × 6 semen samples per season). *P < 0.05; **P < 0.01 and ***P < 0.001.
Discussion
Sheep have a marked reproductive seasonality regulated by melatonin (Chemineau et al. 2008). Apart from its synthesis at the level of the pineal gland, this hormone is also synthesised in many tissues, including those of the reproductive tract (González-Arto et al. 2017). Melatonin exerts its functions, in part, after binding to specific receptors (Zhao et al. 2019), such as MT1, which has several polymorphic sites revealed by restriction enzymes RsaI and MnlI (Messer et al. 1997). Unlike in ewes, the influence of carrying these polymorphisms in rams has not been widely studied. Our group reported differences in reproductive behaviour in young and adult lambs with different genotypes according to these polymorphisms (Abecia et al. 2020), but their possible influence on seminal quality was unknown.
The results of this study showed that there was a main effect of RsaI and MnlI polymorphisms on the percentage of spermatozoa with membrane integrity and viable spermatozoa with low levels of ROS and without PS translocation. Moreover, RsaI polymorphism also influenced percentages of motile and DNA-intact spermatozoa. Thus, the seminal quality of T/T rams was lower than the other genotypes, C/T or C/C. Regarding MnlI polymorphism, higher percentages of viable spermatozoa, viable sperm with low ROS and viable cells without PS translocation were observed in samples from G/A rams. These differences were generally higher in the reproductive than the non-reproductive season, which will be discussed later.
Although the above-mentioned results seemed to suggest that heterozygous G/A males had better semen quality than homozygotes G/G or A/A, it should not be forgotten that the mixed ANOVA analysis revealed a significant interaction between RsaI and MnlI polymorphisms in total motility, viability and viable spermatozoa with low ROS, and a tendency to significance in viable sperm without PS translocation. Furthermore, it had to be borne in mind that T/T males presented worse semen quality than the C/C or C/T, and four of the seven G/G males in this study were also T/T, which could be the reason for the differences between the G/G and G/A samples. Therefore, it was essential to study the effect of the combination of both polymorphisms on seminal quality. The results, considering both polymorphisms simultaneously, pointed to a negative effect on seminal quality of carrying one of the two low-frequency alleles, that is, T or A, in homozygosity. These results were confirmed when the data of those males that did not carry these alleles in homozygosity were grouped and compared with those that did, given that the former did not present differences in semen quality between them. With this approach, we tried to solve the problem of the small number of animals carrying some combinations of alleles. The results revealed that rams carrying the T/T or A/A genotype, regardless of their genotype for the other polymorphism, presented lower seminal quality, especially for viability, viable sperm with low ROS and viable cells without PS translocation. Moreover, differences were more significant for T/T rams than A/A rams, and again during the reproductive season. Samples from T/T males also showed worse sperm motility.
In other species, the melatonin receptor MT1 in spermatozoa appears to be involved in modulating motility (Fujinoki 2008), preservation of sperm viability (Fernández-Alegre et al. 2022) or inhibition of apoptosis (Espino et al. 2011). Therefore, the presence of mutations in the MTNR1A gene in rams could alter the functionality of the MT1 receptor in spermatozoa, decreasing the seminal quality. In this respect, other mutations in the receptor, different from RsaI and MnlI, have only been described in relation to sperm morphology in Sanjabi rams (Kianpoor et al. 2018). Furthermore, we must not forget the existence of melatonin receptor MT1 in the tissues of the male reproductive tract (Casao et al. 2012) so that mutations in them could affect the correct development of spermatogenesis.
It is important to note that, although both polymorphisms are silent mutations (Carcangiu et al. 2009), they have been linked with other mutations present in the MTNR1A gene that induce amino acid changes in the final protein (Calvo et al. 2018; Luridiana et al. 2020). Both the RsaI and the MnlI polymorphisms have been found, in the Rasa Aragonesa breed, to be in linkage disequilibrium with another SNP that causes an amino acid change from an arginine to a cysteine (R336C) (Calvo et al. 2018). Moreover, in Sarda sheep, the MnlI polymorphism is linked to another SNP that produces a change from valine to isoleucine (V220I) (Luridiana et al. 2020), and the functional characterisation of this mutation revealed that it affects the inhibition of the adenylate cyclase, which suggests a potential modification in melatonin signalling in animals carrying the mutation (Trecherel et al. 2010).
In general, differences between genotypes were higher in the reproductive than in the non-reproductive season. This fact could be attributed to differences in the melatonin concentration in seminal plasma between both seasons (Casao et al. 2010), being higher in the reproductive one. Thus, if the melatonin receptor MT1 in spermatozoa from T/T or A/A males is less efficient in melatonin binding or transmitting the signal in the cAMP/PKA pathway, the effects on semen quality would be more evident in this season. Furthermore, our group has recently shown that, during the reproductive season, individuals expressing the two high-frequency alleles for these polymorphisms, both in homo- and heterozygosis, exhibit increased gene expression of the RORA gene (Tobajas et al. 2023). RORA is a gene related to spermatogenesis (Mandal et al. 2018) and testicular structure (Sayed et al. 2019) whose transcription might be controlled by melatonin binding to its receptors (Slominski et al. 2016). Therefore, the mutations in the melatonin receptor MT1 from T/T or A/A rams may affect the expression of RORA in these individuals, which could also explain why T/T or A/A rams show lower seminal quality in this study.
In a previous study, we studied the relationship between the MTNR1A polymorphisms of rams and the fertility rate of the ewes after AI (Abecia et al. 2023). We reported that for the RsaI polymorphism, T/T rams led to a significantly lower fertility rate in the reproductive season than C/C and C/T, with no differences between these two genotypes. For MnlI, significant differences were observed between genotypes, with A/A being associated with lower fertility and G/A with higher fertility. The results of the present study suggest that the observed differences in fertility could be due to differences in seminal quality since TT rams presented lower seminal quality than C/C and C/T, and G/A rams showed higher seminal quality than GG and AA, particularly during the reproductive season.
The results point to the fact that mutations in the MTNR1A gene in rams could decrease their seminal quality. Genotyping of rams based on melatonin receptor 1A polymorphisms could be a powerful tool in male selection for sires in AI or natural mating programs.
Conflicts of interest
The authors declare no conflicts of interest. Jose Alfonso Abecia is an Editor of the journal, but was blinded from the peer review process for this paper.
Declarations of funding
This work was supported by Ministerio de Ciencia e Innovación (Gobierno de España) (PID2020-113239RB-100) and Gobierno de Aragón (A07_23R). V P-D has a predoctoral contract from Diputación General de Aragón (DGA).
References
Abecia JA, Mura MC, Carvajal-Serna M, Pulinas L, Macías A, Casao A, Pérez-Pe R, Carcangiu V (2020) Polymorphisms of the melatonin receptor 1A (MTNR1A) gene influence the age at first mating in autumn-born ram-lambs and sexual activity of adult rams in spring. Theriogenology 157, 42-47.
| Crossref | Google Scholar | PubMed |
Abecia JA, Heredia A, Pérez-Pe R, Casao A, Carcangiu V, Mura MC, Miranda-De la Lama G (2022) Polymorphisms of the melatonin receptor 1A (MTNR1A) gene affect the sexual performance of Rasa Aragonesa rams without changing their social dominance. Journal of Animal Behaviour and Biometeorology 10, 2231.
| Crossref | Google Scholar |
Abecia JA, Macías A, Laviña A, Martín E, Casao A, Canto F, Carvajal-Serna M, Peña-Delgado V, Pérez-Pe R (2023) Polymorphisms of the melatonin receptor 1A (MTNR1A) gene of Rasa Aragonesa rams affect their fertility after artificial insemination. In ‘10th International Sheep Veterinary Congress (ISVC 2023). Animal - Science Proceedings’. Vol. 14, pp. 77–78. doi:10.1016/j.anscip.2023.01.108
Arjoune A, Alsaleh AB, Messaoudi SA, Chelbi H, Jelassi R, Assidi M, Najar T, Haddad B, Sirard MA (2023) Analysis of MTNR1A genetic polymorphisms and their association with the reproductive performance parameters in Two Mediterranean Sheep Breeds. Animals 13, 448.
| Crossref | Google Scholar |
Ax RL, Dally M, Didion BA, Lenz RW, Love CC, Varner DD, Hafez B, Bellin ME (2000) Semen evaluation. ‘Reproduction in farm animals’. (Eds B Hafez, ESE Hafez) pp. 363–375. (John Wiley & Sons, Ltd) doi:10.1002/9781119265306.ch25
Barrett P, Conway S, Jockers R, Strosberg AD, Guardiola-Lemaitre B, Delagrange P, Morgan PJ (1997) Cloning and functional analysis of a polymorphic variant of the ovine Mel 1a melatonin receptor. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1356, 299-307.
| Crossref | Google Scholar |
Calvo JH, Serrano M, Martinez-Royo A, Lahoz B, Sarto P, Ibañez-Deler A, Folch J, Alabart JL (2018) SNP rs403212791 in exon 2 of the MTNR1A gene is associated with reproductive seasonality in the Rasa aragonesa sheep breed. Theriogenology 113, 63-72.
| Crossref | Google Scholar | PubMed |
Carcangiu V, Mura MC, Vacca GM, Pazzola M, Dettori ML, Luridiana S, Bini PP (2009) Polymorphism of the melatonin receptor MT1 gene and its relationship with seasonal reproductive activity in the Sarda sheep breed. Animal Reproduction Science 116, 65-72.
| Crossref | Google Scholar | PubMed |
Carcangiu V, Luridiana S, Vacca GM, Daga C, Mura MC (2011) A polymorphism at the melatonin receptor 1A (MTNR1A) gene in Sarda ewes affects fertility after AI in the spring. Reproduction, Fertility and Development 23, 376-380.
| Crossref | Google Scholar |
Casao A, Cebrián I, Asumpção ME, Pérez-Pé R, Abecia JA, Forcada F, Cebrián-Pérez JA, Muiño-Blanco T (2010) Seasonal variations of melatonin in ram seminal plasma are correlated to those of testosterone and antioxidant enzymes. Reproductive Biology and Endocrinology 8, 59.
| Crossref | Google Scholar | PubMed |
Casao A, Gallego M, Abecia JA, Forcada F, Pérez-Pé R, Muiño-Blanco T, Cebrián-Pérez JÁ (2012) Identification and immunolocalisation of melatonin MT1 and MT2 receptors in Rasa Aragonesa ram spermatozoa. Reproduction, Fertility and Development 24, 953-961.
| Crossref | Google Scholar | PubMed |
Chemineau P, Guillaume D, Migaud M, Thiéry JC, Pellicer-Rubio MT, Malpaux B (2008) Seasonality of reproduction in mammals: intimate regulatory mechanisms and practical implications. Reproduction in Domestic Animals 43(Suppl 2), 40-47.
| Crossref | Google Scholar | PubMed |
Cosso G, Nehme M, Luridiana S, Pulinas L, Curone G, Hosri C, Carcangiu V, Mura MC (2021) Detection of polymorphisms in the mtnr1a gene and their association with reproductive performance in awassi ewes. Animals 11, 583.
| Crossref | Google Scholar |
Delgadillo JA, Hernández H, Abecia JA, Keller M, Chemineau P (2020) Is it time to reconsider the relative weight of sociosexual relationships compared with photoperiod in the control of reproduction of small ruminant females? Domestic Animal Endocrinology 73, 106468.
| Crossref | Google Scholar |
Dubocovich ML, Markowska M (2005) Functional MT1 and MT2 melatonin receptors in mammals. Endocrine 27, 101-110.
| Crossref | Google Scholar | PubMed |
Dubocovich ML, Rivera-Bermudez MA, Gerdin MJ, Masana MI (2003) Molecular pharmacology, regulation and function of mammalian melatonin receptors. Frontiers in Bioscience 8, d1093-d1108.
| Crossref | Google Scholar |
Espino J, Ortiz Á, Bejarano I, Lozano GM, Monllor F, García JF, Rodríguez AB, Pariente JA (2011) Melatonin protects human spermatozoa from apoptosis via melatonin receptor- and extracellular signal-regulated kinase-mediated pathways. Fertil Steril 95, 2290-2296.
| Crossref | Google Scholar | PubMed |
Fernández-Alegre E, Lacalle E, Soriano-Úbeda C, González-Montaña JR, Domínguez JC, Casao A, Martínez-Pastor F (2022) Bos taurus and Cervus elaphus as non-seasonal/seasonal models for the role of melatonin receptors in the spermatozoon. International Journal of Molecular Sciences 23, 6284.
| Crossref | Google Scholar |
Forcada F, Abecia JA, Sierra I (1992) Seasonal changes in oestrus activity and ovulation rate in Rasa Aragonesa ewes maintained at two different body condition levels. Small Ruminant Research 8, 313-324.
| Crossref | Google Scholar |
Fujinoki M (2008) Melatonin-enhanced hyperactivation of hamster sperm. Reproduction 136, 533-541.
| Crossref | Google Scholar | PubMed |
Gillan L, Evans AG, Maxwell WMC (1997) Capacitation status and fertility of fresh and frozen-thawed ram spermatozoa. Reproduction, Fertility and Development 9, 481-487.
| Crossref | Google Scholar | PubMed |
Gimeno-Martos S, Casao A, Yeste M, Cebrián-Pérez JA, Muiño-Blanco T, Pérez-Pé R (2019) Melatonin reduces cAMP-stimulated capacitation of ram spermatozoa. Reproduction, Fertility and Development 31, 420-431.
| Crossref | Google Scholar | PubMed |
González-Arto M, Vicente-Carrillo A, Martínez-Pastor F, Fernández-Alegre E, Roca J, Miró J, Rigau T, Rodríguez-Gil JE, Pérez-Pé R, Muiño-Blanco T, Cebrián-Pérez JA, Casao A (2016) Melatonin receptors MT1 and MT2 are expressed in spermatozoa from several seasonal and nonseasonal breeder species. Theriogenology 86, 1958-1968.
| Crossref | Google Scholar | PubMed |
González-Arto M, Aguilar D, Gaspar-Torrubia E, Gallego M, Carvajal-Serna M, Herrera-Marcos LV, Serrano-Blesa E, Dos Santos Hamilton TR, Pérez-Pé R, Muiño-Blanco T, Cebrián-Pérez JA, Casao A (2017) Melatonin MT₁ and MT₂ receptors in the ram reproductive tract. International Journal of Molecular Sciences 18, 662.
| Crossref | Google Scholar |
Guthrie HD, Welch GR (2006) Determination of intracellular reactive oxygen species and high mitochondrial membrane potential in Percoll-treated viable boar sperm using fluorescence-activated flow cytometry. Journal of Animal Science 84, 2089-2100.
| Crossref | Google Scholar | PubMed |
Harrison RA, Vickers SE (1990) Use of fluorescent probes to assess membrane integrity in mammalian spermatozoa. Journal of Reproduction & Infertility 88, 343-352.
| Crossref | Google Scholar | PubMed |
Kianpoor S, Abdolmohammadi A, Hajarian H, Nikousefat Z, Khamisabadi H (2018) Association of MTNR1A and CYP19 genes polymorphisms with sperm quality and testicular size in Sanjabi breed rams. Annals of Animal Science 18, 699-711.
| Crossref | Google Scholar |
Li X, Darzynkiewicz Z (1995) Labelling DNA strand breaks with BrdUTP. Detection of apoptosis and cell proliferation. Cell Proliferation 28, 571-579.
| Crossref | Google Scholar |
Luridiana S, Cosso G, Pulinas L, Di Stefano MV, Curone G, Carcangiu V, Mura MC (2020) New polymorphisms at MTNR1A gene and their association with reproductive resumption in sarda breed sheep. Theriogenology 158, 438-444.
| Crossref | Google Scholar | PubMed |
Mandal K, Sarkar RK, Sen Sharma S, Jain A, Majumdar SS (2018) Sertoli cell specific knockdown of RAR-related orphan receptor (ROR) alpha at puberty reduces sperm count in rats. Gene 641, 18-24.
| Crossref | Google Scholar |
Martínez-Marcos P, Carvajal-Serna M, Lázaro-Gaspar S, Pérez-Pé R, Muiño-Blanco T, Cebrián-Pérez JA, Casao A (2019) Presence of melatonin-catabolizing non-specific enzymes myeloperoxidase and indoleamine 2,3-dioxygenase in the ram reproductive tract. Reproduction in Domestic Animals 54, 1643-1650.
| Crossref | Google Scholar |
Martínez-Royo A, Lahoz B, Alabart JL, Folch J, Calvo JH (2012) Characterisation of the Melatonin Receptor 1A (MTNR1A) gene in the Rasa Aragonesa sheep breed: Association with reproductive seasonality. Animal Reproduction Science 133, 169-175.
| Crossref | Google Scholar | PubMed |
Messer LA, Wang L, Tuggle CK, Yerle M, Chardon P, Pomp D, Womack JE, Barendse W, Crawford AM, Notter DR, Rothschild MF (1997) Mapping of the melatonin receptor 1a (MTNR1A) gene in pigs, sheep, and cattle. Mammalian Genome 8, 368-370.
| Crossref | Google Scholar | PubMed |
Motulsky HJ, Brown RE (2006) Detecting outliers when fitting data with nonlinear regression – a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics 7, 123.
| Crossref | Google Scholar |
Notter DR, Cockett NE, Hadfield TS (2003) Evaluation of melatonin receptor 1a as a candidate gene influencing reproduction in an autumn-lambing sheep flock. Journal of Animal Science 81, 912-917.
| Crossref | Google Scholar |
Pelletier J, Bodin L, Hanocq E, Malpaux B, Teyssier J, Thimonier J, Chemineau P (2000) Association between expression of reproductive seasonality and alleles of the gene for Mel(1a) receptor in the ewe. Biology of Reproduction 62, 1096-1101.
| Crossref | Google Scholar | PubMed |
Peña FJ, Johannisson A, Wallgren M, Rodríguez-Martínez H (2003) Assessment of fresh and frozen–thawed boar semen using an Annexin-V assay: a new method of evaluating sperm membrane integrity. Theriogenology 60, 677-689.
| Crossref | Google Scholar |
Sayed RKA, Mokhtar DM, Fernández-Ortiz M, Escames G, Acuña-Castroviejo D (2019) Retinoid-related orphan nuclear receptor alpha (RORα)-deficient mice display morphological testicular defects. Laboratory Investigation 99, 1835-1849.
| Crossref | Google Scholar |
Slominski AT, Zmijewski MA, Jetten AM (2016) RORα is not a receptor for melatonin. Bioessays 38, 1193-1194.
| Crossref | Google Scholar |
Starič J, Farci F, Luridiana S, Mura MC, Pulinas L, Cosso G, Carcangiu V (2020) Reproductive performance in three Slovenian sheep breeds with different alleles for the MTNR1A gene. Animal Reproduction Science 216, 106352.
| Crossref | Google Scholar |
Tobajas A, Peña-Delgado V, Carvajal-Serna M, Carcangiu V, Mura MC, Noya A, Abecia JA, Casao A, Pérez-Pe R (2023) Relación entre el polimorfismo RsaI del gen del receptor de melatonina MT1 (MTNR1A) y la expresión génica del receptor nuclear RORA en el testículo de morueco. In Book of abstracts of the ‘45th Annual Meeting of Spanish Society of Biochemistry and Molecular Biology (SEBBM)’, Zaragoza, Spain. (Zaragoza, Spain). Avalaible at https://congresos.sebbm.es/zaragoza2023/wp-content/uploads/LIBRO-RESUMENES-SEBBM-23_Completo-V13.pdf
Trecherel E, Batailler M, Chesneau D, Delagrange P, Malpaux B, Chemineau P, Migaud M (2010) Functional characterization of polymorphic variants for ovine MT1 melatonin receptors: Possible implication for seasonal reproduction in sheep. Animal Reproduction Science 122, 328-334.
| Crossref | Google Scholar | PubMed |
Ward CR, Storey BT (1984) Determination of the time course of capacitation in mouse spermatozoa using a chlortetracycline fluorescence assay. Developmental Biology 104, 287-296.
| Crossref | Google Scholar | PubMed |
Weaver DR, Liu C, Reppert SM (1996) Nature’s knockout: the Mel1b receptor is not necessary for reproductive and circadian responses to melatonin in Siberian hamsters. Molecular Endocrinology 10, 1478-1487.
| Crossref | Google Scholar | PubMed |
Zhao D, Yu Y, Shen Y, Liu Q, Zhao Z, Sharma R, Reiter RJ (2019) Melatonin synthesis and function: evolutionary history in animals and plants. Frontiers in Endocrinology 10, 249.
| Crossref | Google Scholar | PubMed |


