Genomic analyses suggest strong population connectivity over large spatial scales of the commercially important baitworm, Australonuphis teres (Onuphidae)
Amanda Padovan A B I , Rowan C. Chick C , Victoria J. Cole C , Ludovic Dutoit D , Patricia A. Hutchings E F , Cameron Jack G and Ceridwen I. Fraser A H
A B I , Rowan C. Chick C , Victoria J. Cole C , Ludovic Dutoit D , Patricia A. Hutchings E F , Cameron Jack G and Ceridwen I. Fraser A H
A Fenner School of Environment and Society, Building 44, Australian National University, Canberra, ACT 2601, Australia.
B CSIRO, Black Mountain Science and Innovation Park, Clunies Ross Street, Canberra, ACT 2601, Australia.
C New South Wales Department of Primary Industries, Port Stephens Fisheries Institute, Taylors Beach, NSW 2316, Australia.
D Department of Zoology, University of Otago, 340 Great King Street, Dunedin, 9016, New Zealand.
E Australian Museum Research Institute, Australian Museum, 1 William Street, Sydney, NSW 2010, Australia.
F Department of Biological Sciences, Macquarie University, North Ryde, NSW 2109, Australia.
G ANU Bioinformatics Consultancy, John Curtin School of Medical Research, Australian National University, Acton, ACT 2601, Australia.
H Department of Marine Science, University of Otago, PO Box 56, Dunedin, 9054, New Zealand.
I Corresponding author. Email: amanda.padovan@csiro.au
Marine and Freshwater Research 71(11) 1549-1556 https://doi.org/10.1071/MF20044
Submitted: 11 February 2020 Accepted: 27 April 2020 Published: 21 May 2020
Journal Compilation © CSIRO 2020 Open Access CC BY
Abstract
Barriers to dispersal can disrupt gene flow between populations, resulting in genetically distinct populations. Although many marine animals have potential for long-distance dispersal via a planktonic stage, gene flow among populations separated by large geographic distances is not always evident. Polychaetes are ecologically important and have been used as biological surrogates for marine biodiversity. Some polychaete species are used as bait for recreational fisheries, with this demand supporting commercial fisheries for polychaetes to service the retail bait market. However, despite their ecological and economic importance, very little is known about the life history or population dynamics of polychaetes, and few studies have used genetic or genomic approaches to understand polychaete population connectivity. Here, we investigate the population structure of one commonly collected beachworm species used for bait on the eastern coast of Australia, namely, Australonuphis teres, by using genome-wide single-nucleotide polymorphism data. We sampled A. teres from hierarchical nested spatial scales along 900 km of the coast in New South Wales. We identified six genetic groups, but there was no clear geographic pattern of distribution. Our results suggest that there is considerable gene flow among the sampled populations. These high-resolution genomic data support the findings of previous studies, and we infer that oceanographic processes promote genetic exchange among polychaete populations in south-eastern Australia.
Additional keywords: admixture, fisheries, genotype-by-sequencing, management, polychaete, population structure, SNP.
Introduction
Life history, particularly the larval stage, and environmental influences govern biodiversity and population structure in the marine environment (Thorrold 2006; Chan et al. 2018). Understanding population connectivity, size and structure is critical for effective management of marine resources. Pelagic larval duration along with physical and ecological processes can greatly affect the likelihood of successful dispersal and population connectivity for marine species (Thiel and Gutow 2005a, 2005b; Cowen and Sponaugle 2009; Riginos et al. 2011; Kinlan and Gaines 2003). Despite evidence of strong population connectivity in some species (Shulman and Bermingham 1995; Roberts 1997; Cowen et al. 2007), many other marine taxa, even those with apparently high dispersal potential, show surprisingly little evidence for gene flow among disjunct populations (Dibacco et al. 2006; Purcell et al. 2006; Froukh and Kochzius 2007; Marko et al. 2007; Temby et al. 2007; Miller et al. 2009, 2014; Xuereb et al. 2018). There is, thus, a growing recognition that marine systems are not all open and interconnected (Dennis and Hellberg 2010; Karsenti et al. 2011; Iacchei et al. 2013). Defining the scale of connectivity and determining the factors that influence gene flow are critical for understanding population dynamics and genetic structure of marine species, and for developing effective management strategies to mitigate anthropogenic effects on populations and ecosystems (Giangrande et al. 2005; Waples and Punt 2008).
Polychaete worms are common in intertidal ecosystems (Cole et al. 2007, 2017), and are good indicators of species richness and community patterns in benthic invertebrate assemblages (Olsgard and Somerfield 2000; Giangrande et al. 2005). Indeed, polychaetes have been used as biological surrogates for marine biodiversity (Olsgard et al. 2003; Shokri et al. 2009). Among benthic groups, polychaetes are one of the best indicators of environmental disturbance, because there are both sensitive and tolerant species across pristine and heavily disturbed habitats (Olsgard and Somerfield 2000). Polychaetes are important members of the marine food chain; they can act as predators, scavengers and grazers of diverse other organisms, or as prey for a variety of bird, fish and crustacean species (Fauchald and Jumars 1979; Jumars et al. 2015).
In addition to their ecological importance, many species of polychaetes are an important bait resource for recreational fishers (Cole et al. 2018). As with any fisheries resource, management relies on information about stock size and population connectivity (Cowen et al. 2007; Fogarty and Botsford 2007). In Australia, recreational and commercial fishers target beachworms directly for bait or for sale into the bait market respectively. Beachworms, including the ‘kingworm’ or ‘stumpy’ Australonuphis teres (Onuphidae), inhabit high-energy sandy beaches from Maroochydore, Queensland to Lakes Entrance, Victoria (Paxton 1979). Although little is known about their population biology, they are thought to be broadcast spawners, have been recorded to be sexually mature at 420 mm in length (Paxton 1979), and are suspected to breed multiple times a year (Paxton 1986). As adults, dispersal of A. teres is relatively limited (Paxton 1979), with large topographic structures (e.g. headlands and river mouths) that separate sandy beaches acting as physical barriers to dispersal.
Despite their ecological and economic importance, few population genetic (and fewer population genomic) studies have yet been conducted for polychaetes. In one genetic study from the USA, the baitworm Glycera dibranchiata (Glyceridae) was inferred to have little connectivity among populations within an estuary and between intertidal and subtidal populations (Bristow and Vadas 1991). In contrast, in Australia, a recent study of estuarine polychaetes found little genetic differentiation among populations of the nephtyids Aglaophamus australiensis and Nephtys longipes, from which the authors inferred that pelagic larval dispersal is probably mediated by ocean currents in these two species (Smith et al. 2015). However, this latter research was based only on small fragments of mitochondrial and nuclear loci, which were not of a high-enough resolution to enable assessment of whether population connectivity was ongoing or merely recent. The connectivity of populations of polychaetes along Australian coasts, thus, remains largely unknown, yet, such knowledge is of considerable importance for managing the sustainable harvest of beachworms and other polychaete bait species.
Here, we use genomic approaches to investigate population structure of the polychaete worm A. teres at multiple nested spatial scales (i.e. sites separated by kilometres, beaches separated by tens of kilometres, and regions separated by hundreds of kilometres) along 900 km of the New South Wales (NSW) coast in Australia. We generated a single-nucleotide polymorphism (SNP) dataset via genotype-by-sequencing. The three possible findings are as follows: (1) genetic homogeneity in all A. teres samples across spatial scales, which would indicate a single panmictic population and considerable gene flow along the coast, (2) genetically distinct populations of A. teres, indicating negligible gene flow among populations, or (3) a mixed result, with both similarities and differences among populations, indicating some gene flow. This study is one of the first to use large-scale genomic data to assess population connectivity in polychaetes.
Materials and methods
Field methods for sampling Australonuphis teres
Populations of A. teres were sampled in the Eastern Warm Temperate biogeographic zone of the NSW coast. Three bioregions were selected and we designated them as North, Central and South, corresponding to the Tweed–Moreton (28°S–30.5°S), Manning (30.5°S–30.75°S) and Batemans (34.6°S–36.6°S) bioregions respectively. A fully nested design was used to obtain samples of A. teres. In each of the three regions (separated by hundreds of kilometres), two beaches (separated by tens of kilometres), each with two randomly chosen sites (separated by kilometres), 45 individuals were collected. The posterior end (the last 20 mm of the body) was removed and placed in 98% ethanol and stored below –18°C for subsequent analysis. Beaches were selected as those where commercial harvesting of beachworms occurs (>0.6 t year–1 commercial catch over the previous 2 years). Samples of the population of A. teres were obtained through beach sampling, whereby a mesh bag containing a fish frame was dragged along the sand in the swash zone to entice the worms to emerge, and any worms that surfaced were collected by hand.
DNA extraction, library preparation and sequencing
Tissue samples were removed from ethanol, dried and subsampled. DNA was extracted with the Qiagen DNeasy 96 Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The quality and quantity of DNA were evaluated by gel electrophoresis and Qubit fluorometer (Invitrogen, Carlsbad, CA, USA). The 40 highest-yield extractions from each site were selected for sequencing, with a total of 480 individual A. teres being used in library preparation.
A genotyping-by-sequencing (GBS) approach to generate sequence data was used, as applied in similarly non-model systems in the past, and following Fraser et al. (2018a) and references therein. Briefly, GBS library preparation followed standard protocols (Elshire et al. 2011) with minor modifications (Wilson et al. 2019). To each DNA sample, a uniquely barcoded PstI adaptor was added (2.25 ng per sample; Morris et al. 2011). Digestion was performed with 4 U PstI-HF (New England Biolabs, MA, USA) in 1 × CutSmart Buffer, and incubation at 37°C for 2 h. Adapters were ligated with T4 DNA ligase in 1 × ligation buffer (New England Biolabs, MA, USA) and incubated at 16°C for 90 min (with 2 min at 37°C every 30 min) and 80°C for 30 min. Purification was performed using a Qiagen MinElute 96 UF PCR Purification Kit (Qiagen, Hilden, Germany), with elution in 25 µL 1 × TE. Polymerase chain reactions (PCRs) were run on 50-µL volumes containing 10 µL purified DNA, 1 × MyTaqTM HS Master Mix (Bioline, Sydney, Australia) and 1 µM each of PCR primers 5′AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T and 5′CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T (where * indicates phosphorothioation) at 72°C for 5 min, 95°C for 60 s, and 24 cycles of 95°C for 30 s, 65°C for 30 s and 72°C for 30 s, with a final extension step at 72°C for 5 min. Concentrations for each sample were assessed using a LabChip GXII (Caliper Life Sciences, MA, USA) and pooled equimolarly. A 200-base pair (bp) fraction (fragment size 250–450 bp) of the pooled library was separated via electrophoresis on a 1.5% agarose gel. Sequencing of DNA from this size range was performed on one lane of a high-output flowcell in an Illumina NextSeq 500 system (Illumina, CA, USA, 75 bp paired-end). The raw sequence data are available in the NCBI Sequence Read Archive under BioProject: PRJNA512571 (http://www.ncbi.nlm.nih.gov/bioproject/512571).
Data analysis
Sequencing yielded 185 204 387 reads that were, on average, 76 bp long. The quality of the sequences was first assessed using FASTQC (Andrews 2010), and SNPs were extracted using STACKS 2.41 (Catchen et al. 2013). All scripts are available as Supplementary Material to this paper.
Raw sequence reads were demultiplexed and quality filtered using the process_radtags module in STACKS. As there is no reference genome, SNP calling was performed using the denovo_map.pl pipeline, using default parameter settings. After removing low-quality individuals, SNPs with less than 60% of individuals sequenced were discarded as likely collapsed repeats. We explored the data following McGaughran et al. (2019) in vcftools (Danecek et al. 2011) and filtered to have a maximum of 50% of samples with missing genotypes, yielding 853 SNP loci for 273 samples.
The remaining analysis was performed in R, following McGaughran et al. (2019), with all scripts used being available in Supplementary Material. We used several packages, including adegenet (Jombart 2008; Jombart and Ahmed 2011), vcfR (Knaus and Grünwald 2017), poppr (Kamvar et al. 2014, 2015), devtools (Wickham et al. 2019), hierfstat (Goudet 2005), ade4 (Chessel et al. 2004; Dray and Dufour 2007; Bougeard and Dray 2018), pegas (Paradis 2010), ggplot2 and ggbiplot (Wickham 2016), radiator (Gosselin 2019) and SeqVarTools (Gogarten et al. 2019). We calculated pairwise Nei’s distance (Nei 1987) and Weir and Cockerham’s Fst (Weir and Cockerham 1984) values at three population levels, namely, region (n = 3), beach (n = 6) and site (n = 12). We looked at the relationship between geographical (all three levels independently) and both measures of genetic differentiation by using a Mantel test. We used principal component analysis (PCA) to visualise patterns in the genetic data.
To further investigate connectivity and population structure, we investigated partitioning of variance at the three hierarchically sampled geographic levels (i.e. region, beach, site) by using an analysis of molecular variance (AMOVA, Excoffier et al. 1992), using the R packages poppr (Kamvar et al. 2014, 2015), adegenet (Jombart 2008; Jombart and Ahmed 2011) and vcfR (Knaus and Grünwald 2017). To assess statistical significance, a randomisation test was performed using 1000 permutations.
Population-structure analysis was performed using fastSTRUCTURE ver. 2.3.4 (Pritchard et al. 2000; Falush et al. 2003, 2007; Hubisz et al. 2009). We ran fastSTRUCTURE with default settings for between 1 and 12 populations, chose the model complexity to best explain the structure in the data, and produced plots to visualise the admixture analyses.
Results and discussion
Our GBS dataset contained 853 SNPs for 273 A. teres from the eastern coast of Australia. Genetic differentiation among populations in A. teres was small or near negligible across the sampling range. Model complexity of the fastSTRUCTURE analysis suggested six genetic populations, but there was no relationship between ‘genetic populations’ and geographic distribution (Fig. 1). The PCA (Fig. 2) showed no geographic structuring and each component represented less than 4% of the variation.
The lack of strong genetic differentiation was confirmed by pairwise measures of genetic distance between populations showing absent to negligible differentiation among populations along the eastern coast of Australia, using both Nei’s genetic distance and Weir and Cockerham’s Fst (Table 1).
The Mantel test showed no significant correlation between the geographic and genetic distance matrix, Nei’s D –0.153 (P-value 0.672) and Weir and Cockerham’s Fst –0.115 (P-value 0.587), further indicating that there was little genetic differentiation among populations across our sampling range. An AMOVA confirmed limited genetic structure across the sampling range, with most variance being among samples within beaches, and within samples (Table 2).

|
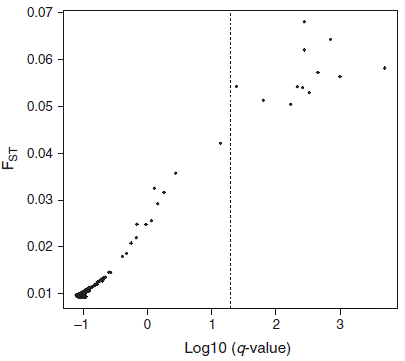
Using BayeScan ver. 2.1 (Foll and Gaggiotti 2008) to try to partition variance between potentially adaptive and truly neutral loci, we found that 12 SNPs showed an excess of differentiation among the three regions (Fig. 3) and, thus, could be under selection. In the absence of a reference genome, this number of bi-allelic variants is not enough to enable us to meaningfully assess genetic structure from a neutral versus adaptive perspective.
|
Population genetic structure of sedentary marine species is generally expected to be shaped by the dispersal ability of their larvae (Levin 2006). Long-lived pelagic larvae can connect populations through migration and gene flow, whereas species with non-dispersive larvae might be expected to have genetically differentiated populations (Kesäniemi et al. 2012). However, there are notable exceptions to these generic expectations; for example, congeneric species with similar life-history traits can show dissimilarities in population structure (Temby et al. 2007; Miller et al. 2009, 2014).
The reproduction and larval development of A. teres is not well understood, but A. teres have been found with over 100 000 eggs in their body cavity (Paxton 1986), and mature gametes have been found throughout the year, suggesting a long spawning season (Paxton 1979). Exposure to a wide variety of oceanographic and biological processes would occur over this period, potentially influencing larval dispersal distance.
With these polychaetes increasingly being used as bait (Cole et al. 2018), there is a growing need to understand their population structure to support sustainable resource-management practices. Our results suggested that despite movement throughout the sample range of this study, each geographic population has a different, but not diagnostic, genetic composition, and there has been bidirectional genetic mixing along the coast. This inference is consistent with previous research along the same region on the eastern coast of Australia, except for benthic estuarine polychaete species, which were inferred to have high levels of gene flow among populations (Smith et al. 2015). Although they live in different habitats, A. teres and the two estuarine species studied by Smith et al. (2015) are likely to be influenced by the same ocean currents.
Most prior studies have found no or little genetic differentiation among polychaete populations (e.g. Shen and Gu 2015; Zaâbi et al. 2015; David et al. 2016). High levels of gene flow were inferred between populations of two polychaetes, Laenereis culveri and Capitella nonatoi, living in lagoons in Brazil, albeit with there being evidence of somewhat restricted gene flow between particular populations (Seixas et al. 2018). However, in a similar area, Nunes et al. (2017) found strong genetic differentiation between Phragmatopoma caudata polychaetes from Florida and those from Brazil, suggesting a biogeographical barrier between these locations. Overall, most research suggests that gene flow can be high in polychaetes, presumably because of planktonic larval dispersal. However, most previous studies have been limited to just one or a few markers, such as, for example, COI (Chatzigeorgiou et al. 2015; David et al. 2016), 18S rRNA (Shen and Gu 2015; Sun et al. 2018), 28S rRNA (Shen and Gu 2015; Sun et al. 2018) and ITS (Nunes et al. 2017; Sun et al. 2018), whereas our study employed a genome-wide approach with hundreds of SNP loci, which enhanced confidence in our inferences.
The eastern coast of Australia appears to have few biogeographical barriers for marine coastal organisms with a high dispersal ability (e.g. Coleman et al. 2013; Smith et al. 2015; Bellgrove et al. 2017). Despite the general southward flow of the East Australian Current (EAC; Coleman et al. 2013), some small pelagic invertebrates have been inferred to have travelled 1000 km north from their release site (Ruello 1975; Montgomery 1990). Northward movement probably results from dispersal with counter-currents and local eddies (Coleman et al. 2011, 2013). These oceanographic currents could help explain the patchy distribution of A. teres, as the eddies, together with other population processes, affect the spread, deposition and survival of pelagic larvae (Levin 2006; Keane and Neira 2008; Chan et al. 2018). Other polychaete worms respond to chemical cues from adults when settling (Hsieh 1994), which could lead to density-dependent processes, including density blocking, limiting gene flow between connected populations (Waters et al. 2013; Fraser et al. 2018b). As the carrying capacity of a beach or site is approached, the rate of successful settlement of additional larvae might diminish, restricting gene flow. In the case of A. teres, the presence of six genetic groups could indicate that past structure that has since been eroded by natural disturbance events (e.g. storms), changes in environmental conditions affecting long-term rates of successful larval dispersal (e.g. changes in currents and temperature regimes), anthropogenic disturbance (e.g. periods of heavy harvesting reducing population densities and supporting successful recruitment), or combinations of the above, allowing the observed diverse lineages to establish at each site. Understanding the factors affecting genetic diversity, and minimising anthropogenic processes that could reduce diversity, should be a priority for the ongoing sustainable management of these beachworm populations. This study is one of the first population genomics studies of polychaetes and has demonstrated the power of SNP analyses in resolving fine-scale population structure. Genomic tools can assist with testing evolutionary and ecological questions for polychaetes globally.
Compliance with ethical standards
The authors declare no conflict of interest. The beachworms were sampled following jurisdictional requirements for biological sample collection and all necessary approvals were obtained before commencing this research.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
This work was funded by the New South Wales Recreational Fishing Saltwater Trust, Project DPIS011 (to RCC). C. I. Fraser was supported by a Rutherford Discovery Fellowship from the Royal Society of New Zealand (UOO1803).
Acknowledgements
Several people assisted with collections and other aspects of the research; particular thanks go to NSW DPI Fisheries staff including Ben Kearney, Matt Timmins, Mitch Burns, Tristan New and Emma LeBrault.
References
Andrews, S. (2010). FASTQC. A quality control tool for high throughput sequence data. Available at https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [verified 5 May 2020].Bellgrove, A., van Rooyen, A., Weeks, A. R., Clark, J. S., Doblin, M. A., and Miller, A. D. (2017). New resource for population genetics studies on the Australasian intertidal brown alga, Hormosira banksii: isolation and characterization of 15 polymorphic microsatellite loci through next generation DNA sequencing. Journal of Applied Phycology 29, 1721–1727.
| New resource for population genetics studies on the Australasian intertidal brown alga, Hormosira banksii: isolation and characterization of 15 polymorphic microsatellite loci through next generation DNA sequencing.Crossref | GoogleScholarGoogle Scholar |
Bougeard, S., and Dray, S. (2018). Supervised multiblock analysis in R with the ade4 package. Journal of Statistical Software 86, 1–17.
| Supervised multiblock analysis in R with the ade4 package.Crossref | GoogleScholarGoogle Scholar |
Bristow, G. A., and Vadas, R. L. (1991). Genetic variability in blood worm (Glycera dibranchiata) populations in the Gulf of Maine. Marine Biology 109, 311–319.
| Genetic variability in blood worm (Glycera dibranchiata) populations in the Gulf of Maine.Crossref | GoogleScholarGoogle Scholar |
Catchen, J., Hohenlohe, P. A., Bassham, S., Amores, A., and Cresko, W. A. (2013). Stacks: an analysis tool set for population genomics. Molecular Ecology 22, 3124–3140.
| Stacks: an analysis tool set for population genomics.Crossref | GoogleScholarGoogle Scholar | 23701397PubMed |
Chan, K. Y. K., Sewell, M. A., and Byrne, M. (2018). Revisiting the larval dispersal black box in the Anthropocene. ICES Journal of Marine Science 75, 1841–1848.
| Revisiting the larval dispersal black box in the Anthropocene.Crossref | GoogleScholarGoogle Scholar |
Chatzigeorgiou, G., Sarropoulou, E., Vasileiadou, K., Brown, C., Faulwetter, S., Kotoulas, G., and Arvanitidis, C. (2015) Potential links between polychaete community structure and population genetics (Hermodice carunculatasmith) patterns in the shallow hard substrates. In ‘11th Panhellenic Symposium on Oceanography and Fisheries’. Mytilene, Lesvos island, Greece. pp. 577–580.
Chessel, D., Dufour, A.-B., and Thioulouse, J. (2004). The {ade4} Package – {I}: one-table methods. R News 4, 5–10.
Cole, V. J., Chapman, M. G., and Underwood, A. J. (2007). Landscapes and life-histories influence colonisation of polychaetes to intertidal biogenic habitats. Journal of Experimental Marine Biology and Ecology 348, 191–199.
| Landscapes and life-histories influence colonisation of polychaetes to intertidal biogenic habitats.Crossref | GoogleScholarGoogle Scholar |
Cole, V. J., Hutchings, P. A., and Ross, P. M. (2017). Predicting biodiversity changes due to loss of bioengineers from an intertidal landscape, a case study from Sydney Harbour. Australian Zoologist 39, 194–206.
| Predicting biodiversity changes due to loss of bioengineers from an intertidal landscape, a case study from Sydney Harbour.Crossref | GoogleScholarGoogle Scholar |
Cole, V. J., Chick, R. C., and Hutchings, P. A. (2018). A review of global fisheries for polychaete worms as a resource for recreational fishers: diversity, sustainability and research needs. Reviews in Fish Biology and Fisheries 28, 543–565.
| A review of global fisheries for polychaete worms as a resource for recreational fishers: diversity, sustainability and research needs.Crossref | GoogleScholarGoogle Scholar |
Coleman, M. A., Roughan, M., Macdonald, H. S., Connell, S. D., Gillanders, B. M., Kelaher, B. P., and Steinberg, P. D. (2011). Variation in the strength of continental boundary currents determines continent-wide connectivity in kelp. Journal of Ecology 99, 1026–1032.
| Variation in the strength of continental boundary currents determines continent-wide connectivity in kelp.Crossref | GoogleScholarGoogle Scholar |
Coleman, M. A., Feng, M., Roughan, M., Cetina-Heredia, P., and Connell, S. D. (2013). Temperate shelf water dispersal by Australian boundary currents: implications for population connectivity. Limnology and Oceanography: Fluids and Environments 3, 295–309.
Cowen, R. K., and Sponaugle, S. (2009). Larval dispersal and marine population connectivity. Annual Review of Marine Science 1, 443–466.
| Larval dispersal and marine population connectivity.Crossref | GoogleScholarGoogle Scholar | 21141044PubMed |
Cowen, R. K., Gawarkiewicz, G., Pineda, J., Thorrold, S. R., and Werner, F. E. (2007). Population connectivity in marine systems: an overview. Oceanography 20, 14–21.
| Population connectivity in marine systems: an overview.Crossref | GoogleScholarGoogle Scholar |
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., Handsaker, R. E., Lunter, G., Marth, G. T., Sherry, S. T., McVean, G., and Durbin, R. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158.
| The variant call format and VCFtools.Crossref | GoogleScholarGoogle Scholar | 21653522PubMed |
David, A. A., Matthee, C. A., Loveday, B. R., and Simon, C. A. (2016). Predicting the dispersal potential of an invasive polychaete pest along a complex coastal biome. Integrative and Comparative Biology 56, 600–610.
| Predicting the dispersal potential of an invasive polychaete pest along a complex coastal biome.Crossref | GoogleScholarGoogle Scholar | 27126982PubMed |
Dennis, A. B., and Hellberg, M. E. (2010). Ecological partitioning among parapatric cryptic species. Molecular Ecology 19, 3206–3225.
| Ecological partitioning among parapatric cryptic species.Crossref | GoogleScholarGoogle Scholar | 20618906PubMed |
Dibacco, C., Levin, L. A., and Sala E. (2006). Connectivity in marine ecosystems: the importance of larval and spore dispersal. In ‘Connectivity Conservation’. (Eds K. Crooks and M. Sanjayan.) pp. 184–212. (Cambridge University Press: Cambridge, UK.)
Dray, S., and Dufour, A. B. (2007). The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software 22, 1–20.
| The ade4 package: implementing the duality diagram for ecologists.Crossref | GoogleScholarGoogle Scholar |
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., and Mitchell, S. E. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379.
| A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species.Crossref | GoogleScholarGoogle Scholar | 21573248PubMed |
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
| 1644282PubMed |
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587.
| 12930761PubMed |
Falush, D., Stephens, M., and Pritchard, J. K. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes 7, 574–578.
| Inference of population structure using multilocus genotype data: dominant markers and null alleles.Crossref | GoogleScholarGoogle Scholar | 18784791PubMed |
Fauchald, K., and Jumars, P. A. (1979). The diet of worms: a study of polychaete feeding guilds. Oceanography and Marine Biology - An Annual Review 17, 193–284.
Fogarty, M. J., and Botsford, L. W. (2007). Population connectivity and spatial management of marine fisheries. Oceanography 20, 112–123.
| Population connectivity and spatial management of marine fisheries.Crossref | GoogleScholarGoogle Scholar |
Foll, M., and Gaggiotti, O. (2008). A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180, 977–993.
| A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective.Crossref | GoogleScholarGoogle Scholar | 18780740PubMed |
Fraser, C. I., Morrison, A. K., Hogg, A. M. C., Macaya, E. C., van Sebille, E., Ryan, P. G., Padovan, A., Jack, C., Valdivia, N., and Waters, J. M. (2018a). Antarctica’s ecological isolation will be broken by storm-driven dispersal and warming. Nature Climate Change 8, 704–708.
| Antarctica’s ecological isolation will be broken by storm-driven dispersal and warming.Crossref | GoogleScholarGoogle Scholar |
Fraser, C. I., Davies, I. D., Bryant, D., and Waters, J. M. (2018b). How disturbance and dispersal influence intraspecific structure. Journal of Ecology 106, 1298–1306.
| How disturbance and dispersal influence intraspecific structure.Crossref | GoogleScholarGoogle Scholar |
Froukh, T., and Kochzius, M. (2007). Genetic population structure of the endemic fourline wrasse (Larabicus quadrilineatus) suggests limited larval dispersal distances in the Red Sea. Molecular Ecology 16, 1359–1367.
| Genetic population structure of the endemic fourline wrasse (Larabicus quadrilineatus) suggests limited larval dispersal distances in the Red Sea.Crossref | GoogleScholarGoogle Scholar | 17391261PubMed |
Giangrande, A., Licciano, M., and Musco, L. (2005). Polychaetes as environmental indicators revisited. Marine Pollution Bulletin 50, 1153–1162.
| Polychaetes as environmental indicators revisited.Crossref | GoogleScholarGoogle Scholar | 16197965PubMed |
Gogarten, S., Zheng, X., and Stilp, A. (2019). SeqVarTools: tools for variant data. R package version 1.25.0. https://github.com/smgogarten/SeqVarTools [verified 5 May 2020].
Gosselin, T. (2019). radiator: RADseq data exploration, manipulation and visualization using R. https://github.com/smgogarten/SeqVarTools [verified 5 May 2020].
Goudet, J. (2005). hierfstat, a package for R to compute and test hierarchical F‐statistics. Molecular Ecology Notes 5, 184–186.
| hierfstat, a package for R to compute and test hierarchical F‐statistics.Crossref | GoogleScholarGoogle Scholar |
Hsieh, H. L. (1994). Larval development and substrate preference at settlement in Pseudopolydora diopatra (Polychaeta: Spionidae). Invertebrate Reproduction & Development 25, 205–214.
| Larval development and substrate preference at settlement in Pseudopolydora diopatra (Polychaeta: Spionidae).Crossref | GoogleScholarGoogle Scholar |
Hubisz, M. J., Falush, D., Stephens, M., and Pritchard, J. K. (2009). Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources 9, 1322–1332.
| Inferring weak population structure with the assistance of sample group information.Crossref | GoogleScholarGoogle Scholar | 21564903PubMed |
Iacchei, M., Ben-Horin, T., Selkoe, K. A., Bird, C. E., García-Rodríguez, F. J., and Toonen, R. J. (2013). Combined analyses of kinship and FST suggest potential drivers of chaotic genetic patchiness in high gene-flow populations. Molecular Ecology 22, 3476–3494.
| Combined analyses of kinship and FST suggest potential drivers of chaotic genetic patchiness in high gene-flow populations.Crossref | GoogleScholarGoogle Scholar | 23802550PubMed |
Jombart, T. (2008). Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405.
| Adegenet: a R package for the multivariate analysis of genetic markers.Crossref | GoogleScholarGoogle Scholar | 18397895PubMed |
Jombart, T., and Ahmed, I. (2011). adegenet 1.3–1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071.
| adegenet 1.3–1: new tools for the analysis of genome-wide SNP data.Crossref | GoogleScholarGoogle Scholar | 21926124PubMed |
Jumars, P. A., Dorgan, K. M., and Lindsay, S. M. (2015). Diet of worms emended: an update of polychaete feeding guilds. Annual Review of Marine Science 7, 497–520.
| Diet of worms emended: an update of polychaete feeding guilds.Crossref | GoogleScholarGoogle Scholar | 25251269PubMed |
Kamvar, Z. N., Tabima, J. F., and Grünwald, N. J. (2014). Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281.
| Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction.Crossref | GoogleScholarGoogle Scholar | 24688859PubMed |
Kamvar, Z. N., Brooks, J. C., and Grünwald, N. J. (2015). Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Frontiers in Genetics 6, 1–10.
| Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality.Crossref | GoogleScholarGoogle Scholar |
Karsenti, E., Acinas, S. G., Bork, P., Bowler, C., de Vargas, C., Raes, J., Sullivan, M., Arendt, D., Benzoni, F., Claverie, J. M., Follows, M., Gorsky, G., Hingamp, P., Iudicone, D., Jaillon, O., Kandels-Lewis, S., Krzic, U., Not, F., Ogata, H., Pesant, S., Reynaud, E. G., Sardet, C., Sieracki, M. E., Speich, S., Velayoudon, D., Weissenbach, J., Wincker, P., Abergel, C., Arslan, D., Audic, S., Aury, J. M., Babic, N., Beaufort, L., Bittner, L., Boss, E., Boutte, C., Brum, J., Carmichael, M., Casotti, R., Chambouvet, A., Chang, P., Chica, C., Clerissi, C., Colin, S., Cornejo-Castillo, F. M., Da Silva, C., De Monte, S., Decelle, J., Desdevises, Y., Dimier, C., Dolan, J., Duhaime, M., Durrieu de Madron, X., d’Ortenzio, F., d’Ovidio, F., Ferrera, I., Garczarek, L., Garet-Delmas, M. J., Gasmi, S., Gasol, J. M., Grimsley, N., Heilig, R., Ignacio-Espinoza, J., Jamet, J. L., Karp-Boss, L., Katinka, M., Khalili, H., Kolber, Z., Le Bescot, N., Le Gofff, H., Lima-Mendez, G., Mahé, F., Mazzocchi, M. G., Montresor, M., Morin, P., Noel, B., Pedro’s-Alió, C., Pelletier, E., Perez, Y., Picheral, M., Piganeau, G., Poirot, O., Poulain, J., Poulton, N., Prejger, F., Prihoda, J., Probert, I., Rampal, J., Reverdin, G., Romac, S., Romagnan, J. B., Roullier, F., Rouviere, C., Samson, G., Santini, S., Sarmento, H., Sciandra, A., Solonenko, S., Stemmann, L., Subirana, L., Sunagawa, S., Tanaka, A., Testor, P., Thompson, A., Tichanné-Seltzer, V., Tirichine, L., Toulza, E., Tozzi, S., Veluchamy, A., and Zingone, A. (2011). A holistic approach to marine eco-systems biology. PLoS Biology 9, e1001177.
| A holistic approach to marine eco-systems biology.Crossref | GoogleScholarGoogle Scholar | 22028628PubMed |
Keane, J. P., and Neira, F. J. (2008). Larval fish assemblages along the south-eastern Australian shelf: linking mesoscale non-depth-discriminate structure and water masses. Fisheries Oceanography 17, 263–280.
| Larval fish assemblages along the south-eastern Australian shelf: linking mesoscale non-depth-discriminate structure and water masses.Crossref | GoogleScholarGoogle Scholar |
Kesäniemi, J. E., Geuverink, E., and Knott, K. E. (2012). Polymorphism in developmental mode and its effect on population genetic structure of a spionid polychaete, Pygospio elegans. Integrative and Comparative Biology 52, 181–196.
| Polymorphism in developmental mode and its effect on population genetic structure of a spionid polychaete, Pygospio elegans.Crossref | GoogleScholarGoogle Scholar | 22576815PubMed |
Kinlan, B. P., and Gaines, S. D. (2003). Propagule dispersal in marine and terrestrial environments: a community perspective. Ecology 84, 2007–2020.
| Propagule dispersal in marine and terrestrial environments: a community perspective.Crossref | GoogleScholarGoogle Scholar |
Knaus, B. J., and Grünwald, N. J. (2017). vcfr: a package to manipulate and visualize variant call format data in R. Molecular Ecology Resources 17, 44–53.
| vcfr: a package to manipulate and visualize variant call format data in R.Crossref | GoogleScholarGoogle Scholar | 27401132PubMed |
Levin, L. A. (2006). Recent progress in understanding larval dispersal: new directions and digressions. Integrative and Comparative Biology 46, 282–297.
| Recent progress in understanding larval dispersal: new directions and digressions.Crossref | GoogleScholarGoogle Scholar | 21672742PubMed |
Marko, P. B., Rogers-Bennett, L., and Dennis, A. B. (2007). MtDNA population structure and gene flow in lingcod (Ophiodon elongatus): limited connectivity despite long-lived pelagic larvae. Marine Biology 150, 1301–1311.
| MtDNA population structure and gene flow in lingcod (Ophiodon elongatus): limited connectivity despite long-lived pelagic larvae.Crossref | GoogleScholarGoogle Scholar |
McGaughran, A., Terauds, A., Convey, P., and Fraser, C. I. (2019). Genome-wide SNP data reveal improved evidence for Antarctic glacial refugia and dispersal of terrestrial invertebrates. Molecular Ecology 28, 4941–4957.
| Genome-wide SNP data reveal improved evidence for Antarctic glacial refugia and dispersal of terrestrial invertebrates.Crossref | GoogleScholarGoogle Scholar | 31596994PubMed |
Miller, K. J., Maynard, B. T., and Mundy, C. N. (2009). Genetic diversity and gene flow in collapsed and healthy abalone fisheries. Molecular Ecology 18, 200–211.
| Genetic diversity and gene flow in collapsed and healthy abalone fisheries.Crossref | GoogleScholarGoogle Scholar | 19076275PubMed |
Miller, K. J., Mundy, C. N., and Mayfield, S. (2014). Molecular genetics to inform spatial management in benthic invertebrate fisheries: a case study using the Australian greenlip abalone. Molecular Ecology 23, 4958–4975.
| Molecular genetics to inform spatial management in benthic invertebrate fisheries: a case study using the Australian greenlip abalone.Crossref | GoogleScholarGoogle Scholar | 25211183PubMed |
Montgomery, S. S. (1990). Movements of juvenile eastern king prawns, Penaeus plebejus, and identification of stock along the east coast of Australia. Fisheries Research 9, 189–208.
| Movements of juvenile eastern king prawns, Penaeus plebejus, and identification of stock along the east coast of Australia.Crossref | GoogleScholarGoogle Scholar |
Morris, G. P., Grabowski, P. P., and Borevitz, J. O. (2011). Genomic diversity in switchgrass (Panicum virgatum): from the continental scale to a dune landscape. Molecular Ecology 20, 4938–4952.
| Genomic diversity in switchgrass (Panicum virgatum): from the continental scale to a dune landscape.Crossref | GoogleScholarGoogle Scholar | 22060816PubMed |
Nei, M. (1987). ‘Molecular evolutionary genetics.’ (Columbia University Press: New York, USA.)
Nunes, F. L. D., Van Wormhoudt, A., Faroni-Perez, L., and Fournier, J. (2017). Phylogeography of the reef-building polychaetes of the genus Phragmatopoma in the western Atlantic Region. Journal of Biogeography 44, 1612–1625.
| Phylogeography of the reef-building polychaetes of the genus Phragmatopoma in the western Atlantic Region.Crossref | GoogleScholarGoogle Scholar |
Olsgard, F., and Somerfield, P. J. (2000). Surrogates in marine benthic investigations: which taxonomic unit to target? Journal of Aquatic Ecosystem Stress and Recovery 7, 25–42.
| Surrogates in marine benthic investigations: which taxonomic unit to target?Crossref | GoogleScholarGoogle Scholar |
Olsgard, F., Brattegard, T., and Holthe, T. (2003). Polychaetes as surrogates for marine biodiversity: lower taxonomic resolution and indicator groups. Biodiversity and Conservation 12, 1033–1049.
| Polychaetes as surrogates for marine biodiversity: lower taxonomic resolution and indicator groups.Crossref | GoogleScholarGoogle Scholar |
Paradis, E. (2010). pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26, 419–420.
| pegas: an R package for population genetics with an integrated–modular approach.Crossref | GoogleScholarGoogle Scholar | 20080509PubMed |
Paxton, H. (1979). Taxonomy and aspects of the life history of Australian beachworms (Polychaeta: Onuphidae). Marine and Freshwater Research 30, 265–294.
| Taxonomy and aspects of the life history of Australian beachworms (Polychaeta: Onuphidae).Crossref | GoogleScholarGoogle Scholar |
Paxton, H. (1986). Generic revision and relationships of the family Onuphidae (Annelida: Polychaeta). Records of the Australian Museum 38, 1–74.
| Generic revision and relationships of the family Onuphidae (Annelida: Polychaeta).Crossref | GoogleScholarGoogle Scholar |
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
| 10835412PubMed |
Purcell, J. F. H., Cowen, R. K., Hughes, C. R., and Williams, D. A. (2006). Weak genetic structure indicates strong dispersal limits: a tale of two coral reef fish. Proceedings. Biological Sciences 273, 1483–1490.
| Weak genetic structure indicates strong dispersal limits: a tale of two coral reef fish.Crossref | GoogleScholarGoogle Scholar |
Riginos, C., Douglas, K. E., Jin, Y., Shanahan, D. F., and Treml, E. A. (2011). Effects of geography and life history traits on genetic differentiation in benthic marine fishes. Ecography 34, 566–575.
| Effects of geography and life history traits on genetic differentiation in benthic marine fishes.Crossref | GoogleScholarGoogle Scholar |
Roberts, C. M. (1997). Connectivity and management of Caribbean coral reefs. Science 278, 1454–1457.
| Connectivity and management of Caribbean coral reefs.Crossref | GoogleScholarGoogle Scholar | 9367956PubMed |
Ruello, N. V. (1975). Geographical distribution, growth and breeding migration of the eastern Australian king prawn Penaus plebejus Hess. Australian Journal of Marine and Freshwater Research 26, 343–354.
| Geographical distribution, growth and breeding migration of the eastern Australian king prawn Penaus plebejus Hess.Crossref | GoogleScholarGoogle Scholar |
Seixas, V. C., Paiva, P. C., and Russo, C. A de M. (2018). Comparative population genetics and demographic history of two polychaete species suggest that coastal lagoon populations evolve under alternate regimes of gene flow. Marine Biology 165, 179.
| Comparative population genetics and demographic history of two polychaete species suggest that coastal lagoon populations evolve under alternate regimes of gene flow.Crossref | GoogleScholarGoogle Scholar |
Shen, P. P., and Gu, J. D. (2015). Genetic population structure of polychaeta Neanthes glandicincta (Nereididae) of the Mai Po Inner Deep Bay Ramsar Site, Hong Kong. Ecotoxicology 24, 1557–1565.
| Genetic population structure of polychaeta Neanthes glandicincta (Nereididae) of the Mai Po Inner Deep Bay Ramsar Site, Hong Kong.Crossref | GoogleScholarGoogle Scholar | 25967938PubMed |
Shokri, M. R., Gladstone, W., and Kepert, A. (2009). Annelids, arthropods or molluscs are suitable as surrogate taxa for selecting conservation reserves in estuaries. Biodiversity and Conservation 18, 1117–1130.
Shulman, M. J., and Bermingham, E. (1995). Early life histories, ocean currents, and the population genetics of Caribbean reef fishes. Evolution 49, 897–910.
| Early life histories, ocean currents, and the population genetics of Caribbean reef fishes.Crossref | GoogleScholarGoogle Scholar | 28564869PubMed |
Smith, L. M., Hutchings, P., and Fraser, C. I. (2015). Molecular evidence supports coastal dispersal among estuaries for two benthic marine worm (Nephtyidae) species in southeastern Australia. Marine Biology 162, 1319–1327.
| Molecular evidence supports coastal dispersal among estuaries for two benthic marine worm (Nephtyidae) species in southeastern Australia.Crossref | GoogleScholarGoogle Scholar |
Sun, Y., Wong, E., Ahyong, S. T., Williamson, J. E., Pat, A., and Kupriyanova, E. K. (2018). Barcoding and multi-locus phylogeography of the globally distributed calcareous tubeworm genus Hydroides Gunnerus, 1768 (Annelida, Polychaeta, Serpulidae). Molecular Phylogenetics and Evolution 127, 732–745.
| Barcoding and multi-locus phylogeography of the globally distributed calcareous tubeworm genus Hydroides Gunnerus, 1768 (Annelida, Polychaeta, Serpulidae).Crossref | GoogleScholarGoogle Scholar | 29906604PubMed |
Temby, N., Miller, K., and Mundy, C. (2007). Evidence of genetic subdivision among populations of blacklip abalone (Haliotis rubra Leach) in Tasmania. Marine and Freshwater Research 58, 733–742.
| Evidence of genetic subdivision among populations of blacklip abalone (Haliotis rubra Leach) in Tasmania.Crossref | GoogleScholarGoogle Scholar |
Thiel, M., and Gutow, L. (2005a). The ecology of rafting in the marine environment. I. the floating substrata. Oceanography and Marine Biology - An Annual Review 42, 181–263.
| The ecology of rafting in the marine environment. I. the floating substrata.Crossref | GoogleScholarGoogle Scholar |
Thiel, M., and Gutow, L. (2005b). The ecology of rafting in the marine environment. II. the rafting organisms and community. Oceanography and Marine Biology - An Annual Review 43, 279–418.
| The ecology of rafting in the marine environment. II. the rafting organisms and community.Crossref | GoogleScholarGoogle Scholar |
Thorrold, S. R. (2006). Ocean ecology: don’t fence me in. Current Biology 16, R638–R640.
| Ocean ecology: don’t fence me in.Crossref | GoogleScholarGoogle Scholar | 16920611PubMed |
Waples, R., and Punt, A. (2008). Integrating genetic data into management of marine resources: how can we do it better? Fish and Fisheries 9, 423–449.
| Integrating genetic data into management of marine resources: how can we do it better?Crossref | GoogleScholarGoogle Scholar |
Waters, J. M., Fraser, C. I., and Hewitt, G. M. (2013). Founder takes all: density-dependent processes structure biodiversity. Trends in Ecology & Evolution 28, 78–85.
| Founder takes all: density-dependent processes structure biodiversity.Crossref | GoogleScholarGoogle Scholar |
Weir, B. S., and Cockerham, C. C. (1984). Estimating F‐statistics for the analysis of population structure. Evolution 38, 1358–1370.
| 28563791PubMed |
Wickham, H. (2016). ‘ggplot2: dlegant graphics for data analysis.’ (Springer-Verlag: New York, USA.)
Wickham, H., Hester, J., and Chang, W. (2019). Package ‘devtools’. https://github.com/r-lib/devtools [verified 5 May 2020].
Wilson, P. B., Streich, J. C., Murray, K. D., Eichten, S. R., Cheng, R., Aitken, N. C., Spokas, K., Warthmann, N., Gordon, S. P., Vogel, J. P., Borevitz, J. O., Liu, S., Bradford, K., Ezrati, S., Budak, H., Lopez, D., Catalan, P., Garvin, D., Vogel, J., Gordon, S., Hazen, S., and Mur, L. (2019). Global diversity of the Brachypodium species complex as a resource for genome-wide association studies demonstrated for agronomic traits in response to climate. Genetics 211, 317–331.
| Global diversity of the Brachypodium species complex as a resource for genome-wide association studies demonstrated for agronomic traits in response to climate.Crossref | GoogleScholarGoogle Scholar | 30446522PubMed |
Xuereb, A., Benestan, L., Normandeau, E., Curtis, J. M. R., Bernatchez, L., and Fortin, M.-J. (2018). Asymmetric oceanographic processes mediate connectivity and population genetic structure, as revealed by RADseq, in a highly dispersive marine invertebrate (Parastichopus californicus). Molecular Ecology 27, 2347–2364.
| Asymmetric oceanographic processes mediate connectivity and population genetic structure, as revealed by RADseq, in a highly dispersive marine invertebrate (Parastichopus californicus).Crossref | GoogleScholarGoogle Scholar | 29654703PubMed |
Zaâbi, S., Metais, I., Gillet, P., Afli, A., and Boumaiza, M. (2015). Preliminary study of population genetic diversity of Hyalinoecia tubicola (Polychaeta: Onuphidae) from the north east coast of Tunisia (western Mediterranean) using random amplified polymorphic DNA markers. Journal of Coastal Zone Management 18, 1–6.
| Preliminary study of population genetic diversity of Hyalinoecia tubicola (Polychaeta: Onuphidae) from the north east coast of Tunisia (western Mediterranean) using random amplified polymorphic DNA markers.Crossref | GoogleScholarGoogle Scholar |





